Abstract
Previous studies have shown that longevity is associated with enhanced cellular stress resistance. This observation supports the disposable soma theory of aging, which suggests that the investment made in cellular maintenance will be proportional to selective pressures to extend lifespan. Maintenance of protein homeostasis is a critical component of cellular maintenance and stress resistance. To test the hypothesis that enhanced protein repair and recycling activities underlie longevity, we measured the activities of the 20S/26S proteasome and two protein repair enzymes in liver, heart and brain tissues of 15 different mammalian and avian species with maximum lifespans (MLSP) ranging from 3 to 30 years. The data set included Snell dwarf mice, in which lifespan is increased by ∼50% compared to their normal littermates. None of these activities in any of the three tissues correlated positively with MLSP. In liver, 20S/26S proteasome and thioredoxin reductase (TrxR) activities correlated negatively with body mass. In brain tissue, TrxR was also negatively correlated with body mass. Glutaredoxin (Grx) activity in brain was negatively correlated with MLSP and this correlation remained after residual analysis to remove the effects of body mass, but was lost when the data were analysed using Felsenstein’s independent contrasts. Snell dwarf mice had marginally lower 20S proteasome, TrxR and Grx activities than normal controls in brain, but not heart tissue. Thus, increased longevity is not associated with increased protein repair or proteasomal degradation capacities in vertebrate endotherms.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-010-9157-5) contains supplementary material, which is available to authorized users.
Keywords: Protein homeostasis, Maximum lifespan, Aging, Protein degradation, Protein repair
Introduction
Vertebrate endotherms display a wide range of maximum lifespans (MLSP), from several years to over a century. Yet aging processes appear strikingly similar amongst endotherm species, suggesting that it may stem from common cellular and molecular mechanisms. It follows that the evolution of increasing MLSP may occur via selection of common key molecular characters; however, we have a limited understanding of what these molecular characters are.
Enhanced resistance to exogenous stressors appears to have co-evolved with longevity (reviewed in Robb et al. 2009). Inter-species comparisons indicate that, with some exceptions, longer-lived species have increased cellular stress resistance (Kapahi et al. 1999; Ogburn et al. 2001; Miller et al. 2010), and comparisons amongst mice harbouring single gene mutations that increase lifespan also reveal this trend (Holzenberger et al. 2003; Salmon et al. 2005; Maynard and Miller 2006; Bokov et al. 2009). A key observation is that inter- and intra-species differences in cellular stress resistance are present in young adults, suggesting that longevity is associated with enhanced stress resistance over the lifespan rather than becoming evident only with advanced age. The positive correlation between cellular stress resistance and MLSP is consistent with the disposable soma theory of lifespan, which suggests that the extent of the investment made in cellular maintenance will be determined by selective pressures to extend lifespan (Kirkwood et al. 2000).
Maintenance of protein homeostasis is a critical component of cellular maintenance and defence against stress (Morimoto 2008). Proteins are susceptible to the oxidative stress that arises directly from exposure to oxidants, or secondarily due to the disruption of metabolic redox pathways. Exposure to oxidative stress can lead to oxidative protein damage that directly alters protein structure, leading to dramatic decreases in their activity and/or aggregation of partially denatured proteins. Aberrant protein aggregation in brain tissue is a key feature of the major neurodegenerative disorders associated with aging (Martinez et al. 2008). To maintain protein function in the face of oxidative stress, cells utilize “protein repair” enzymes, capable of maintaining protein redox homeostasis through the regulation of thiol–disulfide exchange on critical residues, particularly cysteine. Important protein repair enzymes include thioredoxin, thioredoxin reductase and glutaredoxin. Thioredoxin (Trx) is responsible for reducing disulfide bonds on proteins, and thioredoxin reductase (TrxR) reduces the oxidized active site of Trx to maintain its maximal activity. TrxR therefore maintains the thioredoxin cycle—an important component of cellular resistance to oxidative stress (Arner 2009). Caenorhabditis elegans with a TrxR null gene have a reduced mean and maximal lifespan compared to wild type (Miranda-Vizuete et al. 2005). Glutaredoxin (Grx) is another important enzyme for maintaining protein function during oxidative stress by catalyzing S-glutathionylation and deglutathionylation of proteins, protecting thiol groups from oxidation. Lens epithelial cells of Grx gene knockout mice show decreased resistance to oxidative stress (Lofgren et al. 2008).
