Abstract
Allosteric interactions between the DNA- and NTP-binding sites of the Escherichia coli DnaB helicase engaged in the DnaB–DnaC complex and the mechanism of NTP hydrolysis by the complex have been examined using the fluorescence titration, analytical ultracentrifugation, and rapid quench-flow technique. Surprisingly, the ssDNA affinity of the DnaB–DnaC complex is independent of the structure of the phosphate group of the cofactor bound to the helicase. Thus, the DnaC protein eliminates the antagonistic allosteric effect of NTP and NDP on the ssDNA affinity of the enzyme. The protein changes the engagement of the DNA-binding subsites of the helicase in interactions with the nucleic acid, depending on the structure of the phosphate group of the present nucleotide cofactor and profoundly affects the structure of the bound DNA. Moreover, the ssDNA affinity of the helicase in the DnaB–DnaC complex is under the control of the nucleotide-binding site of the DnaC protein. The protein does not affect the NTP hydrolysis mechanism of the helicase. Nevertheless, the rate of the chemical step is diminished in the DnaB–DnaC complex. In the tertiary DnaB–DnaC–ssDNA complex, the ssDNA changes the internal dynamics between intermediates of the pyrimidine cofactor, in a manner independent of the base composition of the DNA, while the hydrolysis step of the purine cofactor is specifically stimulated by the homoadenosine ssDNA. The significance of these results for functional activities of the DnaB–DnaC complex is discussed.
The double-stranded DNA is the most stable conformation of the nucleic acid in the cellular environment, in vivo (1-3). Nevertheless, in processes of DNA replication, recombination, and repair, the single-stranded conformation is the metabolically active intermediate, transiently formed in a reaction catalyzed by a class of enzymes called helicases and fueled by NTP1 hydrolysis (3-8). In the Escherichia coli cell, the DnaB protein is the primary replication helicase, which catalyzes unwinding of the duplex DNA in front of the replication fork (8-14). Moreover, the enzyme is a key ingredient of the preprimosome and primosome, large multiple-protein–DNA molecular machines, responsible for priming DNA synthesis and restarting the stalled replication fork (10, 15-18). It is a rule rather than an exception that a replicative helicase does not act alone, even before engaging the molecular machines of DNA replication. Highly specific interactions between the DnaB helicase and the replication factor, the DnaC protein, allow the enzyme to recognize the origin of replication, oriC, or to enter the preprimosome (10, 15-18). At saturation, six DnaC monomers bind to the helicase, although at physiological concentrations of the DnaC protein the DnaB–DnaC complex exists as a distribution of complexes with different numbers of bound DnaC molecules (19). Nevertheless, in the dominant complex, six DnaC protein molecules assemble into a hexamer on the helicase and are held through positive cooperative interactions [companion paper (DOI 10.1021/bi900050x)] (19).
The DnaB hexamer possesses six nucleotide-binding sites, and the maximum stoichiometry of the nucleotide cofactor binding is preserved in its binary complex with the DnaC protein and the tertiary DnaB–DnaC–ssDNA complex [companion paper (DOI 10.1021/bi900050x)] (20-25). In stationary complexes, i.e., in the absence of NTP hydrolysis, the nucleotide-binding sites accept both NTP and NDP with similar intrinsic affinities, although only NTP or NTP nonhydrolyzable analogues induce the high ssDNA affinity of the helicase. As a result, the allosteric linkage between the nucleotide-binding sites and the ssDNA-binding site increases the affinity of nucleotide triphosphate by 3–4 orders of magnitude, while NDP affinity is unaffected [companion paper (DOI 10.1021/bi900050x)] (26). The nucleotide binding process is effectively split into two phases, with the first three nucleotides binding in the high-affinity phase and the remaining three nucleotides binding in the low-affinity phase (20, 23). Studies of the analogous RepA hexameric helicase of plasmid RSF1010 indicate that the high-affinity phase in nucleotide binding is involved in modulating the interactions of the helicase with the ssDNA (27-29). On the other hand, the DnaC protein has a single nucleotide-binding site, which exclusively binds adenosine nucleotides and does not possess an intrinsic ATPase activity (30-32). Moreover, the DnaC protein exists in equilibrium between two conformational states prior to the nucleotide binding (31, 32). The DnaC protein preserves its nucleotide binding capability both in the binary DnaB–DnaC and tertiary DnaB–DnaC–ssDNA complexes [companion paper (DOI 10.1021/bi900050x)].
The DnaB–DnaC complex is the smallest unit of the specific multiple protein–protein complexes, which control and regulate the DnaB helicase (15-19). The experiments and analyses, described in the companion papers (DOI 10.1021/bi900050x and DOI 10.1021/bi9000529) provide the long-awaited data on interrelationships among the protein–protein, protein–nucleic acid, and protein–nucleotide interactions of both protein components of the DnaB–DnaC complex. However, very little is known about the dynamics of the NTP hydrolysis by the DnaB–DnaC complex. This is in spite of the fact that the DnaC protein is involved in both initiation and elongation stages of the DNA replication, in which the DnaB protein is engaged (15-18). The kinetic mechanisms of hydrolysis by the binary DnaB–DnaC and tertiary DnaB–DnaC–ssDNA complexes are unknown. The effect of the type of cofactor on interactions of the DnaB–DnaC complex with the ssDNA and the allosteric role of the nucleic acid in the NTPase activities of the DnaB–DnaC complex have never been quantitatively addressed.
In this paper, we describe direct analyses of the allosteric interactions between the DNA-binding site and NTP-binding sites of the E. coli DnaB helicase engaged in the DnaB–DnaC complex, and the mechanism of the nucleotide binding and hydrolysis by the DnaB–DnaC complex. Surprisingly, the ssDNA affinity of the DnaB–DnaC complex is independent of the structure of the phosphate group of the nucleotide cofactor. However, the structure of the phosphate group of the nucleotide cofactor determines the engagement of the strong and weak DNA-binding subsites of the helicase in interactions with the nucleic acid. The DnaC protein does not change the major features of the mechanism of NTP hydrolysis by the helicase. The effect of the base composition of the ssDNA in the tertiary DnaB–DnaC–ssDNA complex on the dynamics and energetics of intermediate formation is different from the analogous effect observed for the helicase alone.
MATERIALS AND METHODS
Reagents and Buffers
All solutions were made with distilled and deionized >18 MΩ (Milli-Q Plus) water. All chemicals were reagent grade. Buffer T4 is 50 mM Tris adjusted to pH 8.1 with HCl at 20 °C, containing 100 mM NaCl, 5 mM MgCl2, 10% glycerol, and 1 mM DTT.
DnaB Protein
The E. coli DnaB protein was purified as previously described by us (20-22).
Nucleotides and Nucleic Acids
Radioactive [α-32P]ATP, [α-32P]GTP, and [α-32P]CTP were from Amersham. Cold ATP, GTP, and CTP were from Sigma (25). All ssDNA oligomers were from Midland (Midland, TX) [companion paper (DOI 10.1021/bi9000529)]. The concentrations of the DNA oligomer, labeled at the 5′ end with fluorescein, 5′-Fl-dA(pA)19 and 5′-Fl-dA(pA)9, have been spectrophotometrically determined (19, 27).
Rapid Quench-Flow Kinetics
The single-turnover time courses of NTP hydrolyses by the DnaB helicase were determined using the rapid quench-flow instrument, RQF-3 (KinTek), as we described previously (25) and in the companion paper (DOI 10.1021/bi9000529). The kinetic measurements were performed under single-turnover conditions (DOI 10.1021/bi9000529) (25, 33). The kinetic curves were fitted to extract relaxation times and amplitudes using the nonlinear least-squares routine (Mathematica, Wolfram, IL) with the exponential function defined as (25)
| (1) |
where P(t) is the [α-32P]NDP concentration at time t, P(∞) is the [α-32P]NDP concentration at transient equilibrium, Ai is the amplitude corresponding to the ith relaxation process, λi is the time constant (reciprocal relaxation time, 1/τi) characterizing the ith relaxation process, and n is the number of relaxation processes. All further analyses of the data were performed using Mathematica and Kaleida Graph (Synergy Software, Reading, PA) (25).
Fluorescence Measurements
Steady-state fluorescence titrations were performed using the SLM-AMINCO 8100C instrument, as described in the companion papers (DOI 10.1021/bi900050x and DOI 10.1021/bi9000529) (34-40). The relative fluorescence increase in the ssDNA etheno derivatives, ΔFobs, upon the binding of the DnaB–DnaC complex, is defined as ΔFobs=(Fi−Fo)/Fo, where Fi is the fluorescence of the DNA at a given titration point and Fo is the initial value of the fluorescence of the sample (34-40).
Quantitative Determination of Binding Isotherms and Stoichiometries of the DnaB–DnaC–ssDNA Complexes
The method for quantitatively estimating the total average degree of binding, ΣΘi (number of DnaB–DnaC complexes bound per ssDNA oligomer) and the free DnaB–DnaC complex concentration have been previously described in detail by us [companion paper (DOI 10.1021/bi900050x)] (34-40).
Analytical Ultracentrifugation Measurements
Sedimentation velocity experiments were performed with an Optima XL-A analytical ultracentrifuge (Beckman Inc., Palo Alto, CA), using double-sector charcoal-filled 12 mm centerpieces. Sedimentation velocity scans were collected at the absorption band of the fluorescein marker (485 nm) [companion paper (DOI 10.1021/bi9000529)].
RESULTS
Binding of the ssDNA 20-mer, dεA(pεA)19, to the Total DNA-Binding Site of the DnaB Helicase Engaged in the DnaB–DnaC Complex, in the Presence of ATP Nonhydrolyzable Analogue AMP-PNP
The total DNA-binding site of a protein associating with the DNA is an entire fragment of the nucleic acid, which is blocked (occluded) by the enzyme from accepting another protein molecule (40-42). In the case of the DnaB helicase, the total DNA-binding site comprises a fragment of 20 ± 3 nucleotides (35-38, 41-44). To examine the binding of the ssDNA to the total DNA-binding site of the helicase, engaged in the complex with the DnaC protein, we used the etheno derivative of the homoadenosine ssDNA 20-mer, dA(pA)19, i.e., dεA(pεA)19, analogously to our previous studies of the DnaB helicase (35-38). As discussed in the companion paper (DOI 10.1021/bi9000529), the fluorescence of the etheno derivatives is particularly suitable for examining the ssDNA–protein interactions. This is because the emission intensity of the nucleic acid predominantly depends on its structure and much less on the environment of the fluorophore (45-48).
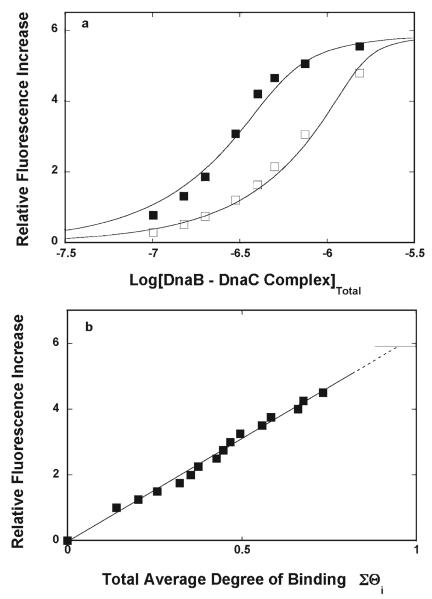
Fluorescence titrations of the ssDNA 20-mer, dεA(pεA)19, with the DnaB–DnaC complex, at two different nucleic acid concentrations, 4.9 × 10−7 M and 1.5 × 10−6 M (oligomer), in buffer T4, containing 1 mM AMP-PNP, are shown in Figure 1a. At the selected concentrations of the DnaB and DnaC proteins, the helicase is at least ~95% saturated with the DnaC [companion paper (DOI 10.1021/bi900050x)]. A slight excess of the DnaC protein over the enzyme does not affect the titration results, as the ssDNA affinity of the DnaC protein is ~3 orders of magnitude lower than the corresponding affinity of the helicase [companion paper (DOI 10.1021/bi900050x)]. Binding of the DnaB–DnaC complex to dεA(pεA)19 causes a strong, ~6-fold increase in the nucleic acid fluorescence, indicating a dramatic change in the nucleic acid structure in the complex (45-48). For the sake of comparison, the relative fluorescence increase of the same ssDNA oligomer, upon binding the DnaB helicase alone, is ~2.6-fold [companion paper (DOI 10.1021/bi900050x)]. The selected nucleic concentrations provide the separation of the titration curves up to a ΔFobs value of ~4. The total average degree of binding, ΣΘi, ofthe DnaB–DnaC complex on the ssDNA 20-mer has been obtained using the quantitative approach described in a companion paper (DOI 10.1021/bi900050x) (40-44, 49).
Figure 1.
