Abstract
We investigated correlated responses in the transcriptomes of longevity-selected lines of Drosophila melanogaster to identify pathways that affect life span in metazoan systems. We evaluated the gene expression profile in young, middle-aged, and old male flies, finding that 530 genes were differentially expressed between selected and control flies when measured at the same chronological age. The longevity-selected flies consistently showed expression profiles more similar to control flies one age class younger than control flies of the same age. This finding is in accordance with a younger gene expression profile in longevity-selected lines. Among the genes down-regulated in longevity-selected lines, we found a clear over-representation of genes involved in immune functions, supporting the hypothesis of a life-shortening effect of an overactive immune system, known as inflammaging. We judged the physiological age as the level of cumulative mortality. Eighty-four genes were differentially expressed between the control and longevity-selected lines at the same physiological age, and the overlap between the same chronological and physiological age gene lists included 40 candidate genes for increased longevity. Among these candidates were genes with roles in starvation resistance, immune response regulation, and several that have not yet been linked to longevity. Investigating these genes would provide new knowledge of the pathways that affect life span in invertebrates and, potentially, mammals.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-010-9162-8) contains supplementary material, which is available to authorized users.
Keywords: Microarray, Aging, Inflammaging, Candidate genes, Drosophila melanogaster, Laboratory evolution
Introduction
Invertebrate models of longevity have contributed to our understanding and knowledge of aging over the last century (Strehler 1961; Klass and Hirsh 1976; Arking and Dudas 1989; Jazwinski 1996; Lithgow 1996) and continue to do so (Partridge 2009). Compared to vertebrate model organisms and humans, invertebrates pose few insuperable ethical issues and can provide fast, and sometimes more accurate, answers to which pathways can be a target for intervention in the aging process. In the present study, we demonstrate contrasting gene expression in three independently replicated longevity-selected Drosophila melanogaster lines and three independent control lines.
The short life span of D. melanogaster (Pearl et al. 1923) allows us to select for increased longevity in numerous generations and to obtain a remarkable response. Generally, increased longevity can be selected one of two ways in Drosophila: based on virgin life span (Zwaan et al. 1995) or mated life span (Luckinbill et al. 1984). We chose to select for mated life span. The flies in the longevity-selected lines could mate throughout life, but only the eggs laid after 50% of the population had died contributed to the next generation. According to evolutionary theories of aging (Medaware 1952; Williams 1957; Hamilton 1966; Kirkwood 1977), organisms should not start to age until reproduction occurs, so this protocol should select for delayed onset of senescence (Luckinbill et al. 1984).
We studied the difference in gene expression between lines selected for mated longevity (LS) and non-selected control (C) lines. We investigated the transcriptional selection response in young, middle-aged, and old flies, and contrasting expression in C and LS lines at the same chronological age and when the same proportion of a cohort had died (same physiological age), elaborating on the design of Curtis et al. (Fig. 1) (Curtis et al. 2007). The genes that had significantly different transcript abundance between the C and LS lines at the same chronological age belonged to one of two categories: candidates for affecting longevity, playing a causal role in the prolonged life span of the LS lines, or biomarkers of physiological age, reflecting the proposed delayed senescence of LS lines. Similarly, the genes that had significantly different transcript abundance at the same physiological age also belonged to one of two categories: candidates for longevity, playing a role in causing the prolonged life span of the LS lines, or biomarkers of chronological age that are not affected by the altered expression of the candidate genes for longevity. The genes that were differentially regulated both at the same chronological and physiological age are the candidate genes for increased longevity. Because these genes should be the primary targets for selection, a cluster analysis was expected to separate gene expression according to selection regime.
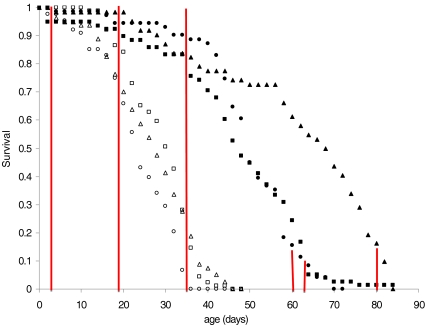
Fig. 1.
Survival plots of C lines (open symbols) and LS lines (closed symbols). Sampling times are depicted by vertical lines. The lifespan curves differ among the lines (logrank test; χ2 = 361, df = 5, P < 0.0001)
We hypothesized that the genes with differential expression between selection regimes at the same chronological age have a younger expression profile in LS lines compared to C lines, i.e., the expression in old LS flies is more similar to the expression in younger control flies than in control flies of the same chronological age. Furthermore, we hypothesized that genes up-regulated in LS lines compared to C lines are genes with expression that decreases with age. In addition, genes down-regulated in LS lines compared to C lines are genes with expression that increases with age.
Results
The effect of selection on longevity
The effect of selection was significant in all tests (Table 1), and the median life span of longevity-selected lines was, on average, 66% longer than the life span of non-selected flies when mated. Selection also significantly increased the 90th percentile of life span (Table 2). Analysis of Gompertz parameters found that the age-independent component of mortality differed between selection regimes, whereas the rate of aging was unaffected by selection. No significant differences were observed in the exponential increase in mortality between lines within experiments (Table 2).
Table 1.
