Abstract
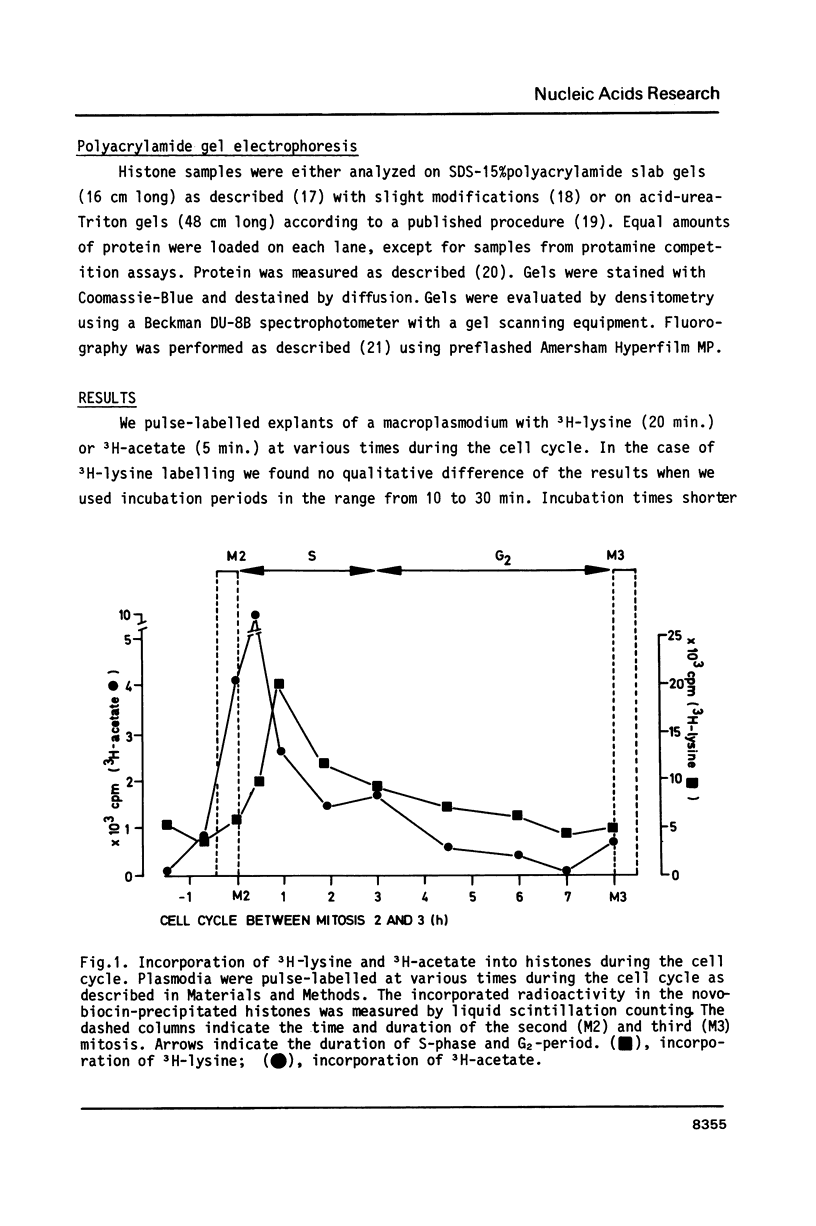
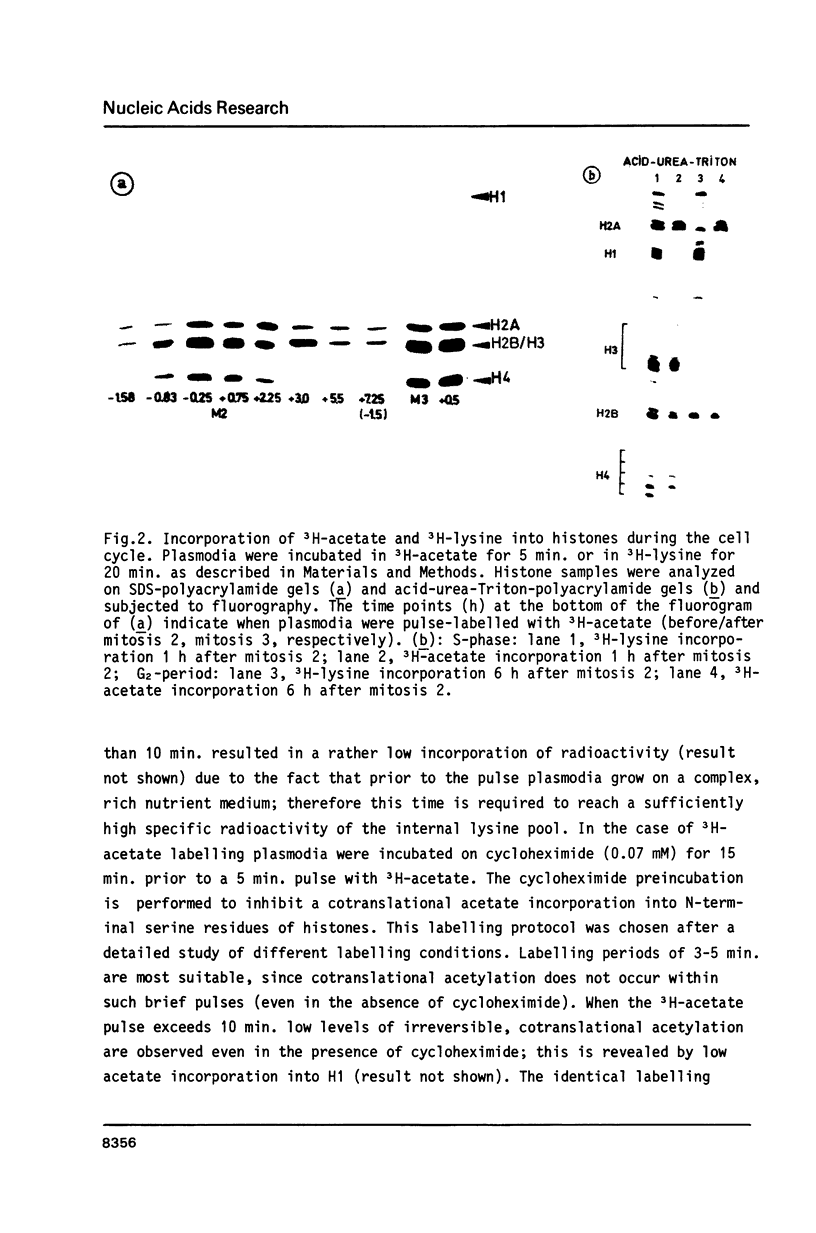
Postsynthetic acetylation of core histones exhibits a peak during S-phase of the Physarum cell cycle. The maximum 3H-acetate incorporation precedes the maximum of histone synthesis. Acetate is incorporated into all core histones during S-phase, but only into H2A and H2B during G2-period. Resolution of acetylated H4-subspecies reveals acetate incorporation into preexisting H4, but not into newly synthesized molecules during mitosis and early S-phase. In a protamine competition assay histones from S-phase chromatin are released at lower protamine concentrations as compared to the lower acetylated G2-chromatin. We demonstrate a preferential release of highly acetylated H4-subspecies at low protamine concentrations. Our results fit into a general model of the relationship between histone acetylation and chromatin assembly. According to this model acetylation of core histones would serve as a signal for displacement of histones from nucleosomes by modulating histone-protein or histone-DNA interactions. We propose that this mechanism operates during DNA-replication and transcription, as well as during other chromatin rearrangements.