(a) Fluorescence titrations (λex = 320 nm; λem = 410 nm) of the etheno derivative of the ssDNA 20-mer, dεA(pεA)19, with the DnaB–DnaC complex in buffer T4 (pH 8.1 and 10 °C), containing 1 mM AMP-PNP, at two different concentrations of the nucleic acid. The concentrations of the ssDNA oligomer were (■) 4.9 × 10−7 M and (□) 1.5 × 10−6 M (oligomer). The concentration of the DnaB–DnaC complex was equal to the concentration of the DnaB hexamer. The concentration of the DnaC protein was kept as a minimal concentration that saturated six DnaC-binding sites on the DnaB helicase >95%. The solid lines are nonlinear, least-squares fits of the titration curves to a single-binding site isotherm (eq 2) with two fitting parameters: K20=2.0 × 107 M−1, and ΔFmax=5.9. (b) Dependence of the relative fluorescence increase, ΔFobs, on ΣΘi (■). The values of ΣΘi have been determined using the quantitative method described in Materials and Methods. The solid line connects the experimental points and does not have a theoretical basis. The dashed line is the extrapolation of the plot to the maximum values of the observed fluorescence increase (ΔFmax = 5.9), marked by the solid horizontal line.
The dependence of the observed relative fluorescence increase, ΔFobs, on the total average degree of binding, ΣΘi, is shown in Figure 1b. The separation of the titration curves allowed us to obtain the values of ΣΘi up to ~0.7 of the DnaB–DnaC complex per 20-mer. The plot in Figure 1b is, within experimental accuracy, linear. Extrapolation to the maximum fluorescence increase (ΔFmax = 5.9 ± 0.3) provides a ΣΘi of 0.9 ± 0.1, showing that at saturation a single DnaB–DnaC complex binds to the ssDNA 20-mer. To extract the binding constant, K20, the plots in Figure 1a have been fitted using a single-site binding model
| (2) |
where [C]F is the free concentration of the DnaB–DnaC complex, i.e., equal to the total concentration of the DnaB hexamer. The obtained K20 value was (2 ± 0.7) × 107 M−1. This value is, within experimental accuracy, the same as the value of the analogous parameter, obtained for the DnaB helicase alone [K20 = (1.5 ± 0.2) × 107 M−1 (see Discussion)] (35-38).
Binding of the ssDNA 20-mer, dεA(pεA)19, to the Total DNA-Binding Site of the DnaB Helicase, Engaged in the DnaB–DnaC Complex, in the Presence of ADP
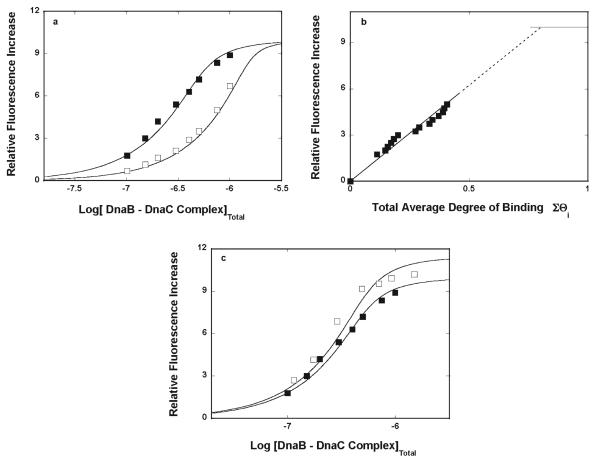
Fluorescence titrations of the ssDNA 20-mer, dεA(pεA)19, with the DnaB–DnaC complex, at two different nucleic acid concentrations, 4.9 × 10−7 M and 1.5 × 10−6 M (oligomer), in buffer T4, containing 1 mM ADP, are shown in Figure 2a. Binding of the DnaB–DnaC complex to the ethenoadenosine ssDNA 20-mer causes an increase in the nucleic acid fluorescence even larger, ~12-fold, than that observed in the presence of the ATP nonhydrolyzable analogue (Figure 1a). The dependence of the observed relative fluorescence increase, ΔFobs, on the total average degree of binding, ΣΘi, is shown in Figure 2b. The separation of the titration curves allowed us to obtain the values of ΣΘi up to ~0.6 of the DnaB–DnaC complex per 20-mer. Extrapolation to the maximum fluorescence increase (ΔFmax = 12 ± 0.5) shows that at saturation, a single DnaB–DnaC complex binds to the ssDNA 20-mer in the presence of ADP (Figure 2b). The titration curves in Figure 2a have been analyzed using a single-binding site model (eq 2). The obtained K20 value of (1.7 ± 0.6) × 107 M−1, for example, is, within experimental accuracy, the same as the value of the binding constant obtained in the presence of AMP-PNP (Figure 1a). This result is very surprising. Under the same solution conditions and in the presence of ADP, the binding constant of the DnaB helicase for dεA(pεA)19, in the absence of the DnaC protein, is (3 ± 0.7) × 104 M−1 and the helicase induces an only ~2.6-fold increase in nucleic acid emission (data not shown) (see Discussion).
Figure 2.
(a) Fluorescence titrations (λex = 320 nm; λem = 410 nm) of the etheno derivative of the ssDNA 20-mer, dεA(pεA)19, with the DnaB–DnaC complex in buffer T4 (pH 8.1 and 10 °C), containing 1 mM ADP, at two different concentrations of the nucleic acid. The concentrations of the ssDNA oligomer were (■) 4.9 × 10−7 M and (□) 1.5 × 10−6 M (oligomer). The concentration of the DnaB–DnaC complex was equal to the concentration of the DnaB hexamer. The concentration of the DnaC protein was kept as a minimal concentration that saturated six DnaC-binding sites on the DnaB helicase >95%. The solid lines are nonlinear, least-squares fits of the titration curves to a single-binding site isotherm (eq 2) with two fitting parameters: K20 =1.7 × 107 M−1, and ΔFmax = 12. (b) Dependence of the relative fluorescence increase, ΔFobs, on the total average degree of binding of the DnaB–DnaC complex on the ssDNA 20-mer, ΣΘi (■). The values of ΣΘi have been determined using the quantitative method described in Materials and Methods. The solid line connects the experimental points and does not have a theoretical basis. The dashed line is an extrapolation of the plot to the maximum values of the observed fluorescence increase (ΔFmax = 12), marked by the solid horizontal line.
Binding of the ssDNA 10-mer, dεA(pεA)9, to the Strong DNA-Binding Subsite of the DnaB Helicase Engaged in the DnaB–DnaC Complex, in the Presence of ATP Nonhydrolyzable Analogue AMP-PNP or ADP
The total DNA-binding site of the DnaB helicase is built of two DNA-binding subsites, strong and weak binding areas located sequentially on the protein molecule, with the strong subsite occluding the 5′ end of the bound nucleic acid (43, 44). To address the effect of the presence of the DnaC protein on the interactions of the ssDNA with the strong DNA-binding subsite of the DnaB helicase, we performed fluorescence titration studies of the binding of the ssDNA 10-mer, dεA(pεA)9, to the DnaB–DnaC complex. Our previous studies have shown that the 10-mer binds exclusively to the strong binding subsite of the enzyme (44). Fluorescence titrations of the ssDNA 10-mer, dεA(pεA)9, with the DnaB–DnaC complex, at two different nucleic acid concentrations, 4.9 × 10−7 M and 1.5 × 10−6 M (oligomer), in buffer T4, containing 1 mM AMP-PNP, are shown in Figure 3a. In the case of the 10-mer, binding of the DnaB–DnaC complex causes an ~10-fold increase in the nucleic acid fluorescence, which is dramatically larger than the ~6-fold increase observed in the presence of the 20-mer, dεA(pεA)9 (Figure 1a). The dependence of the relative fluorescence increase, ΔFobs, on the total average degree of binding, ΣΘi, is shown in Figure 3b. Extrapolation to the maximum fluorescence increase (ΔFmax = 10 ± 0.5) gives a ΣΘi of 0.8 ± 0.1. Thus, at saturation, a single DnaB–DnaC complex binds to the ssDNA 10-mer. The titration curves in Figure 3a have been analyzed using the single-binding site model (eq 2). The obtained K10 value of (2 ± 0.6) × 107 M−1 indicates that the 10-mer, which exclusively binds to the strong DNA-binding subsite of the helicase, has the same value for the binding constant in the presence of AMP-PNP, as obtained for the ssDNA 20-mer, which engages the total DNA-binding site of the enzyme (Figure 1a), although the structures of the bound nucleic acids, as indicated by profoundly different values of ΔFmax, are different (see Discussion).
Figure 3.
(a) Fluorescence titrations (λex = 320 nm; λem = 410 nm) of the etheno derivative of the ssDNA 10-mer, dεA(pεA)9, with the DnaB–DnaC complex in buffer T4 (pH 8.1 and 10 °C), containing 1 mM AMP-PNP, at two different concentrations of the nucleic acid. The concentrations of the ssDNA oligomer were (■) 4.9 × 10−7 M and (□) 1.5 × 10−6 M (oligomer). The concentration of the DnaB–DnaC complex was a minimal concentration that saturated six DnaC-binding sites on the DnaB helicase >95%. The solid lines are nonlinear, least-squares fits of the titration curves to a single-binding site isotherm (eq 2) with two fitting parameters: K20 = 2.0 × 107 M−1, and ΔFmax = 10. (b) Dependence of the relative fluorescence increase, ΔFobs, on the total average degree of binding of the DnaB–DnaC complex on the ssDNA 10-mer, ΣΘi (■). The values of ΣΘi have been determined using the quantitative method described in Materials and Methods. The solid line connects the extrapolation of the plot to the maximum values of the observed fluorescence increase (ΔFmax = 10), marked by the solid horizontal line. (c) Fluorescence titration (λex = 320 nm; λem = 410 nm) of the etheno derivative of the ssDNA 10-mer, dεA(pεA)9, with the DnaB–DnaC complex in buffer T4 (pH 8.1 and 10 °C), containing 1 mM ADP. The concentration of the ssDNA oligomer was 4.9 × 10−6 M (oligomer) (□). For the sake of comparison, the fluorescence titration of the same 10-mer at the same nucleic acid concentration but in the presence of 1 mM AMP-PNP is included in the panel (■) (details in the text).
The fluorescence titration of the etheno derivative of the ssDNA 10-mer, dεA(pεA)9, with the DnaB–DnaC complex in buffer T4 (pH 8.1 and 10 °C), containing 1 mM ADP, is shown in Figure 3c. For the sake of comparison, the fluorescence titration of the same 10-mer, but in the presence of 1 mM AMP-PNP, is included in the panel. As observed for the ssDNA 20-mer, the maximum fluorescence increase for the nucleic acid is larger in the presence of ADP, as compared to that for the sample containing the ATP nonhydrolyzable analogue (Figure 2a). The solid line in Figure 3c is a nonlinear least-squares fit of the titration curve, using the single-binding site model (eq 2), which provides the values of the binding constant: K10 = (2 ± 0.6) × 107 M−1, and ΔFmax = 11.5 ± 1. This is radically different from the values [K10 = (1.5 ± 0.6) × 104 M−1, and ΔFmax ~ 2.3 ± 0.3] determined for binding of the 10-mer, dεA(pεA)9, to the DnaB helicase alone, in the presence of ADP (data not shown). Moreover, it is evident that the presence of ADP does not affect the affinity of the DnaB–DnaC complex for the ssDNA, which exclusively binds to the strong DNA-binding subsite, as compared to the affinity observed in the presence of AMP-PNP. Consequently, in terms of the affinities, the effects of ADP and AMP-PNP on the 10-mer binding are completely analogous to the effects of both cofactors, observed in the case of the binding of the 20-mer, which engages the total DNA-binding site of the helicase, to the binary DnaB–DnaC complex (Figures 1a and 2a). However, unlike the behavior of the 20-mer, the structure of the bound 10-mer seems to be very similar, in the presence of both AMP-PNP and ADP, as indicated by the similar ΔFmax values (see Discussion).
Binding of the ssDNA 20- and 10-mer, dεA(pεA)19 and dεA(pεA)9, Respectively, to the DnaB Helicase, Engaged in the DnaB–DnaC Complex, in the Presence of GDP
In the binding systems described above, both protein components of the DnaB–DnaC complex are engaged in interactions with the nucleotide cofactor. Recall that the nucleotide-binding site of the DnaC protein is highly specific for the adenosine nucleotides (30). The nucleotide-binding sites of the DnaB helicase can accept nucleotides, in a manner independent of the type of the base, although with a preference for purine cofactors (20-23). In the next set of experiments, we address the association of the DnaB–DnaC complex with the ssDNA 20- and 10-mers in the presence of GDP, which exclusively binds to the nucleotide-binding sites of the helicase. In other words, we examine the ssDNA affinity of the DnaB helicase in the binary DnaB–DnaC complex, where the nucleotide-binding site of the DnaC protein is not engaged in interactions with the cofactor.