Results from five independent experiments studying the effects of selection on longevity
| Source | Proportional hazard analysis | ||
|---|---|---|---|
| df | χ2 | P | |
| Linea | 4 | 151.24 | <0.001 |
| Selectiona | 1 | 35.47 | <0.001 |
| Lineb | 4 | 62.11 | <0.001 |
| Selectionb | 1 | 108.93 | <0.001 |
| Line | 4 | 127.18 | <0.001 |
| Selection | 1 | 167.52 | <0.001 |
| Line | 4 | 27.5232 | <0.001 |
| Selection | 1 | 192.708 | <0.001 |
| Line | 4 | 205.33 | <0.001 |
| Selection | 1 | 32.82 | <0.001 |
The proportional hazard analysis of mortality risk was performed with line nested within the selection regime
aMated longevity without antibiotics in the medium
bCorrelated response of virgin longevity with antibiotics in the medium. The remaining experiments investigated mated longevity with antibiotics in the medium
Table 2.
Confidence limits (CLs) of Gompertz parameters and the 90th percentile of life span from five independent experiments studying the effects of selection on longevity
| CLs of the basal mortality rate | CLs of the rate of aging | 90%-tile | |
|---|---|---|---|
| Ca | 0.00602–0.00789 | 0.07072–0.08139 | 46* |
| LSa | 0.00373–0.00533 | 0.06617–0.07668 | 50* |
| Cb | 0.00201–0.00387 | 0.05698–0.07150 | 63* |
| LSb | 0.00075–0.00166 | 0.05831–0.07215 | 72.8* |
| C | 0.01688–0.0339 | 0.03406–0.04689 | 40* |
| LS | 0.00724–0.01095 | 0.03790–0.04393 | 66* |
| C | 0.00402–0.00734 | 0.04532–0.05966 | 60* |
| LS | 0.00165–0.00348 | 0.05257–0.06739 | 67.4* |
| C | 0.0152–0.02520 | 0.0361–0.0564 | 39* |
| LS | 0.0022–0.00417 | 0.04953–0.06234 | 67* |
*P < 0.001; Fisher’s exact test
aMated longevity without antibiotics in the medium
bCorrelated response of virgin longevity with antibiotics in the medium. The remaining experiments investigated mated longevity with antibiotics in the medium
Age-dependent transcriptome changes
We contrasted LS male flies on post-eclosion day 19 (LS19) with flies from day 3 (LS3). Also, we contrasted flies from day 35 (LS35) with LS19 and, finally, flies after 90% cumulative mortality had been reached on days 60–80 (LS60) were compared with LS35. Setting the false discovery rate (FDR) at 0.05 and using a minimum log2(fold-change) (lfc) of 1, the moderated F-statistics in the multiclass analysis revealed that 577 unique genes were differentially expressed with age in LS flies (ESM Table S1). In C lines, contrasting C19 with C3 and C35 with C19 revealed 267 unique genes that changed with age (ESM Fig. S2a, b; ESM Table S1). A highly significant overlap was found for 179 genes differentially expressed with age in lines from both selection regimes (Table 3). Significant overlap was found in both selection regimes for genes that reacted to aging in a previous D. melanogaster study (Landis et al. 2004), as well as genes that respond to heat shock (Kristensen et al. 2005) and selection for increased stress resistance (Sørensen et al. 2007) (Table 3). The significant gene ontology terms (GO terms) are listed in ESM Table S3.
Table 3.
Overlap of gene lists
Numbers above the shaded diagonal denote overlapping genes between lists, below the diagonal are P values, and shaded is the number of unique genes in the lists. The data include genes responding to aging in C (AC) and LS lines (ALS); genes significantly different between C and LS lines at the same chronological age (Ch) and same physiological age (P), and the overlap between the two (OL) (present study); genes responding to aging (Landis) (23); genes responding to a life-span-extending over-expression of MnSOD at the same chronological age (MnSOD Ch) and same physiological age (MnSOD P) (Curtis et al. 2007); genes correlated with variance in life span (Lifespan) and starvation (Starv) in isofemale lines (27); genes responding to selection for increased longevity (Long), starvation resistance (Starv sel), heat knock-down (KD), and by rearing at constant 30°C (Sørensen et al. 2007); and genes responding to a brief heat shock (Heat) (Kristensen et al. 2005)
NS non-significant
The effect of selection on the transcriptome at the same chronological age
Contrasting LS and C lines of the same chronological age (LS3–C3, LS19–C19, and LS35–C35), the moderated F-statistics in the multiclass analysis revealed that 530 unique genes were differentially expressed in LS lines (FDR = 0.05; Table 3 and ESM Table S1). Only these genes were included in further analyses of differences at the same chronological age. The results are summarized in ESM Fig. S2c.
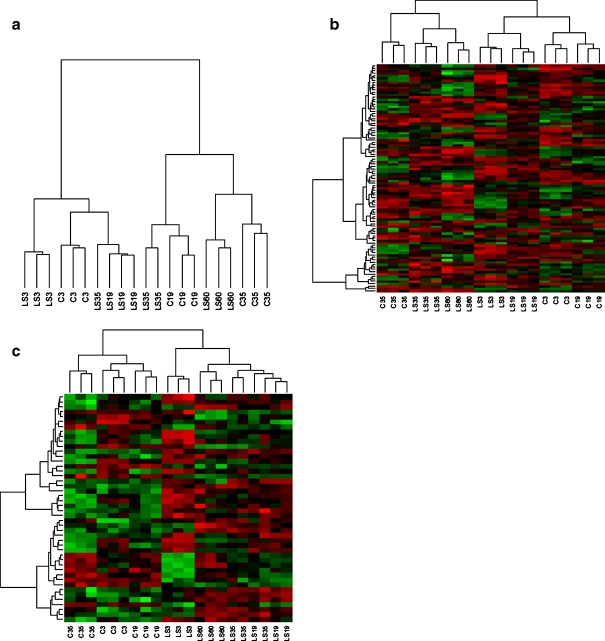
We saw a clear signal of delayed aging in the expression pattern of the genes differentially expressed between selection regimes at the same chronological age. At any given time point, the different lines of each selection regime clustered together, except from one LS35 line, which clustered with the three LS19 lines (Fig. 2a and ESM Fig. S4). The main expression difference was between young (C3, LS3, and LS19) and old flies (C19, C35, LS35, and LS60). Gene expression in the different age classes of LS lines was most similar to the gene expression of C lines from the previous age class (Fig. 2a). All significant probe sets were either up- (343) or down-regulated (248) in all significant contrasts at the same chronological age. All of the overlaps between these contrasts were highly significant (ESM Table S5), and most of the overall significantly differentially expressed probe sets were significant in the LS35–C35 contrast (ESM Fig. S2c).