Full text
PDF




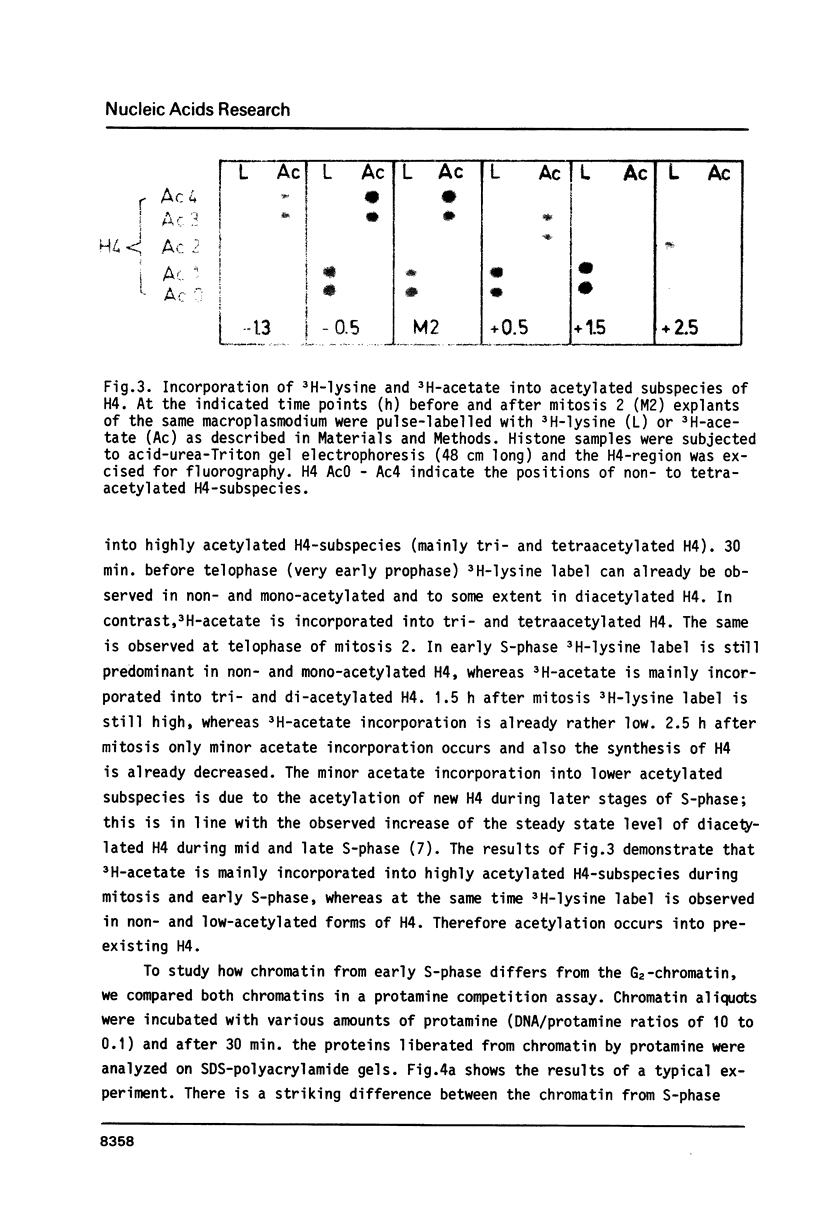

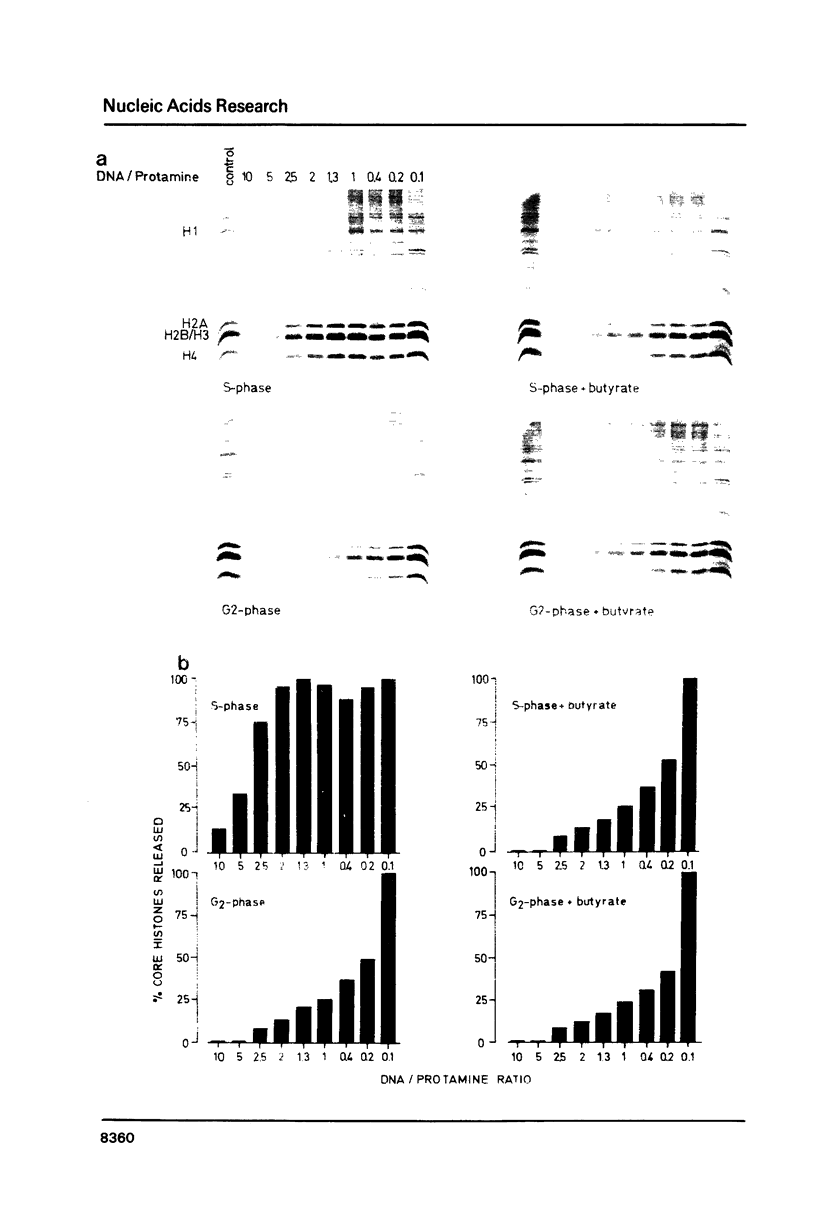






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apriletti J. W., David-Inouye Y., Eberhardt N. L., Baxter J. D. Interactions of the nuclear thyroid hormone receptor with core histones. J Biol Chem. 1984 Sep 10;259(17):10941–10948. [PubMed] [Google Scholar]
- Ausio J., van Holde K. E. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986 Mar 25;25(6):1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- Baer B. W., Rhodes D. Eukaryotic RNA polymerase II binds to nucleosome cores from transcribed genes. Nature. 1983 Feb 10;301(5900):482–488. doi: 10.1038/301482a0. [DOI] [PubMed] [Google Scholar]
- Chicoine L. G., Schulman I. G., Richman R., Cook R. G., Allis C. D. Nonrandom utilization of acetylation sites in histones isolated from Tetrahymena. Evidence for functionally distinct H4 acetylation sites. J Biol Chem. 1986 Jan 25;261(3):1071–1076. [PubMed] [Google Scholar]
- Christensen M. E., Dixon G. H. Hyperacetylation of histone H4 correlates with the terminal, transcriptionally inactive stages of spermatogenesis in rainbow trout. Dev Biol. 1982 Oct;93(2):404–415. doi: 10.1016/0012-1606(82)90127-0. [DOI] [PubMed] [Google Scholar]
- Christensen M. E., Rattner J. B., Dixon G. H. Hyperacetylation of histone H4 promotes chromatin decondensation prior to histone replacement by protamines during spermatogenesis in rainbow trout. Nucleic Acids Res. 1984 Jun 11;12(11):4575–4592. doi: 10.1093/nar/12.11.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Bresnahan D., Thompson S., Sealy L., Chalkley R. Novobiocin precipitates histones at concentrations normally used to inhibit eukaryotic type II topoisomerase. Nucleic Acids Res. 1986 May 12;14(9):3671–3686. doi: 10.1093/nar/14.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bernardin W., Koller T., Sogo J. M. Structure of in-vivo transcribing chromatin as studied in simian virus 40 minichromosomes. J Mol Biol. 1986 Oct 5;191(3):469–482. doi: 10.1016/0022-2836(86)90142-7. [DOI] [PubMed] [Google Scholar]