Although protein repair enzymes play a significant role in protein homeostasis, the majority of protein damage cannot be reversed or repaired, and so compromised proteins must be removed to prevent their aggregation and interference with normal cellular activities (Grune et al. 2004). The 20S/26S proteasome is a multi-subunit proteolytic complex found in the cytosol and nucleus that performs this task. The 20S proteasome degrades oxidatively damaged proteins (Reinheckel et al. 1998; Davies 2001), whereas the 26S proteasome, which is composed of the core 20S barrel with regulatory cap regions on either end, recognizes and degrades ubiquitin-tagged proteins that may or may not be damaged (Jung et al. 2009). In addition to a global role in damaged protein degradation, the nuclear 26S proteasome also degrades ubiquitylated RNA polymerase II associated with transcriptional arrest at sites of DNA damage (Daulney and Tansey 2009) and ubiquitin-tagged apoptotic proteins, thus preventing cell death (Bader and Steller 2009). Ubiquitin is a 76 amino acid peptide that is ligated to damaged proteins through the catalytic activity of ubiquitin ligases, thus targeting them for degradation (Hershko and Ciechanover 1998).
In this study, we test the hypothesis that longer-lived species have superior protein homeostasis mechanisms, manifesting as enhanced protein repair and proteasomal degradation activities. To address this, activities of thioredoxin reductase, glutaredoxin, and the 20S/26S proteasome were measured in liver, heart and brain of 15 vertebrate endotherm species with MLSPs ranging from 3 to 30 years (Table 1). Steady-state ubiquitin levels were measured by western blot in eight species for which ubiquitin’s amino acid sequence was known to be 100% conserved. By using this broad comparative approach with 15 species, we avoid the erroneous assumption that all between-species differences in these measured activities are attributable to differences in MLSP rather than body mass, metabolic rate or another unknown parameter (reviewed by Speakman 2005). We use analysis of residuals to differentiate associations with body mass from those with MLSP or metabolic rate. Felsenstein’s independent contrasts (Felsenstein 1985) are also employed to address problems associated with non-independence of data points (i.e. species) due to shared evolutionary history. Finally, we also measure these same activities in heart and brain of young adult long-lived Snell dwarf mice and their age-matched normal littermates to investigate whether increased lifespan is associated with enhanced protein homeostatic mechanisms in this intra-specific model of longevity.
Table 1.
Sex, age, mass and maximal lifespan (MLSP) of the mammalian and avian species
| Common name | Scientific name | Number and sex | Age | Mass (kg) | MLSP (years)a | Source |
|---|---|---|---|---|---|---|
| Mammalia | ||||||
| Snell mouse (DW/J) | Mus musculus | 6 F (+/dw) | 6–7 months | 0.030 | 2.7 | Jackson Laboratories (Bar Habor, ME, USA) |
| 6 F (dw/dw) | 6–7 months | 0.010 | 3.9 | |||
| C57BL/6Ncr1BR mouse | Mus musculus | 4 F | 4 months | 0.028 | 3.5 | Charles River (Wilmington, MA, USA) |
| 4 M | ||||||
| Syrian hamster | Mesocricetus auratus | Unknown | 4–8 months | 0.130 | 3.9 | Equitech-Bio, Inc. (Kerrville, TX, USA) |
| Norway rat | Rattus norvegicus | 6 M | 3 months | 0.550 | 5 | Charles River (Wilmington, MA, USA) |
| Mongolian gerbil | Meriones unguiculatus | Unknown | 4–8 months | 0.070 | 6.3 | Equitech-Bio, Inc. (Kerrville, TX, USA) |
| 13-lined ground squirrel | Spermophilus tridecemlineatus | 8 F | Unknown | 0.205 | 7 | University of Manitoba Field Research Station (Carmen, MB, Canada |
| 3 M | ||||||
| Rabbit | Oryctolagus cuniculus | 4 F | 3.5 months | 2.60 | 11.8 | Local abattoir (Fort Erie, ON, Canada) |
| 1 M | ||||||
| Guinea pig | Cavia porcellus | Unknown | Unknown | 1.05 | 12 | Rockland (Gilbertsville, PA, USA) |
| Big brown bat | Eptesicus fuscus | 3 F | 6–12 months | 0.025 | 19 | McMaster University Bat Laboratory (Hamilton, ON, Canada) |
| 2 M | ||||||
| Sussex sheep | Ovis aries | 5 F | 6–12 months | 29.5 | 19.6 | Local abattoir (Fort Erie, ON, Canada) |
| White-tailed deer | Odocoileus virginianus | 3 M | 2.5–3.5 years | 110 | 21.6 | Local hunters (Cobden, ON, Canada) |