Fig. 2.
Dendrograms, or heatmaps, of genes differentially expressed with selection for increased mated longevity, showing how lines cluster according to the expression of genes that are differentially expressed (P < 0.05) in C and LS lines. a Genes differentially expressed at the same chronological age, b at the same physiological age, and c genes that overlap between a and b. The diagrams were drawn using the R package Limma (Smyth 2005)
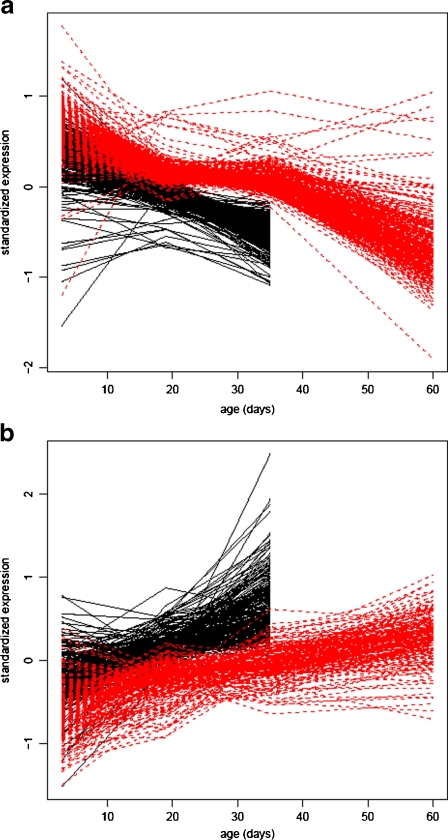
Genes significantly up-regulated in the LS lines compared to C lines on day 35 were generally genes that were down-regulated with age in both C and LS lines (Fig. 3a), and 88% of them were significantly down-regulated (FDR = 0.05, lfc > 0, ESM Fig. S2d). However, age affected the LS lines less than the C lines, especially between days 19 and 35, which gave rise to a significant interaction between selection and age effects (Table 4). In particular, GO terms representing mitosis, meiosis, and biogenesis were over-represented among these genes (ESM Table S3). Genes significantly down-regulated in LS lines on day 35 were those that were generally up-regulated with age in both C and LS lines (Fig. 3b), and 71% of them were significantly up-regulated (FDR = 0.05, lfc = 0, ESM Fig. S2e). Age seemed to affect LS lines less than C lines between days 19 and 35, and there was a significant interaction between selection and age effects (Table 5). GO terms representing immune response, among others, were over-represented among these genes (ESM Table S3).
Fig. 3.
Expression profiles of genes a up-regulated or b down-regulated in LS lines at day 35. Solid lines represent average expression in C lines, dotted lines represent average expression in LS lines
Table 4.
ANOVA results from the analysis of the expression of genes significantly up-regulated in LS lines 35 days post-eclosure compared to the same age in C lines
| Source | dfa | MSb | P |
|---|---|---|---|
| Gene | 207 | 50.2 | <0.0001 |
| Selection | 1 | 140 | <0.0001 |
| Age | 3 | 161 | <0.0001 |
| Gene × sel | 207 | 0.3 | <0.0001 |
| Gene × age | 621 | 0.4 | <0.0001 |
| Gel × age | 2 | 10.6 | <0.0001 |
| Gene × sel × age | 414 | 0.1 | <0.05 |
| Error | 2,912 | 0.1 |
aDegrees of freedom
bMean squares
Table 5.
ANOVA results from the analysis of the expression of genes significantly down-regulated in LS lines 35 days post-eclosure compared to the same age in C lines
| Source | dfa | MSb | P |
|---|---|---|---|
| Gene | 328 | 72 | <0.0001 |
| Selection | 1 | 34.5 | <0.0001 |
| Age | 3 | 415.6 | <0.0001 |
| Gene × sel | 328 | 0.2 | <0.0001 |
| Gene × age | 984 | 0.2 | <0.0001 |
| Sel × age | 2 | 25.6 | <0.0001 |
| Gene × sel × age | 656 | 0.01832 | NS |
| Error | 4,606 | 0.03461 |
NS non-significant
aDegrees of freedom
bMean squares
The 530 genes significantly overlapped with genes previously found to respond to aging (LS lines in present study and Landis et al. 2004) and genes that respond to heat shock exposure (Sørensen et al. 2005) (Table 3). However, no significant overlap was seen with genes that respond to longevity-increasing over-expression of manganese superoxide dismutase (MnSOD) at the same chronological age (Curtis et al. 2007). MnSOD is an important antioxidant in cellular defense against reactive oxygen species (ROS). Also, we found no significant overlap with genes correlated with longevity or starvation resistance in a genome-wide study of 40 isofemale lines (Ayroles et al. 2009) or a diverse set of selection regimes, including increased longevity (Sørensen et al. 2007).