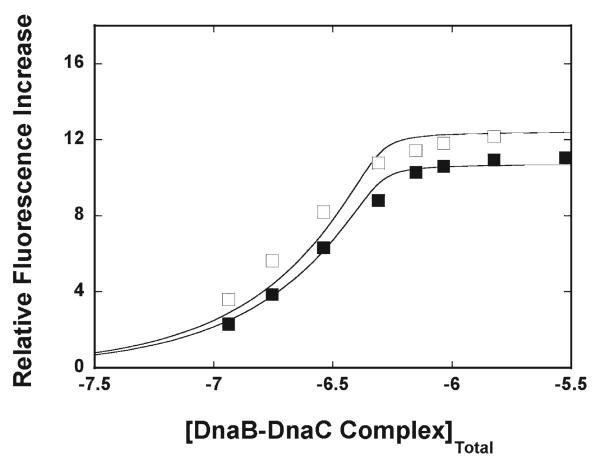
Fluorescence titrations of the ssDNA 20-mer, dεA(pεA)19, and 10-mer, dεA(pεA)9,with the DnaB–DnaC complex are shown in Figure 4. The large increase in the fluorescence emission of both ssDNA oligomers is similar to the analogous large increase in the emission intensity observed in the presence of the ADP; i.e., it is significantly larger than the increase in the nucleic acid fluorescence observed in the presence of the ATP nonhydrolyzable analogue AMP-PNP (Figures 2a,b and 3a,c). Thus, both diphosphate nucleosides exert a similar effect on the structure of the bound nucleic acid in the total DNA-binding site or the strong DNA-binding subsite. The solid lines in Figure 4 are nonlinear least-squares fits of the titration curves, which provide the following values: K20=(2 ± 0.3) × 108 M−1 and ΔFmax=10.7 ± 1.5 for the 20-mer and K10=(2 ± 0.3) 108 M−1 and ΔFmax=12.4 ± 1.5 for the 10-mer. The data indicate that, although the values of the fluorescence intensities are similar to the values observed in the presence of ADP, the values of K20 and K10 are considerably higher in the presence of GDP, which only binds to nucleotide-binding sites of the DnaB helicase (see Discussion).
Figure 4.
Fluorescence titration (λex=320 nm; λem=410 nm) of the etheno derivative of the ssDNA 20-mer, dεA(pεA)19 (■), and the 10-mer, dεA(pεA)9 (□), with the DnaB–DnaC complex in buffer T4 (pH 8.1 and 10 °C), containing 1 mM GDP. The concentrations of both ssDNA oligomers were 4.9 × 10−7M(oligomer). The concentration of the DnaB–DnaC complex was equal to the total concentration of the DnaB hexamer (detail in the text). The solid lines are nonlinear, least-squares fits of the titration curves to a single-binding site isotherm (eq 2) with two fitting parameters: for the 20-mer, K20= 2 × 108 M−1 and ΔFmax=10.7, and for the 10-mer, K10 =2 × 108M−1 and ΔFmax=12.4.
Analytical Sedimentation Velocity Studies of the Binding of the ssDNA to the Total DNA-Binding Site and the Strong DNA-Binding Subsite of the DnaB Helicase, Engaged in the DnaB-DnaC Complex, in the Presence of ADP
The surprisingly high affinity of the DnaB–DnaC complex for the ssDNA in the presence of diphosphate nucleosides has been further addressed, using the analytical sedimentation method (19, 26, 27, 34). We utilized the fact that the sedimentation coefficients of the ssDNA oligomers, 5′-Fl-dA(pA)19 and 5′-Fl-dA(pA)9, which contain fluorescein at their 5′ end, are ~1.3–1.5 S, i.e., much smaller than the sedimentation coefficient of the binary DnaB–DnaC complex (~17 S) (19). Therefore, the sedimentation of the nucleic acids in the tertiary complex can be exclusively monitored at the fluorescein absorption band (485 nm) without any interference from the free protein and nucleotide cofactor absorbances [companion papers (DOI 10.1021/bi900050x and DOI 10.1021/bi9000529)] (19, 26, 27, 34).
Sedimentation velocity profiles of 5′-Fl-dA(pA)19, alone in buffer T4 (pH 8.1 and 10 °C), are shown in Figure 5a. The profiles have been recorded at a rotational speed of 30000 rpm. The total nucleic acid concentration was 7 × 10−7M(oligomer). Clearly, at the applied rotational speed, the sedimentation boundary of the DNA moves very slowly from the meniscus. The situation is different in the presence of the binary DnaB–DnaC complex. Sedimentation velocity profiles of 5′-Fl-dA (pA)19, in the presence of the binary complex in buffer T4 (pH 8.1 and 10 °C), containing 1 mM ADP, are shown in Figure 5b. The total nucleic acid concentration was 7 × 10−7 M(oligomer), while the concentration of the DnaB–DnaC complex was 1.5 × 10−6 M (expressed as the concentration of the DnaB hexamer). Equilibrium binding data discussed above indicate that, at these selected concentrations, the nucleic acid should be almost completely saturated with the binary complex (Figure 2a). Inspection of the profiles in Figure 5b shows that there is, within experimental accuracy, a single moving sedimentation boundary indicating the presence of a single molecular species (19, 26, 27, 34). Thus, as predicted on the basis of the equilibrium titration data, practically, the entire population of the ssDNA oligomer is indeed associated with the binary complex. For the selected concentrations of the macromolecular species involved, the value of the binding constant, which would provide such behavior, must be ≥107 M−1 (Figure 2a,b). The sedimentation velocity scans have been analyzed using the time-derivative approach, which provides an s20,w of 16.9 ± 0.3 S, for the tertiary DnaB–DnaC–20-mer complex [companion paper (DOI 10.1021/bi900050x)] (19).
Figure 5.
(a) Absorption profiles, recorded at 485 nm, of analytical sedimentation velocity runs of the ssDNA 20-mer, 5′-Fl-dA)(pA)19 alone, in buffer T4 (pH 8.1 and 10 °C), containing 1 mM ADP. The total nucleic acid concentration was 7 × 10−7 M (oligomer). (b) Absorption profiles, recorded at 485 nm, of the analytical sedimentation velocity runs of the ssDNA 20-mer, 5′-Fl-dA(pA)19, in the presence of the binary DnaB–DnaC complex, in buffer T4 (pH 8.1 and 10 °C), containing 1 mM ADP. The total nucleic acid and protein concentrations were 7 × 10−7 M(oligomer) DnaB, 1.5 × 10−6 M (hexamer), and 9 × 10×6 M DnaC. (c) Absorption profiles, recorded at 485 nm, of analytical sedimentation velocity runs of the ssDNA -10-mer, 5′-Fl-dA(pA)9 alone, in buffer T4 (pH 8.1 and 10 °C), containing 1 mM ADP. The total nucleic acid concentration was 3 × 10−6 M (oligomer). (d) Absorption profiles, recorded at 485 nm, of analytical sedimentation velocity runs of the ssDNA 10-mer, 5′-Fl-dA(pA)9 in the presence of the binary DnaB–DnaC complex, in buffer T4 (pH 8.1 and 10 °C), containing 1 mM ADP. The total nucleic acid and protein concentrations were 3 × 10−6 M (oligomer) DnaB, 1.5 × 10−6 M (hexamer), and 9 × 10−6 M DnaC. All scans were collected at 30000 rpm.
Analogous sedimentation velocity profiles of the ssDNA 10-mer, 5′-Fl-dA(pA)9, alone in buffer T4 (pH 8.1 and 10 °C), containing 1 mM ADP, are shown in Figure 5c. The total nucleic acid concentration was 3 × 10−6 M (oligomer). As observed for the 20-mer, the sedimentation boundary of the 10-mer moves very slowly from the meniscus. Sedimentation velocity profiles of 5′-Fl-dA(pA)9, in the presence of the binary complex in buffer T4 (pH 8.1 and 10 °C), containing 1 mM ADP, are shown in Figure 5d. The concentration of the DnaB–DnaC complex was 1.5 × 10−6 M (expressed as the concentration of the DnaB hexamer). The value of the binding constant, ~1.7 × 10−7 M−1, obtained in equilibrium studies, indicates that, at the selected total concentrations, the binary complex should be saturated with the 10-mer. In other words, ~50% of the total nucleic acid population should comigrate with the binary complex (Figure 2a,b). The fast-moving boundary in Figure 5d corresponds to the nucleic acid bound in the tertiary complex, and as predicted, it constitutes ~50% of the total absorbance of the DNA. Analysis of the sedimentation velocity scans in Figure 5d, using the time-derivative approach, provides an s20,w of 16.8 ± 0.3 S, for the tertiary DnaB–DnaC–10-mer complex [companion paper (DOI 10.1021/bi900050x)] (19).
Mechanism of DNA-Independent NTP Binding and Hydrolysis at a Single, Noninteracting Site on the DnaB Helicase Engaged in the DnaB–DnaC Complex
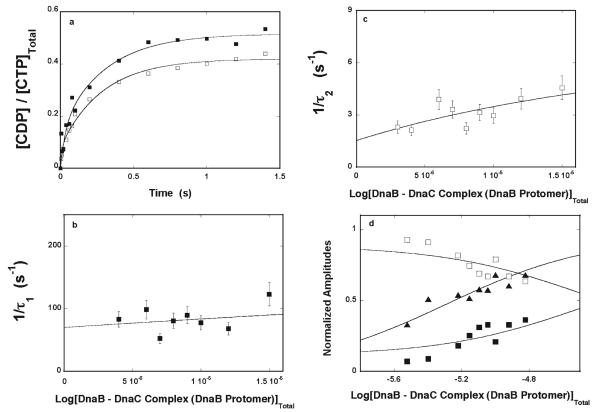
Both the DnaB helicase and the DnaC protein bind adenosine cofactors (20-25, 30-32). Thus, to avoid any possibility of a trivial competitive effect of the DnaC protein on the observed kinetics, the mechanism of the NTP hydrolysis by a single noninteracting site of the DnaB helicase, with the enzyme engaged in the DnaB–DnaC complex, has been examined using CTP and GTP cofactors. A rapid quench-flow kinetic curve of [α-32P]CTP hydrolysis after 1.9 × 10−7 M CTP had been mixed with the DnaB–DnaC complex in buffer T4 is shown in Figure 6a. The concentrations of the DnaB helicase and the DnaC protein were 8 × 10−6 M (hexamer) and 2 × 10−5 M, respectively (final concentrations); i.e., the DnaB protein is >95% saturated with the DnaC protein [companion paper (DOI 10.1021/bi900050x)]. The product formation, [α-32P]CDP, is shown as a molar fraction of the total concentration of [α-32P]CTP. For the sake of comparison, the analogous kinetic curve, obtained for the DnaB protein alone, at the same enzyme concentration, is included in Figure 6a. The two important qualitative aspects of the kinetic curve, already illustrated in the case of the DnaB helicase alone, are even more pronounced for the DnaB–DnaC complex [companion paper (DOI 10.1021/bi9000529)] (25). First, the plateau is significantly lower than the maximum possible value of 1 and is also lower than the corresponding plateau observed for the helicase alone. Thus, the system reaches an effective, transient equilibrium in the active site, which is different from the transient equilibrium observed in the absence of the DnaC protein (25). Second, there is clearly no detectable time lag in the CDP production, which indicates that one simultaneously observes at least two species containing [α-32P]CDP (25).
Figure 6.
(a) Time course of CTP hydrolysis by a single, noninteracting site of the DnaB helicase, engaged in the complex with the DnaC protein (□), in single-turnover experiments, in buffer T4. The produced CDP is shown as a fraction of the total concentration of total concentration of the CTP in the sample. The total concentration of the cofactor was 1.9 × 10−7 M. The concentrations of the DnaB and DnaC proteins were 6.0 × 10−6 M (protomer) and 2.0 × 10−5 M, respectively. For the sake of comparison, the time course of CTP hydrolysis by a single, noninteracting site of the DnaB helicase is included in the panel, obtained at the same cofactor and enzyme concentration (■). The solid line is the nonlinear, least-squares fit of the double-exponential function (eq 1) to the experimental curve. (b) Dependence of the reciprocal of the relaxation time, 1/τ1, for the CTP hydrolysis by the single, noninteracting site of the DnaB helicase, engaged in the complex with the DnaC protein, on the total concentration of the DnaB–DnaC complex, expressed as the concentration of the DnaB protomers. (c) Dependence of the reciprocal of the relaxation time, 1/τ2, for the CTP hydrolysis by the single, noninteracting site of the DnaB helicase, engaged in the complex with the DnaC protein, in buffer T4, on the total concentration of the DnaB–DnaC complex, expressed as the concentration of the DnaB protomers. The error bars in panels b and c are standard deviations determined in three or four independent experiments. (d) Dependence of the normalized total and individual relaxation amplitudes, AT (●), A1 (■), and A2 (□), of CTP hydrolysis by a single, noninteracting site of the DnaB helicase, engaged in the complex with the DnaC protein, on the total concentration of the DnaB–DnaC complex expressed as the concentration of the DnaB protomers. The solid lines in panels b–d are nonlinear, least-squares fits according to the three-step sequential mechanism (eq 3). The rate constants, obtained from the fits of the relaxation times, and the amplitudes are included in Table 1.