- Doenecke D., Gallwitz D. Acetylation of histones in nucleosomes. Mol Cell Biochem. 1982 Apr 30;44(2):113–128. doi: 10.1007/BF00226895. [DOI] [PubMed] [Google Scholar]
- Gröbner P., Loidl P. ADP-ribosyltransferase in isolated nuclei during the cell cycle of Physarum polycephalum. Biochem J. 1985 Nov 15;232(1):21–24. doi: 10.1042/bj2320021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman T. M., DePamphilis M. L., Wassarman P. M. Structure of chromatin at deoxyribonucleic acid replication forks: Okazaki fragments released from replicating SV40 chromosomes by single-strand specific endonucleases are not in nucleosomes. Biochemistry. 1979 Oct 16;18(21):4563–4571. doi: 10.1021/bi00588a017. [DOI] [PubMed] [Google Scholar]
- Herman T. M., DePamphilis M. L., Wassarman P. M. Structure of chromatin at deoxyribonucleic acid replication forks: location of the first nucleosomes on newly synthesized simian virus 40 deoxyribonucleic acid. Biochemistry. 1981 Feb 3;20(3):621–630. doi: 10.1021/bi00506a027. [DOI] [PubMed] [Google Scholar]
- Hüvös P., Sasi R., Fasman G. D. Conformation of control and acetylated HeLa stripped chromatin after reassociation with H1. Biopolymers. 1984 Nov;23(11 Pt 1):2195–2210. doi: 10.1002/bip.360231107. [DOI] [PubMed] [Google Scholar]
- Imai B. S., Yau P., Baldwin J. P., Ibel K., May R. P., Bradbury E. M. Hyperacetylation of core histones does not cause unfolding of nucleosomes. Neutron scatter data accords with disc shape of the nucleosome. J Biol Chem. 1986 Jul 5;261(19):8784–8792. [PubMed] [Google Scholar]
- Johnson E. M., Sterner R., Allfrey V. G. Altered nucleosomes of active nucleolar chromatin contain accessible histone H3 in its hyperacetylated forms. J Biol Chem. 1987 May 25;262(15):6943–6946. [PubMed] [Google Scholar]
- Klug A. From macromolecules to biological assemblies. Nobel Lecture, 8 December 1982. Biosci Rep. 1983 May;3(5):395–430. doi: 10.1007/BF01121953. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Loidl P., Gröbner P. Biosynthesis and posttranslational acetylation of histones during spherulation of Physarum polycephalum. Nucleic Acids Res. 1986 May 12;14(9):3745–3762. doi: 10.1093/nar/14.9.