The effect of selection on the transcriptome at the same physiological age
Contrasting LS and C flies of the same physiological age (LS3–C3, LS35–C19, and LS60–C35), the moderated F-statistics in the multiclass analysis revealed that 84 unique genes were differentially expressed (Table 3 and ESM S1). Only these genes were included in further analyses of differences at the same physiological age. The cluster analysis revealed that, at any given time point, the different lines of each selection regime clustered together (Fig. 2b). The main difference in gene expression was between young (days 3 and 19) and old flies (days 35 and ≥60). The results are summarized in ESM Fig. S2f. All but one of the significant genes were up- (38) or down-regulated (56) in all significant contrasts. GO terms representing adult locomotor behavior and light detection, among others, were over-represented among these genes (ESM Table S3). The 84 genes significantly overlapped (Table 3) with genes that responded to aging in this study but not in the study by Landis et al. (Landis et al. 2004). We also identified significant overlap with genes that respond to selection for increased stress resistance and longevity (Sørensen et al. 2007), but we did not identify significant overlap with genes that respond to life-span-increasing over-expression of MnSOD at the same physiological age (Curtis et al. 2007), to heat shock (Sørensen et al. 2005), or those that correlate with longevity in a genome-wide study of 40 isofemale lines (Ayroles et al. 2009).
Candidate genes for longevity
Comparing the gene lists for contrasting expression in C and LS flies at the same chronological and physiological age revealed an overlap of 46 probe sets representing 40 unique genes (ESM Table S1). Fourteen probe sets were up-regulated and 32 down-regulated in LS lines in both the chronological and physiological comparisons. This overlap was statistically significant (ESM Table S5). The cluster analysis of gene expression in these genes revealed a clear separation of the LS and C lines (Fig. 2c). No significant overlap was found with the candidate genes for life span extension due to increased MnSOD expression as previously found (Curtis et al. 2007). Over-represented GO terms can be found in ESM Table S3.
Discussion
The effect of selection on longevity
A profound response to selection for increased life span was detected in this study (Tables 1 and 2). After 33 generations of selection for mated longevity, the median life span of males was 66% higher compared to control lines, and a difference in longevity was detected in both virgin and mated males with and without antibiotics in the adult food medium. The selection response was not composed of a change in the rate of aging as expressed in mortality doubling time, but it was due to either a decrease in age-independent mortality or a delayed onset of aging that resulted in an increased 90th percentile of life span. This observation is what we would expect if the onset of senescence was delayed in the LS lines compared to the C lines.
The effect of aging on the transcriptome
The highly significant overlap between the genes that respond to aging in the LS and C lines, which was seen in both this study and the study by Landis et al. (Landis et al. 2004) (Table 3), implies that neither selection nor genetic background have a profound impact on the end results of aging as perceived by the transcriptional phenotype. Comparisons with studies of aging in the human (Lu et al. 2004; Rodwell et al. 2004; Zahn et al. 2006) and Caenorhabditis elegans (Lund et al. 2002; Golden et al. 2006) transcriptomes reveal several common GO categories (ESM Table S3 e.g., muscle activity, metal binding, and immune response), which supports the general finding that, even though aging might follow individual trajectories (Kirkwood et al. 2005), there generally is a high degree of overlap in the aging phenotype (Grotewiel et al. 2005; Tarnopolsky et al. 2007).
The effect of selection on the transcriptome at the same chronological age
The fact that the three replicate selection lines cluster together at all time points but one (Fig. 2a) after 33 generations of selection demonstrates that we were able to detect a shared transcriptional response to selection for increased mated longevity.
The genes that were significantly differentially regulated between C and LS lines in these contrasts were either causing the difference in lifespan between selection regimes or showing the downstream effects of these lifespan increasing genes. Genes in both these categories are interesting in an aging context. The probe sets that differed in transcript abundance showed a delayed aging of transcription in LS lines, which can be seen in the clustering of LS flies with C flies from the preceding age group in Fig. 2a. Supporting this pattern, most of the genes significantly up-regulated in LS lines on day 35 are genes that are normally down-regulated with age in both LS and C lines (ESM Fig. S2d). Alternatively, 71% of the genes down-regulated in LS lines on day 35 are genes that are normally up-regulated with age in both LS and C lines (ESM Fig. S2e). Thus, rather than denoting these genes as down- or up-regulated in LS lines compared to C lines, they are genes with expression levels similar to what we detected in younger C flies. The delayed aging can also be seen in Fig. 3a and b, where LS lines have a shallower slope between the second and third time points compared to the C lines. This difference gives rise to a significant interaction between selection regime and age (Tables 4 and 5). Interestingly, most of the genes that are differentially expressed with age did not differ in their age-specific expression between the C and LS lines. This finding indicates that the genome-wide transcriptome of LS lines did not show a collective sign of delayed aging in the LS lines. Therefore, only a subset of the genes differentially expressed with age are affected by selection in these lines. Also, altering the expression of this subset of the age-influenced expression pattern seems to be sufficient for extending life span.
Looking through the list of significant GO terms enriched by genes significantly down-regulated in LS35 compared to C35 (ESM Table S3), the long-lived lines having, overall, a less up-regulated immune response with age seems counterintuitive because a higher resistance against pathogens might be considered beneficial for longevity. However, the age-related up-regulation of genes involved in the immune system might reflect a process that is well documented in both mammals and insects, such as Drosophila, and is known as inflammaging (Zerofsky et al. 2005; Franceschi et al. 2006). The expression of pro-inflammatory genes increases with age, which is hypothesized to be the result of either accumulated exposure to pathogens throughout life (Franceschi et al. 2006) or reflects age-dependent changes in the function of the immune system itself (Zerofsky et al. 2005). We demonstrated an up-regulation of immune response genes consistent with inflammaging in flies that have lived their adult life on medium containing antibiotics and antifungal substances; thus, this study indicates that inflammaging is not due to accumulated exposure to pathogens. Life-span-extending dietary restriction in D. melanogaster can also delay the aging-related increase in immunity-related gene transcription (Pletcher et al. 2005). This result stresses the importance of inflammaging in aging research.