The plot in Figure 6a is clearly biphasic. The solid line in Figure 6a is a nonlinear least-squares fit of the experimental curve using eq 1 with a double-exponential function. The reciprocal relaxation times, 1/τ1 and 1/τ2, as functions of the DnaB–DnaC complex concentration, are shown in panels b and c of Figure 6, respectively (24, 25, 31, 32, 40, 54). The concentration of the complex is expressed as DnaB protomers [companion paper (DOI 10.1021/bi9000529)]. The values of 1/τ1 are considerably higher than those observed for the DnaB helicase alone [companion paper (DOI 10.1021/bi9000529)]. Nevertheless, 1/τ1 is not significantly dependent upon the DnaB concentration, while the values of 1/τ2 show only a slight dependence upon the concentration of the DnaB–DnaC complex. The lack of a time lag and the observed behavior of the relaxation times indicate that both relaxation processes are intramolecular isomerization transitions (25). The dependencies of the individual amplitudes, A1 and A2, of the observed relaxation steps on the concentration of the DnaB–DnaC complex, expressed as the DnaB protomers, are shown in Figure 6d. The absolute values of individual amplitudes are normalized, i.e., expressed as fractions of the total amplitude, Ai/ΣAi. The total amplitude is normalized to the total concentration of the CTP in the sample. The behavior of the amplitudes is analogous to the behavior of the same parameters observed for the DnaB helicase alone [companion paper (DOI 10.1021/bi9000529)]. The slowest relaxation process (amplitude A2) dominates the observed kinetic process at low concentrations of the DnaB–DnaC complex. Both the total amplitude, AT, and the amplitude of the faster normal mode, A1, slowly increase with the concentration of the complex. Such behavior of the relaxation amplitudes confirms that both relaxation processes reflect intramolecular transitions (24, 25, 31, 32, 40, 54).
The behavior of the purine cofactor, GTP, is different. A rapid quench-flow kinetic curve of [α-32P]GTP hydrolysis, after 1.9 × 10−7 M CTP had been mixed with the DnaB–DnaC complex in buffer T4, is shown in Figure 7a. The concentrations of the DnaB helicase and the DnaC protein were 5 × 10−6 M (hexamer) and 2 × 10−5 M, respectively (final concentrations), which ensure that the helicase is predominantly in the complex with the DnaC protein [see above and a companion paper (DOI 10.1021/bi900050x)]. The kinetic curve, obtained for the helicase protein alone, at the same enzyme concentration, is also included in Figure 7a. The plateau of the plot is lower than the maximum possible value of 1 and is also lower than the plateau observed for the enzyme alone (see above). Nevertheless, the plateau reaches the value of ~0.72, as compared to only ~0.54 in the case of the pyrimidine cofactor (Figure 7a). The two phases of product formation are clearly seen in the plot in Figure 7a, although they are less pronounced as observed for CTP. Nevertheless, the absence of a time lag in the appearance of GDP indicates that simultaneously at least two species containing [α-32P]GDP are observed in the experiment [companion paper (DOI 10.1021/bi9000529)] (25). The solid line in Figure 7a is a nonlinear least-squares fit of the experimental curve using eq 1 with a double-exponential function, which provides an excellent description of the experimental curve.
Figure 7.
(a) Time course of GTP hydrolysis by a single, noninteracting site of the DnaB helicase, engaged in the complex with the DnaC protein (□), in single-turnover experiments, in buffer T4. The produced GDP is shown as a fraction of the total concentration of the GTP in the sample. The total concentration of the cofactor was 1.9 × 10−7 M. The concentrations of the DnaB and DnaC proteins were 5.0 × 10−6 M (protomer) and 2.0 × 10−5 M, respectively. For the sake of comparison, the time course of GTP hydrolysis by a single, noninteracting site of the DnaB helicase is included in the panel, obtained at the same cofactor and enzyme concentration (■). The solid line is a nonlinear, least-squares fit of the double-exponential function (eq 1) to the experimental curve. (b) Dependence of the reciprocal of the relaxation time, 1/τ1, for the GTP hydrolysis by the single, noninteracting site of the DnaB helicase, engaged in the complex with the DnaC protein, in buffer T4, on the total concentration of the DnaB–DnaC complex, expressed as the concentration of the DnaB protomers. (c) Dependence of the reciprocal of the relaxation time, 1/τ2,for the GTP hydrolysis by the single, noninteracting site of the DnaB helicase, engaged in the complex with the DnaC protein, in buffer T4, on the total concentration of the DnaB-DnaC complex, expressed as the concentration of the DnaB protomers. The error bars in panels b and c are standard deviations determined in three or four independent experiments. (d) Dependence of the normalized total and individual relaxation amplitudes, AT (●), A1 (■), and A2 (□), of GTP hydrolysis by a single, noninteracting site of the DnaB helicase, engaged in the complex with the DnaC protein, in buffer T4, on the total concentration of the DnaB–DnaC complex, which is expressed as the total concentration of the DnaB protomers. The solid lines in panels b–d are nonlinear, least-squares fits according to the three-step sequential mechanism (eq 3). The rate constants, obtained from the fits of the relaxation times, and amplitudes are included in Table 1.
The reciprocal relaxation times, 1/τ1 and 1/τ2, as functions of the concentration of the DnaB–DnaC complex, expressed as DnaB protomers, are shown in panels b and c of Figure 7, respectively (see above). The behavior of the relaxation times is analogous to the behavior observed for the CTP cofactor, indicating that both relaxation processes are isomerization steps (24, 25, 31, 32, 40, 54). This is further confirmed by the dependence of the normalized total, AT, and individual amplitudes, A1 and A2, of the observed relaxation processes on the concentration of the DnaB–DnaC complex, expressed as the DnaB protomers, shown in Figure 7d (see above). The slowest relaxation process (amplitude A2) dominates the observed kinetic process over a much smaller range of concentrations of the DnaB–DnaC complex, as compared to CTP. As a result, the amplitude of the fast relaxation process, A1, approaches the amplitude of the low process, A2, at a higher concentration of the DnaB–DnaC complex. The behavior of the amplitudes indicates that internal equilibria among the intermediates of the reaction are different from the analogous equilibria in the CTP hydrolysis reaction (Figure 6d).
The obtained data indicate that the simplest mechanism of the CTP and GTP hydrolysis by the DnaB–DnaC complex is described by the general kinetic equation [companion paper (DOI 10.1021/bi9000529)] as DnaB–DnaC
| (3) |
This is a sequential three-step mechanism in which the bimolecular association of the cofactor with the active site of one of the DnaB subunits (H + NTP ↔ T H-NTP) is followed by two reversible first-order transitions, NTP hydrolysis [H-NTP ↔ T (H-NDP · Pi)1] and isomerization of the helicase–product complex [(H-NDP · Pi)1 ↔ (H-NDP 3 Pi)2] (25). In other words, the NTP hydrolysis by the DnaB helicase engaged in the DnaB–DnaC complex proceeds by the same kinetic mechanism that was observed for the helicase alone [companion paper (DOI 10.1021/bi9000529)]. Moreover, the kinetic mechanism is the same for hydrolysis of the purine and pyrimidine cofactors by the DnaB–DnaC complex. The solid lines in Figures 6b–d and 7b–d are nonlinear least-squares fits of the relaxation times and amplitudes according to the proposed mechanism (eq 3). The fits were initiated by numerical, nonlinear least-squares fitting of the individual relaxation times and amplitudes, using the matrix projection operator approach (24, 25, 31, 32, 40, 54). Simultaneous global fitting of both the relaxation times and the amplitudes further refined the obtained values of the equilibrium and rate constants included in Table 1.
Table 1.
Thermodynamic and Kinetic Parameters of the NTP Hydrolysis by the DnaB-DnaC Complex, in a Single Turnover, in the Absence of Various ssDNA Oligomers, in Buffer T4 (pH 8.1 and 20 °C) (see the text for details)a
| NTP complex | K1 (M−1) | K2 | K3 | k2 (s−1) | k−2 (s−1) | k3 (s−1) | k−3 (s−1) |
|---|---|---|---|---|---|---|---|
| CTP | |||||||
| DanB–DanC | (7.5 ± 2.3) × 104 | 0.5 ± 0.1 | 3.8 ± 1.3 | 36 ± 5 | 80 ± 12 | 13.2 ± 2 | 3.5 ± 0.5 |
| DanB–DanC–dA(pA)19 | (9.1 ± 3.0) × 103 | 3.1 ± 1.1 | 5.8 ± 1.9 | 190 ± 37 | 60 ± 12 | 10.0 ± 1.5 | 1.8 ± 0.3 |
| DanB–DanC–dC(pC)19 | (6.0 ± 2.0) × 103 | 4.0 ± 1.3 | 18.6 ± 6 | 300 ± 45 | 75 ± 11 | 13.0 ± 2.1 | 0.7 ± 0.1 |
| DanB–DanC–dT(pa)19 | (3.1 ± 1.0) × 103 | 6.4 ± 2.1 | 4.3 ± 1.4 | 380 ± 57 | 59 ± 9 | 9.0 ± 1.4 | 2.1 ± 0.4 |
| GTP | |||||||
| DanB–DanC | (1.7 ± 0.5) × 104 | 5.6 ± 1.8 | 5.5 ± 1.7 | 150 ± 23 | 27 ± 5 | 6.0 ± 0.9 | 1.1 ± 0.1 |
| DanB–DanC–dA(pA)19 | (9.3 ± 3.1) × 103 | 9.6 ± 0.7 | 5.1 ± 1.3 | 1200 ± 180 | 125 ± 19 | 5.6 ± 0.8 | 1.1 ± 0.1 |
| DanB–DanC–dC(pC)19 | (2.9 ± 0.9) × 104 | 2.0 ± 0.7 | 22.5 ± 7.1 | 180 ± 27 | 90 ± 14 | 18.0 ± 2.9 | 0.8 ± 0.1 |
| DanB–DanC–dT(pT)19 | (2.7 ± 0.8) × 104 | 2.5 ± 0.8 | 15.0 ± 5 | 130 ± 20 | 53 ± 8 | 10.5 ± 1.6 | 0.7 ± 0.1 |
Errors are standard deviations determined using three or four independent experiments.
In the case of the pyrimidine cofactor, CTP, the presence of the DnaC protein strongly increases the value of K1 to ≈7.5 × 104 M−1, which characterizes the binding step and formation of the first intermediate, H-NTP, as compared to the K1 of ≈9.1 103 M−1, observed for the free helicase [companion paper (DOI 10.1021/bi9000529)]. In fact, in the presence of the DnaC protein, the affinity of the pyrimidine cofactor is considerably higher than that observed for the purine cofactor, GTP, while in the absence of the replication factor, the corresponding affinities are very similar [companion paper (DOI 10.1021/bi9000529)]. On the other hand, the K2 value of ≈0.5, characterizing the chemical hydrolysis step [H-CTP T↔(H-CDP· Pi)1] is significantly lower for the DnaB–DnaC complex than the K2 of ≈8.4 observed in the absence of the DnaC protein [companion paper DOI 10.1021/bi9000529)]. In the presence of the DnaC protein, the K2 value of ≈5.6 for the purine cofactor, GTP, is also lower than the value of ~12.2 determined for the DnaB helicase alone [companion paper (DOI 10.1021/bi9000529)]. Nevertheless, the values of K2 for both cofactors indicate that there is no significant release of the free energy of the nucleotide hydrolysis in the chemical step by the DnaB–DnaC complex (55-57). The forward rate constant, k2, characterizing the hydrolysis step, is smaller for both cofactors than that observed for the helicase alone, indicating that the presence of the DnaC protein slows the rate of NTP hydrolysis by the enzyme. Nonetheless, the faster forward rate of hydrolysis of the purine cofactor, as compared to that of the pyrimidine cofactor, is preserved in the DnaB–DnaC complex (Table 1). The presence of the DnaC protein significantly slows the dynamics and lowers the partial equilibrium constant, K3,of the intramolecular transition [(H-NDP· Pi)1↔(H-NDP·Pi)2] for the pyrimidine cofactor, CTP, while it does not affect the same transition in the case of the purine nucleotide, GTP, as compared tothe free enzyme (see Discussion).
Nucleic Acid Specificity in the NTP Hydrolysis Reaction Catalyzed by the Tertiary DnaB–DnaC–ssDNA Complex
Examination of the NTP hydrolysis by the DnaB helicase–ssDNA complex required a high nucleic acid concentration in the sample, because the enzyme has a low ssDNA affinity in the absence of ATP or ATP nonhydrolyzable analogues (35-38, 41-44). On the other hand, as we described above, the DnaB–DnaC complex has the affinity for the ssDNA 20-mer characterized by a K20 binding constant of ~1–2 × 107 M−1, unaffected by the type of cofactor or even the absence of the nucleotides. At the concentration of the ssDNA 20-mers, 6.5 × 10−5 M (oligomer), used in the studies of the DnaB helicase alone, the enzyme engaged in the DnaB–DnaC complex is almost completely saturated with the DNA. Although such a high concentration of the nucleic acid is not necessary in the case of the DnaB–DnaC complex, for direct comparisons, we used the same concentration of various ssDNA 20-mers in the analyses described below, as applied in the studies of the helicase alone [companion paper (DOI 10.1021/bi9000529)].