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl P., Gröbner P. Histone synthesis during the cell cycle of Physarum polycephalum. Synthesis of different histone species is not under a common regulatory control. J Biol Chem. 1987 Jul 25;262(21):10195–10199. [PubMed] [Google Scholar]
- Loidl P., Loidl A., Puschendorf B., Gröbner P. Lack of correlation between histone H4 acetylation and transcription during the Physarum cell cycle. 1983 Sep 29-Oct 5Nature. 305(5933):446–448. doi: 10.1038/305446a0. [DOI] [PubMed] [Google Scholar]
- Loidl P., Loidl A., Puschendorf B., Gröbner P. RNA polymerase activity and template activity of chromatin after butyrate induced hyperacetylation of histones in Physarum. Nucleic Acids Res. 1984 Jul 11;12(13):5405–5417. doi: 10.1093/nar/12.13.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Nickol J. M., Felsenfeld G., Rau D. C. Histone hyperacetylation has little effect on the higher order folding of chromatin. Nucleic Acids Res. 1983 Jun 25;11(12):4065–4075. doi: 10.1093/nar/11.12.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithieux G., Alquier C., Roux B., Rousset B. Interaction of tubulin with chromatin proteins. H1 and core histones. J Biol Chem. 1984 Dec 25;259(24):15523–15531. [PubMed] [Google Scholar]
- Mohberg J., Rusch H. P. Isolation of the nuclear histones from the Myxomycete, Physarum polycephalum. Arch Biochem Biophys. 1969 Nov;134(2):577–589. doi: 10.1016/0003-9861(69)90320-8. [DOI] [PubMed] [Google Scholar]
- Muller S., Erard M., Burggraf E., Couppez M., Sautière P., Champagne M., Van Regenmortel M. H. Immunochemical detection of changes in chromatin subunits induced by histone H4 acetylation. EMBO J. 1982;1(8):939–944. doi: 10.1002/j.1460-2075.1982.tb01275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness P. J., Labhart P., Banz E., Koller T., Parish R. W. Chromatin structure along the ribosomal DNA of Dictyostelium. Regional differences and changes accompanying cell differentiation. J Mol Biol. 1983 May 25;166(3):361–381. doi: 10.1016/s0022-2836(83)80090-4. [DOI] [PubMed] [Google Scholar]