The most prominently up-regulated GO terms were associated with cell division, both meiosis and mitosis (ESM Table S3). A comparably higher fecundity in old age in long-lived females was shown previously (Service 1989), which conceivably could also manifest in increased meiotic activity in males. However, the cells in adult Drosophila are mostly amitotic, and it would be interesting to investigate the tissue-specific expression pattern of the up-regulated mitotic genes. The midgut of Drosophila is maintained by the proliferation of stem cells, and the increased expression of mitotic genes might predominately be located there.
The effect of selection on the transcriptome at the same physiological age
Not nearly as many probe sets were significantly differentially transcribed between the selection regimes at the same physiological age compared to the former analysis (84 vs. 530). This finding was expected as the genes that can function as biomarkers of physiological age in our lines should not be significantly different when contrasting transcription in lines with the same cumulative mortality. The generality of the selection response was supported by the three genetically independent selection lines clustering together at any given time point (Fig. 2b). However, even though we did not find a significant overlap with the genes that respond to selection for increased heat resistance (Sørensen et al. 2007), the same functional group (phototransduction and vision) tops the list of significant GO terms in both studies (Sørensen et al. 2007). This functional group was also differentially expressed with age in this study, as well as in two studies comparing the transcriptional profiles of heads from young and old flies (Kim et al. 2005; Girardot et al. 2006).
Candidate genes for longevity
A cluster analysis of the expression of the 40 candidate genes for longevity revealed a clear separation of the selection regimes (Fig. 2c), which is what we expected to see if we had indentified the genes causing the difference in life span. The separation of selection regimes was not a passive result stemming from the contrast design (LS–C) naturally selecting genes that differ in expression between the selection regimes. This fact is evident in the cluster analysis of genes from both the same chronological and physiological age, in which the largest difference was between young and aged flies (Fig. 2a, b). Although the significant GO terms were too general (e.g., nucleotide binding and regulation of signal transduction ESM Table S3) to give much of an indication of the mechanisms behind the delayed aging in LS lines, many of the candidate genes are associated with relevant processes, such as photolyase, the putative function of which is to repair damaged DNA (NCBI Entrez Gene: http://www.ncbi.nlm.nih.gov/gene). In addition, two of the candidate genes, CG9812 (Harbison et al. 2005) and glycerol-3-phosphate dehydrogenase (Oudman et al. 1994), were previously associated with starvation resistance. Selection for increased starvation resistance sometimes leads to increased longevity (Baldal et al. 2006). Despite this connection between starvation resistance and longevity, we found no significant overlap between the candidate genes we identified and genes associated with selection for increased starvation resistance (Sørensen et al. 2007). However, there was a significant overlap with probe sets associated with variation in starvation resistance among isofemale lines (Table 3), but this overlap consisted of only two genes (Table 3). The functions of the 40 candidate genes are currently being studied in our laboratory.
Comparisons with other studies
The studies that are of most relevance to our results are those by Ayroles et al. (Ayroles et al. 2009), who linked the expression of several genes to life span variation in isofemale lines, and Curtis et al. (Curtis et al. 2007), who investigated gene expression in flies with genetically up-regulated MnSOD expression. We did not find any significant overlap with the genes that correlated with life span in the first study. One possible reason for this discrepancy is that the standing genetic variation in natural populations might not reflect the realized potential for evolution after several generations of intense selection. Another reason could be that the genetic background of our studies might influence the genes that show variation in the population sampled by Ayroles et al. or that responded to selection (present study).
We did not find a significant overlap between genes that were differently transcribed in C and LS lines at the same chronological or physiological age and the genes that responded to the over-expression of MnSOD at the same chronological or physiological age (Table 3). Also, from the functional groups of genes significantly over-represented at the same physiological age in the present study, only very general GO terms overlapped between the two studies: nucleotide binding and signal transduction. The delayed aging in our LS lines seems to have occurred through different mechanisms than MnSOD.
Selection for increased mated life span resulted in replicate lines that retained a young gene expression profile longer than control lines. We identified 40 genes that might be responsible for the increased longevity of the selected lines. Future experiments will reveal if these genes represent pathways that can increase life span independent of sex, genetic background, and the species investigated.
Experimental procedures
Origin of flies and selection regime
The replicate selection and control lines originated from a mass population of flies from mixed geographic origin. The origin of the flies and the set-up of the mass population are described in more detail elsewhere (Bubliy and Loeschcke 2005). The mass population was maintained as one interbreeding population for four generations before three replicate selection and control lines were established. Each replicate line was maintained in five culture bottles with a minimum population of 60 pairs each, with a total population of 300 pairs. The five bottles within a replicate line were mixed each generation to reduce the effect of drift. The longevity selection took place every other generation; after emergence, the flies were placed in food vials and transferred to new vials every second day until approximately 50% of the flies were dead. For the first generation of selection the process took 4 weeks; after 33 generations, 50% mortality was reached after approximately 6.5 weeks. The surviving flies were allowed to start the next generation. The LS lines were first selected for 16 generations at 25°C, interspaced with 16 unselected generations at 25°C. The flies were kept unselected for 10 generations at 20°C, thereafter selection was resumed at 25°C for 17 generations interspaced with 18 unselected generations, also at 25°C. Three replicate lines of the control regime were kept on standard agar–sugar–yeast–oatmeal medium under standard laboratory conditions and allowed to reproduce within a week of eclosure at 25°C for 73 generations. The flies were then kept at 20°C for 10 generations. When the selection of LS lines resumed, the C lines were also moved back to 25°C and completed 70 generations before the experiments. The flies used for experimentation were offspring from an unselected generation to avoid any cross-generational effects of the selection procedure (Watson and Hoffmann 1996; Hoffmann et al. 1997; Hercus and Hoffmann 2000). All flies experienced a light/dark cycle of 12/12 h.