A rapid quench-flow kinetic curve of [α-32P]CTP hydrolysis, after 1.9 × 10−7 M CTP had been mixed with the DnaB–DnaC complex in buffer T4, in the presence of the ssDNA 20-mer, dA(pA)19, is shown in Figure 8a. The final concentrations of the DnaB helicase, the DnaC protein, and the 20-mer were 7 × 10−6 M (hexamer), 2 × 10−5 M, and 6.5 × 10−5 M (oligomer), respectively. The kinetic curve, obtained in the absence of the nucleic acid, at the same helicase and DnaC protein concentrations, is also included in Figure 8a. Unlike the dramatic effect of the ssDNA on the DnaB helicase alone [companion paper (DOI 10.1021/bi9000529)], the presence of dA(pA)19 only moderately affects the kinetics of the NTP hydrolysis by the DnaB–DnaC complex. The total amplitude, AT, of the product formation is increased, as compared to the binary complex. Nonetheless, the total amplitude is much lower than the maximum value of 1, indicating the presence of the rapid equilibria in the active site of the enzyme (25). As previously observed for all examined systems, there is no time lag in the product formation. The solid line in Figure 8a is a nonlinear least-squares fit of the experimental curve using eq 1 with a double-exponential function, to extract the relaxation times and amplitudes of the examined relaxation process. The dependencies of the reciprocal of the relaxation times, 1/τ1 and 1/τ2, on the tertiary complex concentration are shown in panels b and c of Figure 8, respectively.
Figure 8.
(a) Time course of CTP hydrolysis by a single, noninteracting site of the DnaB helicase, engaged in the complex with the DnaC protein (■), in single-turnover experiments, in buffer T4, in the presence of the ssDNA 20-mer, dA(pA)19. The total concentration of the cofactor was 1.9 × 10−7 M. The concentrations of the DnaB helicase and the DnaC protein were 7.0 × 10−6 M (hexamer) and 2.0 × 10−5 M, respectively. The concentration of the ssDNA 20-mer was 6.5 × 10−5 M (oligomer). For the sake of comparison, the time course of CTP hydrolysis by a single, noninteracting site of the DnaB helicase engaged in the DnaB–DnaC complex is included in the panel, obtained at the same cofactor and protein concentrations (□). The solid line is a nonlinear, least-squares fit of the double-exponential function (eq 1) to the experimental curve. (b) Dependence of the reciprocal of the relaxation time, 1/τ1, for the CTP hydrolysis by the single, noninteracting site of the DnaB helicase, engaged in the tertiary DnaB–DnaC–ssDNA complex on the total concentration of the tertiary complex, expressed as the concentration of the DnaB protomers. (c) Dependence of the reciprocal of the relaxation time, 1/τ2, for the CTP hydrolysis by the single, noninteracting site of the DnaB helicase, engaged in the tertiary DnaB–DnaC–ssDNA complex, on the total concentration of the tertiary complex, expressed as the concentration of the DnaB protomers. The error bars in panels b and c are standard deviations determined in three or four independent experiments. (d) Dependence of the normalized, total, and individual relaxation amplitudes, AT (▲), A1 (■), and A2 (□), of CTP hydrolysis by a single, noninteracting site of the DnaB helicase, engaged in the tertiary DnaB–DnaC–ssDNA complex on the total concentration of the tertiary complex, which is expressed as the total concentration of the DnaB protomers. The solid lines in panels b–d are nonlinear least-squares fits according to the three-step sequential mechanism (eq 3). The rate constants, obtained from the fits of relaxation times, and amplitudes are included in Table 1.
The values of 1/τ1 and 1/τ2 are similar to the analogous values observed for the binary complex (Figure 6b,c). However, the plots obtained for the tertiary complex do not show any hyperbolic behavior, indicating that the affinity of the nucleotide binding step is significantly lower than that determined for the binary complex (see below). The dependence of the normalized total, AT, and individual amplitudes, A1 and A2, of the observed relaxation processes on the logarithm of the tertiary complex concentration, expressed as the DnaB protomers, is shown in Figure 8d. The plots are analogous to the behavior of the amplitudes shown in Figure 6d for the binary complex. The behavior of the amplitudes, together with the lack of a time lag in observed kinetics, indicates that both relaxation processes reflect intramolecular transitions of the formed complex, and eq 3 describes the simplest mechanism of the observed reaction [companion paper (DOI 10.1021/bi9000529)] (25). The solid lines in Figure 8b–d are nonlinear least-squares fits of the relaxation times and amplitudes according to the mechanism defined by eq 3. Analogous experiments have been performed for different ssDNA 20-mers, dC(pC)19 and dT(pT)19, and the purine cofactor, GTP (data not shown). The obtained rate and equilibrium constants, for all examined systems, are included in Table 1.
In the case of the pyrimidine cofactor, CTP, the value of the partial equilibrium constant, K1, is considerably decreased for all ssDNA oligomers, as compared to that of the DnaB–DnaC complex in the absence of the nucleic acid. On the other hand, the decrease in the values of K1 is compensated by a considerable increase of the partial equilibrium constants, K2, characterizing the hydrolysis step, which is also similar for all examined tertiary complexes (Table 1). Nevertheless, K2 remains small, indicating that CTP hydrolysis in the active site is accompanied by a small change in free energy (see Discussion). The values of K3 are significantly increased, as compared to that of the binary DnaB–DnaC complex, only in the case of dC(pC)19. The presence of the nucleic acids dramatically increases the forward rate constant, k2, of the CTP hydrolysis by the DnaB–DnaC complex. However, the strong stimulatory effect of dA(pA)19 on the CTP hydrolysis step, observed in the case of the DnaB helicase alone [companion paper (DOI 10.1021/bi9000529)], is absent in the case of the DnaB–DnaC complex. In fact, the values of k2 are higher for dC(pC)19 and dT(pT)19 than for the homoadenosine, ssDNA 20-mer (Table 1). Also, the presence of the DNA has little effect on the dynamics of the transition [(H-CDP · Pi)1 ↔ (H-CDP · Pi)2] following the hydrolysis step.
In the case of the purine cofactor, GTP, the values of the partial equilibrium constants, K1 and K2, for the tertiary DnaB–DnaC–ssDNA complex remain similar to the values observed for the binary DnaB–DnaC complex (Table 1). As a result, there is a free energy change, accompanying the binding of the purine cofactor to the tertiary complex, significantly larger than that observed for the pyrimidine cofactor (see Discussion). Also, unlike the case of the pyrimidine cofactor, the values of K3 are significantly increased, as compared to that of the DnaB–DnaC complex in the absence of the nucleic acid, both for dC(pC)19 and for dT(pT)19. However, the difference between CTP and GTP is particularly striking in the effect of different nucleic acids on the dynamics of the hydrolysis step. In the case of dA (pA)19, the forward rate constant, k2, is increased by a factor of ~8, as compared to that of the binary complex, reaching values of ~1200 s−1, while k2 remains little changed in the case of dC(pC)19 and dT(pT)19 (Table 1). Thus, the strong stimulatory effect of the homoadenosine oligomer on the hydrolysis step, observed before for the DnaB helicase–dA(pA)19 complex [companion paper (DOI 10.1021/bi9000529)], is preserved for the purine cofactor in the tertiary DnaB–DnaC–dA(pA)19 complex (see Discussion). Nevertheless, only the presence of dC(pC)19 significantly affects the dynamics of the step following the hydrolysis [(H-GDP · Pi)1 ↔ (H-GDP · Pi)2], with the forward rate constant, k3, increased by a factor of ~3, as compared to that of the binary complex (see Discussion).
DISCUSSION
The ssDNA Affinity of the DnaB Helicase, Engaged in the DnaB–DnaC Complex, Is Independent of the Structure of the Phosphate Group of the Cofactor
A common rule in interactions of a replicative helicase with the ssDNA is that the enzyme acquires the high ssDNA affinity only in the presence of NTP or the NTP nonhydrolyzable analogues (4-6, 58). The DnaB helicase is not an exception to this rule. In the presence of NTP nonhydrolyzable analogues, the affinity of the enzyme for the ssDNA increases by ~3–4 orders of magnitude, as compared to the ssDNA affinity in the absence of the cofactor (26, 35-38, 41-44). The effect of ADP is much more modest with the affinity of the enzyme increased by only ~1 order of magnitude. Nevertheless, in the E. coli cell, the DnaB helicase enters its physiological activities associated with the replication factor, the DnaC protein. A striking result obtained in this work is that the ssDNA affinity of the total DNA-binding site and the strong DNA-binding subsite of the helicase, engaged in the DnaB–DnaC complex, is independent of the structure of the phosphate group of the cofactor bound to the helicase and the replication factor (Figures 1 and 2). In the presence of AMP-PNP or ADP, the values of the obtained binding constants (K20 ~ 2 × 107 M−1, and K10 ~ 2 × 107 M−1) are, within experimental accuracy, the same and are similar to the values of the corresponding parameters observed for the helicase alone in the presence of AMP-PNP (35-37).
There are several major functional aspects of these results. Recall that the binary DnaB–DnaC complex is the entity that must recognize the ssDNA conformation at the replication origin, oriC, and in the formation of the preprimosome (15-18, 59). The obtained data show that the DnaB–DnaC complex can efficiently recognize the single-stranded conformation of the nucleic acid in a manner independent of the structure of the phosphate group of the cofactor. Thus, the DnaC protein eliminates, to a great extent, the dependence of the DnaB helicase on the type of nucleotide cofactors for the association with ssDNA. By the same token, the protein diminishes the competitive effect of the negative allosteric effector, NDP, on the interactions of the helicase with the ssDNA, while leaving the positive allosteric effect of NTP unaffected. The DnaB helicase alone is only a moderately processive enzyme in the dsDNA unwinding reaction (60, 61). This is due to the increased propensity of the helicase to dissociate from the nucleic acid during NTP hydrolysis (60, 61). Because the replication of the chromosomal DNA is a very processive reaction, this result indicates that the processivity of the helicase must be controlled by the interactions of the helicase with other protein ingredients of the replication apparatus (6, 59). Elimination of the negative allosteric effect of NDP on the enzyme–ssDNA affinity by the DnaC protein indicates that DnaC can be one of those protein factors.
On the other hand, the antagonistic allosteric effect of NTP and NDP on the affinity of the enzyme for the ssDNA conformation has been invoked as being intimately involved in the mechanical translocation on the nucleic lattices and the dsDNA unwinding reaction (4-6). This is because the switch from NTP to NDP in the nucleotide-binding site of the helicase, which increases the affinity of the enzyme alone for the ssDNA conformation by ~3 or more orders of magnitude, would provide a large part of the free energy necessary to perform the required mechanical translocation. However, recall that in this work, we examine the DnaB–DnaC complex, which contains six DnaC molecules (19). The observed effect of the DnaC protein on the ssDNA affinity in the presence of nucleoside tri- versus diphosphate strongly suggests that, when six DnaC molecules are associated with the helicase, the tertiary DnaB–DnaC–ssDNA complex will have an increased efficiency of ssDNA recognition but a diminished efficiency of mechanical translocation and dsDNA unwinding processes (see below).
The Nucleotide-Binding Site of the DnaC Protein Provides Additional Control of the ssDNA Affinity of the Helicase in the DnaB–DnaC Complex
Because the DnaC protein binds only adenosine nucleotides, in the presence of GDP, only the DnaB helicase is associated with the nucleotide cofactor (30-32), yet the ssDNA affinities of the total DNA-binding site and the strong DNA-binding subsite of the enzyme, engaged in the DnaB–DnaC complex, are higher by 1 order of magnitude, in the presence of GDP, as compared to that of the binary complex in which both protein components are saturated with AMP-PNP or ADP (Figure 4a). Recall that the DnaC protein exists in equilibrium between two conformational states, with only one state capable of binding nucleotides (30-32). DnaB helicase preferentially stabilizes the DnaC conformation, which does not bind ADP, particularly, in the presence of the ssDNA [companion paper (DOI 10.1021/bi900050x)]. Thus, in the presence of GDP, the DnaC protein in the tertiary complex is predominantly in the conformational state that does not bind cofactors. The data obtained in this work indicate that, in this state, the DnaC protein acts as a positive allosteric effector on the ssDNA affinity of the helicase, both for the total DNA-binding site and for the strong DNA-binding subsite of the enzyme.