| Domestic dog | Canis familiaris | 3 M | 2–8 years | 11.0 | 24 | Equitech-Bio, Inc. (Kerrville, TX, USA) |
| Yorkshire/Hampshire pig | Sus scrofa | 2 F | 6 months | 99.9 | 27 | Local abattoir (Fort Erie, ON, Canada) |
| 3 M | ||||||
| Black angus/Charlet cow | Bos taurus | 4 F | 1.5–2.5 years | 379.5 | 30 | Local abattoir (Fort Erie, ON, Canada) |
| 1 M | ||||||
| Aves | ||||||
| Japanese quail | Coturnix japonica | 2 F | 4–5 months | 0.264 | 6 | Cro Quail Farms Inc (St. Anne’s, ON, Canada) |
| 2 M | ||||||
| Zebra finch | Taeniopygia guttata | 3 F | 1 year | 0.017 | 14.5 | Trent University Animal Care Facility (Peterborough, ON, Canada) |
| 3 M | ||||||
aMLSP data from de Magalhães et al. (2005)
Materials and methods
Materials
Chemicals were purchased from Bioshop (Burlington, ON, Canada) and Sigma Aldrich (Oakville, ON, Canada; including Fluka and Caledon). BioRad protein dye was purchased from BioRad Laboratories (Hercules, CA, USA). Proteasome peptide Suc-LLVY-AMC, proteasome inhibitor MG-132 and AMC calibration standard were purchased from Enzo Life Sciences (Plymouth Meeting, PA, USA). Anti-ubiquitin primary antibody (P-4D1) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-mouse (goat) secondary antibody was purchased from Rockland Immunochemicals (Gilbertsville, PA, USA). Prestained broad range protein marker was obtained from BioLabs (New England, MA, USA).
Animals
Between 3 and 6 individuals of each of 13 mammalian and 2 avian species were used in this study (Table 1). Species were selected on the bases of (1) phylogenetic position, (2) MLSP, (3) routine diet and (4) availability. With respect to phylogenetic position, we confined our analysis to species with a similar overall body plan and physiology, i.e. vertebrate endotherms. Within this constraint, we sampled species within and between orders. We also attempted to maximize the range of MLSP represented by species included in the study. We also included species with exceptionally long lifespans for their body mass (big brown bat, zebra finch). The nature of the study meant that we were unable to directly control for differences in diet specific to a given species; however, our species collection includes graminovores, nucivores, insectivores, omnivores and carnivores, and no trends were noted that related to dietary preference. We also included wild-caught species where possible to control for domestication effects. All animals were healthy young adults, and in most cases, the exact age and sex of the animal were known.
Tissue collection
Normal C57BL mice were euthanized by cervical dislocation following which brain, heart and liver tissues were immediately excised, flash-frozen in liquid nitrogen and stored at −80°C. Snell normal and dwarf mice were anaesthetized with isoflurane, injected with a mixture of 50 μg/g ketamine and 5 μg/g xylazine before tissue excision within 30–45 min post-mortem. These tissues were flash-frozen in liquid nitrogen and stored at −80°C. Norway rats were euthanized with sodium pentabarbitol sodium injection (Euthanyl®; 2 μg/g body wt), and brain, heart and liver tissues were removed and flash-frozen in liquid nitrogen and stored at −80°C. Tissues from 13-lined ground squirrels were collected at the University of Western Ontario (London, ON, Canada) as described in Page et al. (2009). Active (i.e. non-hibernating) animals were euthanized by Euthanyl overdose (270 mg/ml, 0.2 ml/100 g body wt), and brain, heart and liver tissues were rapidly removed, flash-frozen in liquid nitrogen, and stored at −80°C. Big brown bats, collected from the wild and housed in a captive research colony at McMaster University (Hamilton, ON, Canada), were euthanized using 0.6 mg/g body wt sodium pentabarbitol followed by decapitation. Bat brain, heart and liver tissues were excised and flash-frozen in liquid nitrogen and stored at −80°C. Zebra finch and Japanese quail tissues were collected at Trent University (Peterborough, ON, Canada). Birds were euthanized with 2.0 (finch) or 0.6 (quail) mg/g body wt sodium pentabarbitol (Euthasol®), followed by decapitation. Bird brain, heart and liver tissues were removed and flash-frozen in liquid nitrogen and stored at −80°C. Guinea pigs were euthanized, and heart, brain and liver tissues rinsed in 1× phosphate-buffered saline (PBS) and