- Oliva R., Mezquita C. Histone H4 hyperacetylation and rapid turnover of its acetyl groups in transcriptionally inactive rooster testis spermatids. Nucleic Acids Res. 1982 Dec 20;10(24):8049–8059. doi: 10.1093/nar/10.24.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M., Chalkley R. Histone acetylation increases the solubility of chromatin and occurs sequentially over most of the chromatin. A novel model for the biological role of histone acetylation. J Biol Chem. 1982 Jul 10;257(13):7336–7347. [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Sogo J. M., Ness P. J., Widmer R. M., Parish R. W., Koller T. Psoralen-crosslinking of DNA as a probe for the structure of active nucleolar chromatin. J Mol Biol. 1984 Oct 5;178(4):897–919. doi: 10.1016/0022-2836(84)90318-8. [DOI] [PubMed] [Google Scholar]
- Sogo J. M., Stahl H., Koller T., Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986 May 5;189(1):189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- Waterborg J. H., Matthews H. R. Patterns of histone acetylation in Physarum polycephalum. H2A and H2B acetylation is functionally distinct from H3 and H4 acetylation. Eur J Biochem. 1984 Jul 16;142(2):329–335. doi: 10.1111/j.1432-1033.1984.tb08290.x. [DOI] [PubMed] [Google Scholar]
- Waterborg J. H., Matthews H. R. Patterns of histone acetylation in the cell cycle of Physarum polycephalum. Biochemistry. 1983 Mar 15;22(6):1489–1496. doi: 10.1021/bi00275a025. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Palter K., Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975 Sep;6(1):85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Zweidler A. Resolution of histones by polyacrylamide gel electrophoresis in presence of nonionic detergents. Methods Cell Biol. 1978;17:223–233. [PubMed] [Google Scholar]
- Zwierzina H., Loidl A., Fuith L. C., Helliger W., Puschendorf B., Grunicke H. Depression of histone acetylation by alkylating antitumor agents in murine cells. Cancer Res. 1984 Aug;44(8):3336–3339. [PubMed] [Google Scholar]