The DnaC Protein Strongly and Differently Affects the Structure of the DNA Associated with the Total DNA-Binding Site of the DnaB Helicase in the DnaB–DnaC Complex, in the Presence of an ATP Nonhydrolyzable Analogue, As Compared to ADP
Another startling result is the dramatic effect of the DnaC protein on the structure of the ssDNA associated with the DnaB helicase in the DnaB–DnaC complex. In the presence of AMP-PNP, binding of the DnaB helicase to the ethenoadenosine 20-mer, dεA(pεA)19, induces an ~2.6-fold increase in nucleic acid fluorescence [companion paper (DOI 10.1021/bi9000529)] (35-38, 44). Under the same solution conditions, binding of the DnaB–DnaC complex induces a much larger, ~6-fold, increase in 20-mer fluorescence (Figure 1a). As we pointed out in a companion paper (DOI 10.1021/bi9000529), the fluorescence of ethenoadenosine is strongly quenched in etheno oligomers, as compared to that of free εAMP (45-48). A dynamic model, in which the motion of adjacent ethenoadenosines leads to quenching via intramolecular collision, has been proposed as the quenching mechanism (47). Thus, structural separation of the bases and/or increased viscosity of the environment lead to a decreased base mobility and a diminished fluorescence quenching.
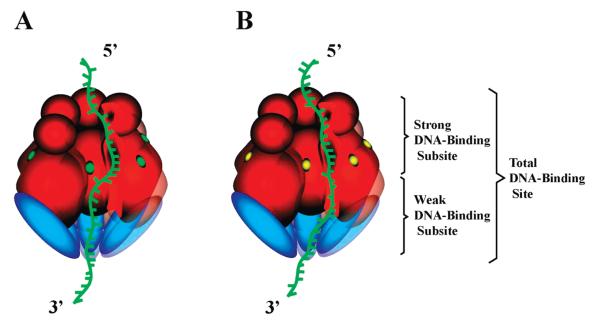
Because at the applied excitation wavelength (320 nm) only ethenoadenosines are excited, the observed increase in the emission intensity of dεA(pεA)19 must result from an increase in the quantum yield of the nucleic acid in the complex with the DnaB–DnaC complex. Therefore, the dramatic increase in the fluorescence emission of dεA(pεA)19, upon binding to the DnaB–DnaC complex in the presence of AMP-PNP, indicates a significant increase in the degree of restriction of the base mobility and/or separation of the adjacent bases of the nucleic acid in the tertiary complex. An even larger effect is observed in the presence of ADP, where the fluorescence emission of dεA(pεA)19 is increased ~12-fold, i.e., twice as much as observed in the case of the ATP analogue. The simplest interpretation of this result is that the same structural changes of the nucleic acid, responsible for the observed fluorescence increase of dεA(pεA)19, are induced in the presence AMP-PNP and ADP. However, in the presence of ADP, the total DNA-binding site of the helicase in the DnaB–DnaC complex becomes engaged in interactions with the nucleic acid; i.e., both the strong and weak DNA-binding subsites interact in a similar fashion with the ssDNA and undergo a similar structural transition. Notice that the DnaC protein is bound to the large 33 kDa domain of each of the DnaB subunits where the weak DNA-binding site is located (19). The data strongly suggest that the DnaC exerts a specific effect on the weak DNA-binding subsite, through the structural changes of the large 33 kDa domain [companion paper (DOI 10.1021/bi900050x)].
On the other hand, in the presence of AMP-PNP, the strong DNA-binding subsite predominantly interacts with the ssDNA (22, 24, 25, 37, 38, 44). Thus, only half of the 20-mer, ~10 nucleotides, is occluded by the strong DNA-binding subsite ~and undergoes a major structural transition, i.e., an increase in base separation and/or a decrease in base mobility (45-48). The remaining half of the bound 20-mer would only weakly interact with the weak DNA-binding subsite of the enzyme, which does not induce any significant conformational changes in the bound DNA (62). As a result, an ~2-fold lower relative fluorescence increase for the nucleic acid should be observed, as compared to that in the presence of ADP, as experimentally observed. The tertiary DnaB–DnaC–ssDNA complexes, in the presence of AMP-PNP or ADP, indicated with a different engagement of the ssDNA in the interactions with the total ssDNA-binding site, are schematically shown in Figure 9.
Figure 9.
Schematic representation of engagement of ssDNA in interactions with the total DNA-binding site of the DnaB helicase in the tertiary DnaB–DnaC–ssDNA complex in the presence of AMP-PNP and ADP. The total ssDNA-binding site occludes ~20 nucleotides of the DNA. In the presence of AMP-PNP (A), the nucleic acid in the tertiary complex predominantly engages the strong DNA-binding subsite, which is located in the vicinity of the 12 kDa domain and occludes ~10 nucleotides the 5′ end of the bound nucleic acid. In the presence of ADP (B), the nucleic acid engages both the strong and weak DNA-binding subsites, located in the 33 kDa domain of the helicase, to which the DnaC protein is bound. The green and yellow ovals indicate the location of the NTP-binding site saturated with NTP and ADP, respectively.
The DnaC Protein Strongly Affects the Structure of the DNA Associated with the Strong DNA-Binding Subsite of the DnaB Helicase in the DnaB–DnaC Complex, in a Manner Independent of the Structure of the Phosphate Group of the Nucleotide Cofactor
The behavior of the ssDNA 10-mer, dεA(pεA)9, associated with the DnaB–DnaC complex is different from the behavior of the 20-mer discussed above. Recall that the 10-mer binds exclusively to the strong DNA-binding subsite of the DnaB helicase (44). Similarly, ~10- and ~11-fold increases in 10-mer fluorescence are observed in the presence of AMP-PNP and ADP, respectively (Figure 3c). First, these data indicate that the structure of the ssDNA, bound to the strong DNA-binding subsite of the helicase, engaged in the DnaB–DnaC complex, is very different from the analogous structure of the nucleic acid in the complex with the helicase alone, where an only ~2-fold increase in the etheno derivative fluorescence is observed (26). As discussed above, the data on the binding of the DnaB–DnaC complex to the 20-mer in the presence of AMP-PNP strongly suggest that the DnaC exerts a specific effect on the weak DNA-binding subsite. However, the results on the 10-mer binding indicate that the bound DnaC protein specifically affects the entire total DNA-binding site, possibly eliminating the functional distinction between the subsites in the presence of ADP.
Second, the seemingly different behavior of the bound 10-mer, as compared to the 20-mer, which engages the total DNA-binding site, corroborates the conclusions reached in the previous section (see above). In the presence of both AMP-PNP and ADP, the same conformational state of the bound nucleic acid in the strong DNA-binding subsite is induced in the DnaB–DnaC complex. The 10-mer is completely engaged in interactions with the helicase in the strong DNA-binding subsite, and this conformational state is characterized by ~11-fold increase in 10-mer fluorescence (see above). On the other hand, in the presence of AMP-PNP, the 10-mer is too short to engage the weak DNA-binding subsite. As a result, the fluorescence increase of the 10-mer is similar in the presence of both AMP-PNP and ADP and is very similar to the fluorescence increase of the 20-mer, fully engaged in interactions with the total DNA-binding site of the helicase in the DnaB–DnaC complex, in the presence of ADP, as experimentally observed.
The Single, Noninteracting Site on the DnaB Helicase in the Binary DnaB–DnaC Complex Hydrolyzes NTP with the Same Kinetic Mechanism as the DnaB Helicase Alone, in a Manner Independent of the Type of Base of the Nucleotide Cofactor
Analyses of the single-turnover experiments of the CTP and GTP hydrolyses by the binary DnaB–DnaC complex using the relaxation kinetics method have been based on the same principles that were discussed for the DnaB helicase alone in a companion paper (DOI 10.1021/bi9000529). The total amplitude of the kinetic curves of the NTP binding and hydrolysis by the binary complex is significantly lower than 100% of the total available concentrations of the cofactors, indicating that in the time range of the single-turnover experiments, the entire relaxation process can be treated as a reversible reaction (25). The absence of a time lag provides a strong clue that at least two intermediates containing the product of the reaction, NDP, are experimentally observed (25). All kinetic curves required the double-exponential function to represent the time course of the reaction. Consequently, the kinetic mechanism of the NTP hydrolysis by a single, noninteracting site on the DnaB helicase, engaged in the DnaB–DnaC complex, is a three-step sequential process, described by eq 3, as determined for the helicase alone [companion paper (DOI 10.1021/bi9000529)]. Moreover, the mechanism is independent of the type of the base of the nucleotide cofactor, and its sequential nature indicates that the binary complex is predominantly in a single conformational state prior to nucleotide binding [companion paper (DOI 10.1021/bi9000529)]. Therefore, the DnaC protein does not change the major features of the mechanism of the NTP hydrolysis by the helicase but rather modulates them (see below).
The Binary DnaB–DnaC Complex Has a Diminished Rate of Chemical NTP Hydrolysis, Which Deepens the Difference between the Rate of Hydrolysis of the Purine and That of the Pyrimidine Cofactor, As Compared to That of the Helicase Alone
Unlike the DnaB helicase alone [companion paper (DOI 10.1021/bi9000529)], the binary DnaB–DnaC complex has a higher affinity for the pyrimidine cofactor, CTP, than for GTP (Table 1). Nevertheless, the affinity difference is compensated by a drop of more than 1 order of magnitude in the value of the partial equilibrium constant, K2, as compared to that of the helicase alone, in the case of CTP, while K2, in the case of GTP, remains similar to the value obtained for the enzyme alone. The diminished value of K2 for CTP results from a decrease in the forward rate constant, k2, from ~83 s−1 in the case of the enzyme alone to ~36 s−1 for the DnaB–DnaC complex and a strongly increased backward rate constant, k−2, from ~9.9 s−1, characterizing the free helicase, to ~80 s−1, characterizing the binary complex. Analogous values for the purine cofactor, GTP, are k2 ~ 220 s−1 and k−2 ~ 21 s−1 for the enzyme alone and k2 ~ 150s−1 and k−2 ~ 27 s−1 for the enzyme in the binary complex (Table 1). Thus, the DnaB–DnaC complex hydrolyzes NTPs, particularly the pyrimidine cofactor, at a significantly slower rate than the helicase alone. Moreover, in a manner independent of any kinetic model, the efficiency of the hydrolysis by the binary complex is clearly diminished, as already shown by the much lower total amplitude of all kinetic curves, as compared to the free enzyme (Figures 6a and 7a). Recall that the binary complex is the entity that recognizes the origin of replication, oriC, and the preprimosome complex (15-18). Thus, prior to these processes involving the binary complex, the intrinsic NTPase activity of the helicase is diminished by the DnaC protein. Furthermore, the dynamics of the global conformational changes, controlled by the NTP–NDP switch in the nucleotide-binding site of the helicase, already observed for the analogous RepA hexameric helicase, will be significantly slower in the case of the binary complex, particularly for the pyrimidine cofactors, as compared to the helicase alone, indicating that the role of the DnaC is to preferentially select the state of the DnaB helicase with less dynamic flexibility, necessary for the ssDNA recognition, but less efficient in the free energy transduction (see below) (27).
In the Tertiary Complex, the ssDNA Reverses Most of the DnaC Effect on the NTPase Activity of the DnaB Helicase Observed in the Binary Complex and Dramatically Changes the Internal Energetics between Intermediates, Particularly, for the Pyrimidine Cofactor, As Compared to the Helicase Alone
The kinetic mechanism (eq 3) of NTP hydrolysis is preserved in the tertiary DnaB–DnaC–ssDNA complex. Surprisingly, the affinity of the tertiary DnaB–DnaC–ssDNA complex for the pyrimidine cofactor is lower by ~1 order of magnitude, as compared to that of the binary DnaB–DnaC complex, for all examined ssDNA oligomers (Table 1). The affinity is also significantly lower than that observed for the helicase alone [companion paper (DOI 10.1021/bi9000529)]. This is, in part, compensated by the strong increase in the partial equilibrium constant, K2, as compared to that of the binary complex. Thus, the presence of the nucleic acid in the tertiary complex reverses most of the effect of the DnaC protein observed in the binary complex. Moreover, the striking difference between the effect of dA(pA)19 and the effect of dC(pC)19 and dT(pT)19 on the hydrolysis step [H-NTP ↔ (H-NDP · Pi)1] observed for the enzyme alone is missing. In fact, in the presence of the adenosine homo-oligomer, forward rate constant k2 ~ 190 s−1, while in the presence of dT(pT)19, k2 ~ 380 s−1 (Table 1). The corresponding parameters for the free enzyme are as follows: k2 ~ 400 s−1, and k2 ~ 114 s−1, respectively [companion paper (DOI 10.1021/bi9000529)]. In the case of the purine cofactor, GTP, the presence of the nucleic acid has an only modest effect on the partial equilibrium constant, K1. Although the specific stimulatory effect of dA(pA)19 on the GTP hydrolysis step is clearly present, the effect is much less pronounced than that observed for the enzyme alone (Table 1).