immediately frozen in liquid nitrogen and shipped to Brock University (St. Catharines, ON, Canada) on dry ice. Similarly excised tissues from euthanized dogs, hamsters and gerbils were immediately frozen in liquid nitrogen and shipped to Brock University (St. Catharines, ON, Canada) on dry ice. Tissue from domesticated livestock was collected at a local abattoir during normal processing, with the exception of rabbits which were collected during processing from a local farmer. In all cases, brain, heart and liver tissues were collected from these animals within approximately 30 min post-mortem. Due to the size of organs from domesticated livestock and rabbits, tissue samples were isolated from the following locations: brain cortex, right cardiac ventricle and tip of the left liver lobe. Tissues were frozen on-site in dry ice then brought to Brock University (St. Catharines, ON, Canada) where they were stored at −80°C. Fresh white-tailed deer tissue (collected within 1 h post-mortem) was obtained by hunters in Cobden (ON, Canada), following which brain, heart and liver tissues were collected and frozen at −20°C for 1 month, and then transferred to Brock University (St. Catharines, ON, Canada) where they were stored at –80°C. Tissue sampling sites were unspecified for the white-tailed deer samples.
Preparation of tissue homogenates
Brain, heart and liver tissues were homogenized using a PowerGen 125 Homogenizer (Fisher Scientific, Ottawa, Ontario, Canada) in ice cold lysis buffer (50 mM 4-2-hydroxyethyl-1-piperazineethanesulfonic acid (HEPES), 20 mM KCl, 0.1 mM ethylenediamine tetraacetic acid (EDTA), 1 mM DTT, 5% glycerol and 0.1% NP40) at full speed for 3 cycles of 10 s with 10 s rest between each cycle. Following homogenization, samples were centrifuged at 10,000×g for 20 min. Protein concentration was determined using the Bradford technique with a BioRad protein assay kit and homogenates were aliquoted and stored at −80°C.
20S and 26S proteasome activity assays
Assays for 20S and 26S proteasome activities were performed as described by Rodgers and Dean (2003) at room temperature (23°C) using a Varian spectrofluorometer. Conditions for the 20S proteasome assay were 25 mM HEPES pH 7.5, 0.5 mM EDTA, 25 μM peptide and 100 μg sample. The peptide Suc-LEU-LEU-VAL-TYR was used to measure the chymotryptic activity of the proteasome. Activity of the 26S proteasome was measured in the same manner except that samples were pre-incubated in 5 mM adenosine triphosphate (ATP) for 10 min (ATP was retained throughout the activity assay) to stimulate 26S proteasome activity. Both assays were conducted with an excitation wavelength of 360 nm and an emission detection wavelength of 460 nm. Activities were determined by measuring the release of the AMC fluorophore from the peptide–AMC complex for 15 min. Parallel assays were run with 0.2 mM of the specific proteasome inhibitor MG-132 to determine non-proteasomal AMC release. This non-specific activity was minor, but was subtracted from the rate measured in the absence of the inhibitor. The standard curve for proteasome calculations used a serial dilution of 8 μM AMC calibration standard to obtain the relationship between arbitrary fluorescence units (AFU) and [AMC]. Specific proteasome activity was calculated by interpolation from the standard curve according to the equation: 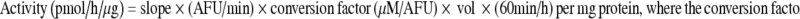
Quantification of protein ubiquitylation
Brain, heart and liver tissue homogenates (50 μg) were separated by SDS-PAGE and electrotransferred to a PVDF membrane. The membranes were incubated in a blocking solution, consisting of 5% skim milk in 1× PBS, for 45 min at room temperature. Following blocking, the membranes were incubated with anti-ubiquitin antibody (1:250) overnight at 4°C in solution containing 5% skim milk in 1× PBS-T and then incubated with anti-mouse secondary antibody for 1 h at room temperature. The membranes were scanned using an Odyssey infrared visual system (LI-COR Biosciences). Ubiquitin level quantification was performed using Odyssey Imaging Software, Version 1.0. The intensity for each sample was compared to an internal standard control which simultaneously resolved within every gel. Band intensity represents intensity of the entire lane from top to bottom of the membrane.