Longevity
Longevity was recorded in five independent experiments. In three of the experiments, mated longevity with antibiotics in the medium was investigated. Virgin longevity with antibiotics in the medium and mated longevity without antibiotics in the medium was studied in one experiment each. The flies were set-up in 200 ml bottles under uncrowded conditions (allowing 10 pairs to lay eggs for 24 h). At the start of emergence, the bottles were emptied and flies collected at less than 12 h old under light CO2 anesthesia. For investigating mated longevity, we collected six vials per replicate line, 15 males and 15 females per vial. To investigate virgin life span, we collected three vials per replicate line, 30 males per vial. Every second day, the flies were transferred to new food vials with 4 ml of standard Drosophila oatmeal–sugar–yeast–agar medium, which contained antibiotics in four of the experiments (alternating between ampicillin 0.1 g/l and doxycycline 0.25 g/l). The number and sex of dead flies were scored at transfer. The number of vials was gradually reduced as deaths occurred, with surviving flies being kept as close to a density of 30 as possible.
Sampling for gene expression analysis
Flies were set-up, collected, and transferred following the same protocol as the longevity assay. Samples were frozen in liquid nitrogen at the same time of day (3 p.m.) on days 3, 19, and 35 after collection to prevent genes with a circadian rhythm from disturbing the results (Fig. 1). Average cumulative mortality in the LS lines at the three time points was 0%, 8%, and 20%, and 0%, 20%, and 90% in the three C lines. Flies from the longevity-selected lines were also sampled after 90% of the original cohort had died (days 60–80 depending on the line, Fig. 1). The sexes were separated over ice and 15 males from each combination of line and age were stored at -80°C.
Life span analysis
The longevity data was analyzed using the proportional hazard analysis function of JMP 7.0. Gompertz parameters were fitted in the program Winmodest (Promislow et al. 1999), which compensated for sub-threshold mortality using maximum likelihood methods. We defined maximum life span as the 90th percentile of life span using quantile regression (Redden et al. 2004). We tested for selection effects on maximum life span using Fisher’s exact test, which at worst is overly conservative (Wang et al. 2004).
RNA extraction and array hybridization
RNA extraction and hybridization was performed as previously described (Sørensen et al. 2005). We used Affymetrix arrays containing 18,953 probes representing more than 14,000 unique genes. The array data was analyzed using R (version 2.9.0; http://www.r-project.org/) based applications. The normalization of the expression values for the samples was performed separately using the Robust Multi-array Average (RMA) algorithm (Irizarry et al. 2003) as implemented in the Affy package (version 1.22.1). Differential expression of each gene was assessed using linear modeling and empirical Bayes methods, which were implemented using the R package Limma (version 2.18.2) (Smyth 2005). We then tested for identical gene expression profiles between treatment groups. The experiment included two main factors, line (C, control; and LS, longevity) and day (3, 19, 35, >60). In order to identify genes underlying increased longevity, we compared the lines at the same chronological age (LS3-C3, LS19-C19, and LS35-C35) and at the same physiological age (LS3-C3, LS35-C19, and LS60-C35). Genes responding to aging were identified separately in the control (C19-C3 and C35-C19) and longevity (LS19–LS3, LS35–LS19, and LS60–LS35) lines. A nested F-test (Smyth 2005) was used to identify genes that were generally different in more than one contrast. We set the significance levels at P = 0.05 and, for assessing the effects of aging, minimum lfc was 1. Multiple testing was adjusted for by controlling the FDR. Genes with significantly different expression profiles that had been assigned to GO categories were used to test for GO enrichment, for both the general line effect and the line-specific aging effect. These tests were run as functional annotation clustering using the DAVID bioinformatics database (Dennis et al. 2003). Dendrograms of the expression patterns and samples were computed by hierarchical clustering using the Euclidean distance measure and complete linkage as an agglomerative method as implemented in R. Dendrograms were combined with a heat map displaying color-coded expression signal intensities for each gene/sample. Genes found to be differentially expressed in our study were compared to several other expression studies. The probability that the overlap of differentially expressed genes is different from the number expected by chance was calculated using Monte Carlo simulations. In each simulation, the gene list within each treatment was permutated and the overlap of induced genes determined.
An analysis of variance was used to assess the overall treatment effects on the expression of genes significantly up- or down-regulated in LS lines on day 35. The treatments in the model included the effects of age, gene, and selection, taking into account the three-way interaction terms. The treatment effects were tested using an F-test.
Electronic supplementary materials
Below is the link to the electronic supplementary material.