Both the binary and the tertiary complexes can contain a different number of bound DnaC molecules (19). Nevertheless, the complex containing six DnaC molecules dominates the population of the complex in the concentration range of the DnaC protein in vivo (19). While the AT-rich regions of oriC will dramatically increase the NTP hydrolysis rate of the bound DnaB helicase alone [companion paper (DOI 10.1021/bi9000529)], the DnaC protein, bound to the enzyme, will act antagonistically to diminish the NTPase activity of the helicase, in terms of both the rate of the process and its efficiency, through changes in the helicase affinity for the nucleotide cofactors and their type, and through changes of the stimulatory effect of the homoadenosine sequences (62, 63). The physiological role of such control exerted on the helicase is still unknown. Nevertheless, the data indicate that in the elongation steps of the DNA replication, the tertiary complex will have different rates of mechanical translocation, depending on the type of bound cofactor and the DNA sequence, similar to the case of the DnaB helicase alone (60, 61). However, the behavior of the tertiary complex will be different from the analogous behavior of the helicase in the absence of the DnaC protein. The helicase alone will accelerate on sequences containing large fractions of adenosines and slows on sequences containing cytidines or thymines in the presence of any nucleotide cofactor (60, 61). The DnaB–DnaC complex will be faster on sequences containing cytidines or thymines in the presence of the pyrimidine cofactor and will accelerate on sequences containing adenosines, but only in the presence of a purine cofactor.
The Tertiary DnaB–DnaC–ssDNA Complex Has a Diminished Free Energy Transduction Capability, As Compared to That of the DnaB Helicase Alone
As previously mentioned [companion paper (DOI 10.1021/bi9000529)], one of the fundamental properties of a motor protein is that the free energy of NTP hydrolysis in the active site must be significantly lower than the free energy of hydrolysis of free NTP, i.e., ΔGNTP° ~ −7.5 kcal/mol, to achieve the required efficiency of the accumulation of energy in the complex (55-57, 65). In the case of the tertiary complex, the average change in the free energy of the NTP hydrolysis in the active site (ΔG2°) approaches −0.9 kcal/mol (20 °C), both for CTP and for GTP (Table 1). Thus, the tertiary complex fulfills one of the fundamental energetic conditions of a motor protein. The value of the partial equilibrium constant, K1, which characterizes the binding of the nucleoside triphosphates to the tertiary complex, is surprisingly low (Table 1). However, the very fast and efficient dsDNA unwinding reaction, catalyzed by the DnaB helicase alone, strongly suggests that the ADP affinity for the nucleotide-binding site of the DnaB helicase during NTP hydrolysis is much lower, if at all, than that observed in the stationary complex [companion paper (DOI 10.1021/bi9000529)] (60, 61). The same should be true for the tertiary complex, if engaged in the same unwinding reaction.
Taking the above considerations into account, we find the average free energy change accompanying the CTP binding to the DnaB–DnaC–ssDNA complex (20 °C) is then (ΔGCTP)1° ≈ −5.1 kcal/mol (Table 1). The following CTP hydrolysis contributes ΔG2° ≈ −0.9 kcal/mol, and the subsequent conformational transition releases ΔG3° ≈ −1.3 kcal/mol. Therefore, the total amount of free energy released in the CTP binding and hydrolysis by the tertiary complex is ΔGT° ≈ −7.3 kcal/mol, i.e., less than ΔGNTP° ≈ −7.5 kcal/mol, of the free nucleotide hydrolysis. Therefore, these data strongly suggest that the tertiary complex, containing six DnaC protein molecules, does not accumulate enough free energy to perform an efficient free energy transduction in the presence of the pyrimidine cofactor, CTP. Analogous calculations, using the data for GTP binding and hydrolysis (Table 1), provide ΔGT° ≈ −8.3 kcal/mol, indicating that the complex stores approximately −0.8 kcal/mol of the free energy of the GTP binding and hydrolysis. Although the DnaB helicase unwinds only a single base pair in a single catalytic event, the value of approximately −0.8 kcal/mol is still not enough to unwind a single base pair of the dsDNA, which requires ~2–2.5 kcal/mol (66). The approximate nature of these calculations is unavoidable. Nevertheless, they suggest that a capability free energy transduction by the tertiary complex could exist in the presence of the purine cofactor.
The results indicate that the tertiary DnaB–DnaC–ssDNA complex containing six DnaC protein molecules has a much weaker, if any, free energy transduction capability than the DnaB helicase alone, particularly in the presence of the pyrimidine cofactor [companion paper (DOI 10.1021/bi9000529)]. Free energy transduction should not be necessary for binding to the oriC or the preprimosome assembly by the DnaB–DnaC complex, as these are recognition processes. Nevertheless, recall that the excess of the DnaC protein inhibits the dsDNA unwinding by the DnaB helicase, which can occur only if tertiary complexes other than the one containing six DnaC molecules participate in the catalysis (64, 65). Moreover, the NTP hydrolysis analyses of the tertiary DnaB–DnaC–ssDNA complex corroborate very well the thermodynamic analyses of the same stationary complex [(DOI 10.1021/bi900050x)], which already strongly suggest that the tertiary complex, with six bound DnaC molecules, is not the major tertiary complex involved in the unwinding reaction. Rather, the examined tertiary complex, containing six DnaC molecules, seems to be a part of the transient, control system of the helicase activity and/or involved in the molecular pump activity of the DnaB helicase (7, 11, 67). The functional studies of DnaB–DnaC complexes, containing different numbers of bound DnaC molecules, are currently being addressed in our laboratory.
ACKNOWLEDGMENT
We to thank Gloria Drennan Bellard for reading the manuscript.
Footnotes
This work was supported by National Institutes of Health Grant R01 GM-46679 (to W.B.).
Abbreviations: NTP, ribonucleoside triphosphate; GTP, guanosine triphosphate; GDP, guanosine diphosphate; CTP, cytidine triphosphate; CDP, cytidine diphosphate; Tris, tris(hydroxymethyl)aminomethane; AMP-PNP, β,γ-imidoadenosine 5′-triphosphate; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; εA, ethenoadenosine; DTT, dithiothreitol.
REFERENCES
- 1.Bloomfield VA, Crothers DM, Tinoco I., Jr. Structure, Properties, and Functions. University Science Books; Sausalito, CA: 1999. Nucleic Acids; pp. 259–334. [Google Scholar]
- 2.Craig ME, Crothers DM, Doty P. Relaxation kinetics of Dimer Formation by Self Complementary Oligonucleotides. J. Mol. Biol. 1971;62:383–401. doi: 10.1016/0022-2836(71)90434-7. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg A, Baker TA. DNA Replication. Freeman; San Francisco: 1992. pp. 355–378. [Google Scholar]
- 4.Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: Structure and properties of isolated helicases. Q. Rev. Biophys. 2002;35:431–478. doi: 10.1017/s0033583502003852. [DOI] [PubMed] [Google Scholar]
- 5.Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: Integration of helicases into cellular processes. Q. Rev. Biophys. 2003;36:1–69. doi: 10.1017/s0033583502003864. [DOI] [PubMed] [Google Scholar]
- 6.Lohman TM, Bjorson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 7.West SC. DNA helicases: New breeds of translocating motors and molecular pumps. Cell. 1996;86:177–180. doi: 10.1016/s0092-8674(00)80088-4. [DOI] [PubMed] [Google Scholar]
- 8.Enemark EJ, Joshua-Tor L. On Helicases and Other Motor Proteins. Curr. Opin. Struct. Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeBowitz JH, McMacken R. The Escherichia coli dnaB Replication Protein Is a DNA Helicase. J. Biol. Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- 10.Baker TA, Funnell BE, Kornberg A. Helicase Action of dnaB Protein During Replication from the Escherichia coli Chromosomal Origin in Vitro. J. Biol. Chem. 1987;262:6877–6885. [PubMed] [Google Scholar]
- 11.Reha-Krantz LJ, Hurwitz J. The dnaB Gene Product of Escherichia coli, I. Purification, Homogeneity, and Physical Properties. J. Biol. Chem. 1978;253:4043–4050. [PubMed] [Google Scholar]
- 12.Reha-Krantz LJ, Hurwitz J. The dnaB Product of Escherichia coli. II. Single Stranded DNA-dependent Ribonucleoside Triphosphatase Activity. J. Biol. Chem. 1978;253:4051–4057. [PubMed] [Google Scholar]
- 13.Marians KJ. Prokaryotic DNA replication. Annu. Rev. Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 14.Bujalowski W. Expanding the Physiological Role of the Hexameric Helicases. Trends Biochem. Sci. 2003;28:116–118. doi: 10.1016/S0968-0004(03)00034-3. [DOI] [PubMed] [Google Scholar]
- 15.Arai K-I, Kornberg A. Mechanism of dnaB Protein Action. IV. General Priming of DNA Replication by dnaB Protein and Primase Compared with RNA Polymerase. J. Biol. Chem. 1981;256:5267–5272. [PubMed] [Google Scholar]
- 16.Tongu K, Peng H, Marians KJ. Identification of a Domain of E. coli Primase Required for Functional Interaction with the DnaB Helicase at the Replication Fork. J. Biol. Chem. 1994;269:4675–4682. [PubMed] [Google Scholar]
- 17.Jones JM, Nakai H. Duplex Opening by Primosome Protein PriA for Replisome Assembly on a Recombination Intermediate. J. Mol. Biol. 1999;289:503–515. doi: 10.1006/jmbi.1999.2783. [DOI] [PubMed] [Google Scholar]
- 18.Wahle E, Lasken RS, Kornberg A. The dnaB-dnaC Replication Protein Complex of Escherichia coli. I. Formation and Properties. J. Biol. Chem. 1989;264:2463–2468. [PubMed] [Google Scholar]
- 19.Wahle E, Lasken RS, Kornberg A. The dnaB-dnaC Replication Protein Complex of Escherichia coli. II. Role of the Complex in Mobilizing dnaB Functions. J. Biol. Chem. 1989;264:2469–2475. [PubMed] [Google Scholar]
- 20.Skarstad K, Wold S. The speed of the Escherichia coli fork in vivo depends on the DnaB-DnaC ratio. Mol. Microbiol. 1995;17:825–831. doi: 10.1111/j.1365-2958.1995.mmi_17050825.x. [DOI] [PubMed] [Google Scholar]
- 21.Marians KJ. PriA: At the crossroads of DNA replication and recombination. Prog. Nucleic Acid Res. Mol. Biol. 1999;63:39–67. doi: 10.1016/s0079-6603(08)60719-9. [DOI] [PubMed] [Google Scholar]
- 22.Galletto R, Jezewska MJ, Bujalowski W. Interactions of the Escherichia coli DnaB Helicase Hexamer with the Replication Factor the DnaC Protein. Effect of Nucleotide Cofactors and the ssDNA on Protein-Protein Interactions and the Topology of the Complex. J. Mol. Biol. 2003;329:441–465. doi: 10.1016/s0022-2836(03)00435-2. [DOI] [PubMed] [Google Scholar]
- 23.Bujalowski W, Klonowska MM. Negative Cooperativity in the Binding of Nucleotides to Escherichia coli Replicative Helicase DnaB Protein. Interactions with Fluorescent Nucleotide Analogs. Biochemistry. 1993;32:5888–5900. doi: 10.1021/bi00073a023. [DOI] [PubMed] [Google Scholar]
- 24.Bujalowski W, Klonowska MM. Close Proximity of Tryptophan Residues and ATP-Binding Site in Escherichia coli Primary Replicative Helicase DnaB Protein. Molecular Topography Studies. J. Biol. Chem. 1994;269:31359–31371. [PubMed] [Google Scholar]
- 25.Bujalowski W, Klonowska MM. Structural Characteristics of the Nucleotide Binding Site of the E. coli primary replicative Helicase DnaB Protein. Studies with Ribose and Base-Modified Fluorescent Nucleotide Analogs. Biochemistry. 1994;33:4682–4694. doi: 10.1021/bi00181a028. [DOI] [PubMed] [Google Scholar]