Enzyme activity assays
Thioredoxin reductase activity assay
The activity of TrxR was assayed essentially as in Arner et al. (1999) in a solution containing 50 mM potassium phosphate buffer, 1 mM EDTA, 0.2 mg/ml, 0.2 mM reduced nicotinamide adenine dinucleotide phosphate (NADPH) and protein sample (50–100 μg). The reaction was initiated by the addition of 2.5 mM DTNB and measured at 412 nm. Parallel assays were performed with 0.5 mM of the specific thioredoxin reductase inhibitor dichloronitrobenzene (DCNB) to determine non-proteasomal AMC release. Although, the non-specific activity was negligible, this activity was subtracted from the rate measured in the absence of the inhibitor.
Glutaredoxin activity assay
Grx activity was assayed essentially as described in Kumar and Holmgren (1999). The reaction buffer contained 0.1 M Tris–HCl, 2 mM EDTA, 100 μg/ml BSA, 0.4 mM NADPH, 1 U/ml glutathione reductase, 10 mM GSH and 50 μg of protein. A background change in absorbance was monitored prior to the addition of 2-hydroxydisulfide, which initiated the reaction.
Statistical analyses
Raw data were natural logarithm (Ln)-transformed prior to correlation analyses. Residuals were calculated from simple linear regression of the dependent variable of interest on body mass to remove the confounding effect of body mass. Significance of the coefficient of correlation was used to determine if the line was different from horizontal, and significance was based on t values for one-tailed tests. A p value of 0.05 was considered significant. Felsenstein’s phylogenetically independent contrasts (Felsenstein 1985) were calculated using PDAP. The phylogenetic tree (Fig. S1) was constructed from four phylogenies from which branch length estimates were taken (Stuart et al. 2002; Springer and Murphy 2007; Gorbunova et al. 2008; Hackett et al. 2008; Prasad et al. 2008); however, the analysis was repeated with all branch lengths set to one, as with a speciation model of character change (Price 1997). Statistical analyses within the intra-species context (Snell dwarf mice) used a two-tailed t test to compare normal mice and Snell dwarf mice. A p value of 0.05 was considered significant.
Results
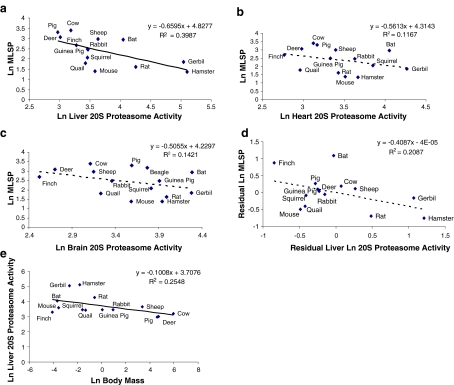
Activity of the 20S proteasome was measured in liver, heart and brain tissues of 3–6 individual young adult specimens from each of the 13 mammalian and 2 avian species. The 20S proteasome activity data were natural logarithm (Ln)-transformed and plotted against Ln-transformed MLSP data for each species (Fig. 1a–c). A correlational analysis revealed a significant inverse relationship between the two parameters in liver, but not in heart or brain. To test whether the negative correlation between 20S proteasome activity in liver and MLSP (Fig. 1a) was actually driven by body mass, we plotted the residuals of the body mass versus MLSP correlation against the residuals of the body mass versus 20S proteasome activity (Fig. 1d). This analysis yielded no significant correlation, indicating the absence of a relationship between 20S proteasome activity and body mass. The direct correlation of liver 20S proteasome activity with body mass, which is significant (p < 0.05), is shown in Fig. 1e (see also Table 3). Because basal metabolic rate is inversely correlated with body mass, we hypothesized that liver 20S proteasome activity may be influenced by mass-specific metabolic rate. This was addressed using mass-specific metabolic rate data from AnAge (de Magalhães et al. 2005). Residual analysis using these data indicated no relationship between mass-specific metabolic rate and 20S proteasome activity (Table S1).
Fig. 1.
Correlation between 20S proteasome activity and MLSP. a 20S proteasome activity is negatively correlated with MLSP in liver, but not in heart (b) or brain (c). Analysis of the residuals of 20S proteasome activity plotted against residual of MLSP (d) shows the absence of a statistically significant correlation. e Statistically significant correlation of liver 20S proteasome activity with body mass (p < 0.05). All values are natural log-transformed and each data point represents the mean from duplicate measurement from three to eight individuals of a given species
Table 3.