(XLS 561 kb)
(PDF 130 kb)
(XLS 283 kb)
(PDF 92.1 kb)
(DOC 43.0 kb)
Acknowledgments
The authors are grateful to Doth Andersen and Marie Rosenstand Hansen for technical assistance, to Mogens Kruhøffer, AROS Applied Biotechnologies, for RNA extraction and hybridization, to Suresh Rattan, Jesper G Sørensen, Torsten N Kristensen, Corneel Vermeulen, and Janneke Wit and anonymous reviewers for helpful comments on the manuscript, and to the Danish Natural Sciences Research Council (frame and center grants to VL), Villum Kann Rasmussen Foundation (VL), Lundbeck Foundation and Carlsbergfondet (stipend to PSa), and the European Commission, within the 6th Framework Program (grant to PSø, contract no. FOOD-CT-2006-016250).
References
- Arking R, Dudas SP. Review of genetic investigations into the aging processes of Drosophila. J Am Geriatr Soc. 1989;37:757–773. doi: 10.1111/j.1532-5415.1989.tb02240.x. [DOI] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, Rollmann SM, Duncan LH, Lawrence F, Anholt RRH, Mackay TFC. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldal EA, Brakefield PM, Zwaan BJ. Multitrait evolution in lines of Drosophila melanogaster selected for increased starvation resistance: the role of metabolic rate and implications for the evolution of longevity. Evolution. 2006;60:1435–1444. [PubMed] [Google Scholar]
- Bubliy OA, Loeschcke V. Correlated responses to selection for stress resistance and longevity in a laboratory population of Drosophila melanogaster. J Evol Biol. 2005;18:789–803. doi: 10.1111/j.1420-9101.2005.00928.x. [DOI] [PubMed] [Google Scholar]
- Curtis C, Landis GN, Folk D, Wehr NB, Hoe N, Waskar M, Abdueva D, Skvortsov D, Ford D, Luu A, Badrinath A, Levine RL, Bradley TJ, Tavare S, Tower J. Transcriptional profiling of MnSOD-mediated lifespan extension in Drosophila reveals a species-general network of aging and metabolic genes. Genome Biol. 2007;8(12):R262. doi: 10.1186/gb-2007-8-12-r262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4, doi:R60 [PubMed]
- Franceschi C, Capri M, Monti D, Giunta S, Olivieri F, Sevini F, Panouraia MP, Invidia L, Celani L, Scurti M, Cevenini E, Castellani GC, Salvioli S. Inflammaging and anti-inflammaging: a systemic perspective on aging and longevity emerged from studies in humans. Mech Ageing Dev. 2006;128:92–105. doi: 10.1016/j.mad.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Girardot F, Lasbleiz C, Monnier V, Tricoire H (2006) Specific age related signatures in Drosophila body parts transcriptome. BMC Genomics 7, doi:6910.1186/1471-2164-7-69 [DOI] [PMC free article] [PubMed]
- Golden TR, Hubbard A, Melov S. Microarray analysis of variation in individual aging C. elegans: approaches and challenges. Exp Gerontol. 2006;41:1040–1045. doi: 10.1016/j.exger.2006.06.034. [DOI] [PubMed] [Google Scholar]
- Grotewiel MS, Martin I, Bhandari P, Cook-Wiens E. Functional senescence in Drosophila melanogaster. Ageing Res Rev. 2005;4:372–397. doi: 10.1016/j.arr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Hamilton WD. Moulding of senescence by natural selection. J Theor Biol. 1966;12:12–45. doi: 10.1016/0022-5193(66)90184-6. [DOI] [PubMed] [Google Scholar]
- Harbison ST, Chang S, Kamdar KP, Mackay TFC. Quantitative genomics of starvation stress resistance in Drosophila. Genome Biol. 2005;6:15. doi: 10.1186/gb-2005-6-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercus MJ, Hoffmann AA. Maternal and grandmaternal age influence offspring fitness in Drosophila. Proc R Soc Lond Ser B-Biol Sci. 2000;267:2105–2110. doi: 10.1098/rspb.2000.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Dagher H, Hercus M, Berrigan D. Comparing different measures of heat resistance in selected lines of Drosophila melanogaster. J Insect Physiol. 1997;43:393–405. doi: 10.1016/S0022-1910(96)00108-4. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM. Longevity, genes, and aging. Science. 1996;273:54–59. doi: 10.1126/science.273.5271.54. [DOI] [PubMed] [Google Scholar]
- Kim SN, Rhee JH, Song YH, Park DY, Hwang M, Lee SI, Kim JE, Gim BS, Yoon JH, Kim YJ, Kim-Ha J. Age-dependent changes of gene expression in the Drosophila head. Neurobiol Aging. 2005;26:1083–1091. doi: 10.1016/j.neurobiolaging.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Kirkwood TBL. Evolution of aging. Nature. 1977;270:301–304. doi: 10.1038/270301a0. [DOI] [PubMed] [Google Scholar]
- Kirkwood TBL, Feder M, Finch CE, Franceschi C, Globerson A, Klingenberg CP, LaMarco K, Omholt S, Westendorp RGJ. What accounts for the wide variation in life span of genetically identical organisms reared in a constant environment? Mech. Ageing Devel. 2005;126:439–443. doi: 10.1016/j.mad.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Klass M, Hirsh D. Non-aging developmental variant of Caenorhabditis elegans. Nature. 1976;260:523–525. doi: 10.1038/260523a0. [DOI] [PubMed] [Google Scholar]