- 26.Jezewska MJ, Kim U-S, Bujalowski W. Interactions of Escherichia coli Primary Replicative Helicase DnaB Protein with Nucleotide Cofactors. Biophys. J. 1996;71:2075–2086. doi: 10.1016/S0006-3495(96)79406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bujalowski W, Jezewska MJ. Kinetic Mechanism of Nucleotide Cofactor Binding to Escherichia coli Replicative Helicase DnaB Protein. Stopped-Flow Kinetic Studies Using Fluorescent, Ribose-, and Base-Modified Nucleotide Analogs. Biochemistry. 2000;39:2106–2122. doi: 10.1021/bi992413m. [DOI] [PubMed] [Google Scholar]
- 28.Rajendran S, Jezewska MJ, Bujalowski W. Multiple-step Kinetic Mechanism of DNA-independent ATP Binding and Hydrolysis by Escherichia coli Replicative Helicase DnaB Protein: Quantitative Analysis Using the Rapid Quench-Flow Method. J. Mol. Biol. 2000;303:773–795. doi: 10.1006/jmbi.2000.4124. [DOI] [PubMed] [Google Scholar]
- 29.Bujalowski W, Klonowska MM, Jezewska MJ. Oligomeric Structure of Escherichia coli Primary Replicative Helicase DnaB Protein. J. Biol. Chem. 1994;269:31350–31358. [PubMed] [Google Scholar]
- 30.Marcinowicz A, Jezewska MJ, Bujalowski W. Multiple Global Conformational States of the Hexameric RepA Helicase of Plasmid RSF1010 With Different ssDNA-Binding Capabilities Are Induced By Different Numbers of Bound Nucleotides. Analytical Ultracentrifugation and Dynamic Light Scattering Studies. J. Mol. Biol. 2008;375:386–408. doi: 10.1016/j.jmb.2007.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jezewska MJ, Lucius AL, Bujalowski W. Binding of Six Nucleotide Cofactors to the Hexameric Helicase RepA Protein of Plasmid RSF1010. I. Direct Evidence of Cooperative Interactions Between the Nucleotide-Binding Sites of a Hexameric Helicase. Biochemistry. 2005;44:3865–3876. doi: 10.1021/bi048037+. [DOI] [PubMed] [Google Scholar]
- 32.Jezewska MJ, Lucius AL, Bujalowski W. Binding of Six Nucleotide Cofactors to the Hexameric Helicase RepA Protein of Plasmid RSF1010. II. Base Specificity, Nucleotide Structure, Magnesium, and Salt Effect on the Cooperative Binding of the Cofactors. Biochemistry. 2005;44:3877–3890. doi: 10.1021/bi048036h. [DOI] [PubMed] [Google Scholar]
- 33.Galletto R, Rajendran S, Bujalowski W. Interactions of Nucleotide Cofactors with the Escherichia coli Replication Factor DnaC Protein. Biochemistry. 2000;39:12959–12969. doi: 10.1021/bi0012484. [DOI] [PubMed] [Google Scholar]
- 34.Galletto R, Bujalowski W. The E. coli Replication Factor DnaC Protein Exists in Two Conformations with Different Nucleotide Binding Capabilities. I. Determination of the Binding Mechanism Using ATP and ADP Fluorescent Analogues. Biochemistry. 2002;41:8907–8920. doi: 10.1021/bi0201264. [DOI] [PubMed] [Google Scholar]
- 35.Galletto R, Bujalowski W. Kinetics of the E. coli Replication Factor DnaC Protein–Nucleotide Interactions. II. Fluorescence Anisotropy and Transient, Dynamic Quenching Stopped-Flow Studies of the Reaction Intermediates. Biochemistry. 2002;41:8921–8934. doi: 10.1021/bi020127w. [DOI] [PubMed] [Google Scholar]
- 36.Johnson KA. Rapid quench kinetic analysis of polymerases, adenosine triphosphatases, and enzyme intermediates. Methods Enzymol. 1995;249:38–61. doi: 10.1016/0076-6879(95)49030-2. [DOI] [PubMed] [Google Scholar]
- 37.Jezewska MJ, Bujalowski W. Global Conformational Transitions in E. coli Primary Replicative DnaB Protein Induced by ATP, ADP and Single-Stranded DNA Binding. J. Biol. Chem. 1996;271:4261–4265. doi: 10.1074/jbc.271.8.4261. [DOI] [PubMed] [Google Scholar]
- 38.Bujalowski W, Jezewska MJ. Interactions of Escherichia coli Primary Replicative Helicase DnaB Protein with Single-Stranded DNA. The Nucleic Acid Does Not Wrap Around the Protein Hexamer. Biochemistry. 1995;34:8513–8519. doi: 10.1021/bi00027a001. [DOI] [PubMed] [Google Scholar]
- 39.Jezewska MJ, Kim U-S, Bujalowski W. Binding of Escherichia coli Primary Replicative Helicase DnaB Protein to Single-Stranded DNA. Long-Range Allosteric Conformational Changes within the Protein Hexamer. Biochemistry. 1996;35:2129–2145. doi: 10.1021/bi952345d. [DOI] [PubMed] [Google Scholar]
- 40.Jezewska MJ, Kim U-S, Bujalowski W. Binding of Escherichia coli Primary Replicative Helicase DnaB Protein to Single-Stranded DNA. Long-Range Allosteric Conformational Changes within the Protein Hexamer. Biochemistry. 1996;35:2129–2145. doi: 10.1021/bi952345d. [DOI] [PubMed] [Google Scholar]
- 41.Jezewska MJ, Bujalowski W. A General Method of Analysis of Ligand Binding to Competing Macromolecules Using the Spectroscopic Signal Originating from a Reference Macromolecule. Application to Escherichia coli Replicative Helicase DnaB Protein-Nucleic Acid Interactions. Biochemistry. 1996;35:2117–2128. doi: 10.1021/bi952344l. [DOI] [PubMed] [Google Scholar]
- 42.Bujalowski W, Jezewska MJ. Spectrophotometry & Spectrofluorimetry. In: Gore MG, editor. A Practical Approach. Oxford University Press; New York: 2000. pp. 141–165. [Google Scholar]
- 43.Bujalowski W. Thermodynamic and Kinetic Methods of Analyses of Protein–Nucleic Acid Interactions. From Simpler to More Complex Systems. Chem. Rev. 2006;106:556–606. doi: 10.1021/cr040462l. [DOI] [PubMed] [Google Scholar]
- 44.McGhee JD, von Hippel PH. Theoretical aspects of DNA–protein interactions: Cooperative and noncooperative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 45.Bujalowski W, Lohman TM, Anderson CF. On the Cooperative Binding of Large Ligands to a One-Dimensional Homogeneous Lattice: The Generalized Three-State Lattice Model. Biopolymers. 1989;28:1637–1643. doi: 10.1002/bip.360280912. [DOI] [PubMed] [Google Scholar]
- 46.Jezewska MJ, Rajendran S, Bujalowski W. Strand Specificity in the Interactions of Escherichia coli Primary Replicative Helicase DnaB Protein with Replication Fork. Biochemistry. 1997;36:10320–10326. doi: 10.1021/bi970712a. [DOI] [PubMed] [Google Scholar]
- 47.Jezewska MJ, Rajendran S, Bujalowski W. Complex of Escherichia coli Primary Replicative Helicase DnaB Protein with a Replication Fork. Recognition and Structure. Biochemistry. 1998;37:3116–3136. doi: 10.1021/bi972564u. [DOI] [PubMed] [Google Scholar]
- 48.Jezewska MJ, Rajendran S, Bujalowska D, Bujalowski W. Does ssDNA Pass Through the Inner Channel of the Protein Hexamer in the Complex with the E. coli DnaB Helicase? Fluorescence Energy Transfer Studies. J. Biol. Chem. 1998;273:10515–10529. doi: 10.1074/jbc.273.17.10515. [DOI] [PubMed] [Google Scholar]
- 49.Jezewska MJ, Rajendran S, Bujalowski W. Functional and Structural Heterogeneity of the DNA Binding of the E. coli Primary Replicative Helicase DnaB Protein. J. Biol. Chem. 1998;273:9058–9069. doi: 10.1074/jbc.273.15.9058. [DOI] [PubMed] [Google Scholar]
- 50.Ledneva RK, Razjivin AP, Kost AA, Bogdanov AA. Interaction of tobacco mosaic virus protein with synthetic polynucleotides containing a fluorescent label: Optical properties of poly(A,εA) and poly(C,eC) copolymers and energy migration from the tryptophan to 1,N6-ethenoadenosine or 3,N4-ethenocytosine residues in RNP. Nucleic Acids Res. 1977;5:4226–4243. doi: 10.1093/nar/5.11.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Secrist JA, Bario JR, Leonard NJ, Weber G. Fluorescent Modification of Adenosine-Containing Coenzymes. Biological Activities and Spectroscopic Properties. Biochemistry. 1972;11:3499–3506. doi: 10.1021/bi00769a001. [DOI] [PubMed] [Google Scholar]
- 52.Baker BM, Vanderkooi J, Kallenbach NR. Base Stacking in a Fluorescent Dinucleotiside Monophosphate: eApeA. Biopolymers. 1978;17:1361–1372. [Google Scholar]
- 53.Tolman GL, Barrio JR, Leonard NJ. Chloroacetaldehyde-Modified Dinucleoside Phosphates. Dynamic Fluorescence Quenching and Quenching due to Intramolecular Complexation. Biochemistry. 1974;13:4869–4878. doi: 10.1021/bi00721a001. [DOI] [PubMed] [Google Scholar]
- 54.Lohman TM, Bujalowski W. Thermodynamic Methods for the Model-Independent Determination of Equilibrium Binding Isotherms for Protein-DNA Interactions, Using Spectroscopic Approaches to Monitor the Binding. Methods Enzymol. 1991;208:258–290. doi: 10.1016/0076-6879(91)08017-c. [DOI] [PubMed] [Google Scholar]
- 55.Bernasconi CF. Relaxation Kinetics. Academic Press; New York: 1976. [Google Scholar]
- 56.Gutfreund H. Kinetics for the Life Sciences. Cambridge University Press; Cambridge, U.K.: 1995. pp. 197–224. [Google Scholar]
- 57.Hammes GG, Schimmel PR. Kinetics and Mechanism. II. Academic Press; New York: 1970. The Enzymes; pp. 67–114. [Google Scholar]
- 58.Bujalowski W, Jezewska MJ. Kinetic Mechanism of the Single-Stranded DNA Recognition by Escherichia coli Replicative Helicase DnaB Protein. Application of the Matrix Projection Operator Technique to Analyze Stopped-Flow Kinetics. J. Mol. Biol. 2000;295:831–852. doi: 10.1006/jmbi.1999.3378. [DOI] [PubMed] [Google Scholar]
- 59.Jencks WP. The Utilization of Binding Energy in Coupled Vectorial Processes. Adv. Enzymol. 1980;51:75–106. doi: 10.1002/9780470122969.ch2. [DOI] [PubMed] [Google Scholar]
- 60.Bagshaw CR, Trentham DR. The Characterization of Myosin-Product Complexes and of Product-Release Steps during the Magnesium Ion-Dependent Adenosine Triphosphatase Reaction. Biochem. J. 1974;141:331–349. doi: 10.1042/bj1410331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abeles RH, Frey PA, Jencks WP. Biochemistry. Jones & Bartlett; 1992. pp. 791–822. [Google Scholar]
- 62.Jezewska MJ, Galletto R, Bujalowski W. Interactions of the RepA helicase hexamer of plasmid RSF1010 with the ssDNA. Quantitative Analysis of Stoichiometries, Intrinsic Affinities, Cooperativities, and Heterogeneity of the Total ssDNA-binding Site. J. Mol. Biol. 2004;343:115–136. doi: 10.1016/j.jmb.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 63.Margulies C, Kaguni JM. Ordered and Sequential Binding of DnaA Protein to oriC, the Chromosomal Origin of Escherichia coli. J. Biol. Chem. 1996;271:17035–17040. doi: 10.1074/jbc.271.29.17035. [DOI] [PubMed] [Google Scholar]
- 64.Galletto R, Jezewska MJ, Bujalowski W. Unzipping Mechanism of the Double-Stranded DNA Unwinding by a Hexameric Helicase. Quantitative Analysis of the Rate of the dsDNA Unwinding, Processivity and Kinetic Step-Size of the Escherichia coli DnaB Helicase Using Rapid Quench-Flow Method. J. Mol. Biol. 2004;343:83–99. doi: 10.1016/j.jmb.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 65.Galletto R, Jezewska MJ, Bujalowski W. Unzipping Mechanism of the Double-Stranded DNA Unwinding by a Hexameric Helicase. The Effect of the 3′ Arm and the Stability of the dsDNA on the Unwinding Activity of the Escherichia coli DnaB Helicase. J. Mol. Biol. 2004;343:101–114. doi: 10.1016/j.jmb.2004.07.056. [DOI] [PubMed] [Google Scholar]
- 66.Marcinowicz A, Jezewska MJ, Bujalowski PJ, Bujalowski W. The Structure of the Tertiary Complex of the RepA Hexameric Helicase of Plasmid RSF1010 with the ssDNA and Nucleotide Cofactors in Solution. Biochemistry. 2007;46:13279–13296. doi: 10.1021/bi700729k. [DOI] [PubMed] [Google Scholar]
- 67.Klotz IM. Energy Changes in Biochemical Reactions. Academic Press; New York: 1967. pp. 52–60. [Google Scholar]
- 68.Allen GC, Jr., Kornberg A. Fine Balance in the Regulation of DnaB Helicase by DnaC Protein in Replication in Escherichia coli. J. Biol. Chem. 1991;266:22096–22101. [PubMed] [Google Scholar]
- 69.Allen GC, Jr., Dixon NE, Kornberg A. Strand Switching of a Replicative DNA Helicase Promoted by the E. coli Primosome. Cell. 1993;74:713–722. doi: 10.1016/0092-8674(93)90518-u. [DOI] [PubMed] [Google Scholar]
- 70.Cocco S, Monasson R, Marko JF. Force and Kinetic Barriers to unzipping of the DNA Double Helix. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8608–8613. doi: 10.1073/pnas.151257598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaplan DL, O'Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]