- Kristensen TN, Sørensen P, Kruhoffer M, Pedersen KS, Loeschcke V. Genome-wide analysis on inbreeding effects on gene expression in Drosophila melanogaster. Genetics. 2005;171:157–167. doi: 10.1534/genetics.104.039610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis GN, Abdueva D, Skvortsov D, Yang JD, Rabin BE, Carrick J, Tavare S, Tower J. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101:7663–7668. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow GJ. Invertebrate gerontology: the age mutations of Caenorhabditis elegans. Bioessays. 1996;18:809–815. doi: 10.1002/bies.950181007. [DOI] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Luckinbill LS, Arking R, Clare MJ, Cirocco WC, Buck SA. Selection for delayed senescence in Drosophila melanogaster. Evolution. 1984;38:996–1003. doi: 10.2307/2408433. [DOI] [PubMed] [Google Scholar]
- Lund J, Tedesco P, Duke K, Wang J, Kim SK, Johnson TE. Transcriptional profile of aging in C. elegans. Curr Biol. 2002;12:1566–1573. doi: 10.1016/S0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- Medaware PB. An unsolved problem in biology. London: K.H. Lewis; 1952. [Google Scholar]
- Oudman L, Delden W, Kamping A, Bijlsma R. Starvation resistance in Drosophila melanogaster in relation to the polymorphisms at the adh and alpha-gpdh loci. J Insect Physiol. 1994;40:709–713. doi: 10.1016/0022-1910(94)90098-1. [DOI] [Google Scholar]
- Partridge L. Some highlights of research on aging with invertebrates, 2009. Aging Cell. 2009;8:509–513. doi: 10.1111/j.1474-9726.2009.00498.x. [DOI] [PubMed] [Google Scholar]
- Pearl R, Parker SL, Gonzalez BM. Experimental studies on the duration of life VII. The Mendelian inheritance of duration of life in crosses of wild type and quintuple stocks of Drosophila melanogaster. Am Nat. 1923;57:153–192. doi: 10.1086/279913. [DOI] [Google Scholar]
- Pletcher SD, Libert S, Skorupa D. Flies and their golden apples: the effect of dietary restriction on Drosophila aging and age-dependent gene expression. Ageing Res Rev. 2005;4:451–480. doi: 10.1016/j.arr.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Promislow DEL, Tatar M, Pletcher S, Carey JR. Below threshold mortality: implications for studies in evolution, ecology and demography. J Evol Biol. 1999;12:314–328. doi: 10.1046/j.1420-9101.1999.00037.x. [DOI] [Google Scholar]
- Redden DT, Fernandez JR, Allison DB. A simple significance test for quantile regression. Stat Med. 2004;23:2587–2597. doi: 10.1002/sim.1839. [DOI] [PubMed] [Google Scholar]
- Rodwell GEJ, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang LL, Xiao WZ, Mindrinos M, Crane E, Segal E, Myers BD, Brooks JD, Davis RW, Higgins J, Owen AB, Kim SK. A transcriptional profile of aging in the human kidney. Nature. 2004;2:2191–2201. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service PM The effect of mating status on lifespan, egg-laying, and starvation resistance in Drosophila melanogaster in relation to selection on longevity. J Insect Physiol. 1989;35:447–452. doi: 10.1016/0022-1910(89)90120-0. [DOI] [Google Scholar]
- Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and computational biology solutions using R and bioconductor. New York: Springer; 2005. [Google Scholar]
- Sørensen JG, Nielsen MM, Kruhoffer M, Justesen J, Loeschcke V. Full genome gene expression analysis of the heat stress response, in Drosophila melanogaster. Cell Stress Chaperones. 2005;10:312–328. doi: 10.1379/CSC-128R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen JG, Nielsen MM, Loeschcke V. Gene expression profile analysis of Drosophila melanogaster selected for resistance to environmental stressors. J Evol Biol. 2007;20:1624–1636. doi: 10.1111/j.1420-9101.2007.01326.x. [DOI] [PubMed] [Google Scholar]
- Strehler BL. Studies on the comparative physiology of aging. 2. On the mechanism of temperature life-shortening in Drosophila melanogaster. J Gerontol. 1961;16:2–12. [Google Scholar]
- Tarnopolsky M, Phillips S, Parise G, Varbanov A, DeMuth J, Stevens P, Qu A, Wang F, Isfort R. Gene expression, fiber type, and strength are similar between left and right legs in older adults. J Gerontol Ser A-Biol Sci Med Sci. 2007;62:1088–1095. doi: 10.1093/gerona/62.10.1088. [DOI] [PubMed] [Google Scholar]
- Wang CX, Li Q, Redden DT, Weindruch R, Allison DB. Statistical methods for testing effects on "maximum lifespan". Mech Ageing Dev. 2004;125:629–632. doi: 10.1016/j.mad.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Watson MJO, Hoffmann AA. Acclimation, cross-generation effects, and the response to selection for increased cold resistance in Drosophila. Evolution. 1996;50:1182–1192. doi: 10.2307/2410659. [DOI] [PubMed] [Google Scholar]
- Williams GC. Pleiotropy, natural-selection, and the evolution of senescence. Evolution. 1957;11:398–411. doi: 10.2307/2406060. [DOI] [Google Scholar]
- Zahn JM, Sonu R, Vogel H, Crane E, Mazan-Mamczarz K, Rabkin R, Davis RW, Becker KG, Owen AB, Kim SK. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:1058–1069. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerofsky M, Harel E, Silverman N, Tatar M. Aging of the innate immune response in Drosophila melanogaster. Aging Cell. 2005;4:103–108. doi: 10.1111/j.1474-9728.2005.00147.x. [DOI] [PubMed] [Google Scholar]
- Zwaan B, Bijlsma R, Hoekstra RE. Direct selection on life-span in Drosophila melanogaster. Evolution. 1995;49:649–659. doi: 10.2307/2410318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
(XLS 561 kb)
(PDF 130 kb)
(XLS 283 kb)
(PDF 92.1 kb)
(DOC 43.0 kb)