Statistical analysis of enzyme activities as correlates of body mass
| Enzyme activity | Correlation (R2) | Slope | p value |
|---|---|---|---|
| Liver 20S proteasome activity | 0.2548 | −0.1008 | <0.05 |
| Heart 20S proteasome activity | 0.1520 | −0.0583 | >0.05 |
| Brain 20S proteasome activity | 0.0866 | −0.0501 | >0.1 |
| Liver 26S proteasome activity | 0.2237 | −0.075 | <0.05 |
| Brain 26S proteasome activity | 0.0178 | −0.0196 | >0.1 |
| Liver TrxR Activity | 0.2142 | −0.0629 | <0.05 |
| Heart TrxR activity | 0.1462 | −0.0315 | >0.05 |
| Brain TrxR activity | 0.3026 | −0.0503 | <0.05 |
| Liver Grx activity | 0.1571 | −0.0489 | <0.1 |
| Heart Grx activity | 0.0040 | −0.0066 | >0.1 |
| Brain Grx activity | 0.0984 | −0.0143 | >0.1 |
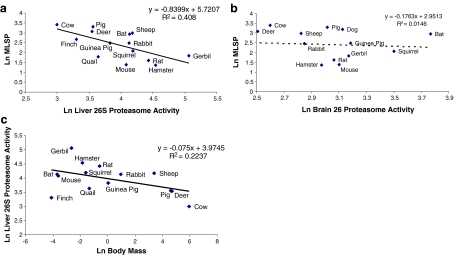
The 20S proteasome plays a key role in the degradation of oxidatively damaged proteins that are not ubiquitin-tagged (Reinheckel et al. 1998; Davies 2001). Conversely, ubiquitin-tagged proteins are degraded by the 26S proteasome (Jung et al. 2009). To estimate the capacity for degrading ubiquitin-tagged proteins, 26S proteasome activity was measured in liver and brain tissue samples, generating similar results to 20S proteasome activity. The activity of the 26S proteasome was negatively correlated (p < 0.05) with MLSP in liver tissue (Fig. 2a), but we observed no correlation between 26S proteasome activity and MLSP in brain tissue (Fig. 2b). Again, analysis of the residuals of liver 26S proteasome activity revealed no correlation with MLSP or mass-specific metabolic rate, and the same results were found using Felsenstein’s independent contrasts (FIC) analysis (Table 2). Thus, liver 26S proteasome activity also correlates negatively with body mass (shown in Fig. 2c, Table 3).
Fig. 2.
Correlation between 26S proteasome activity and MLSP. 26S proteasome activity correlates negatively with MLSP (p < 0.025) in liver (a) but not brain (b). The trend in (a) is driven by the correlation of liver 26S proteasome activity with body mass (c). All values are natural log-transformed and each data point represents the mean from duplicate measurement from three to eight individuals of a given species
Table 2.
Statistical analysis of linear regressions: enzyme activity as a function of lifespan
| Enzyme activity | Correlation (R2) | Slope | p value |
|---|---|---|---|
| Liver 20S proteasome | |||
| Residual | 0.2087 | −0.4087 | >0.05 |
| Residual + FIC | 0.00003 | 0.0044 | >0.05 |
| Heart 20S proteasome | |||
| Residual | 0.0023 | 0.0539 | >0.05 |
| Residual + FIC | 0.0143 | 0.1164 | >0.05 |
| Brain 20S proteasome | |||
| Residual | 0.0072 | −0.0809 | >0.05 |
| Residual + FIC | 0.0013 | −0.0353 | >0.05 |
| Liver 26S proteasome | |||
| Residual | 0.0004 | −0.0227 | >0.05 |
| Residual + FIC | 0.0125 | −0.0984 | >0.05 |
| Brain 26S proteasome | |||
| Residual | 0.0046 | −0.0728 | >0.05 |
| Residual + FIC | 0.0493 | −0.3133 | >0.05 |
| Liver TrxR | |||
| Residual | 0.1306 | −0.5568 | >0.05 |
| Residual + FIC | 0.0095 | −0.1635 | >0.05 |
| Heart TrxR | |||
| Residual | 0.0834 | −0.5849 | >0.05 |
| Residual + FIC | 0.0003 | 0.0386 | >0.05 |
| Brain TrxR | |||
| Residual | 0.0016 | −0.0823 | >0.05 |
| Residual + FIC | 0.0385 | −0.3822 | >0.05 |
| Liver Grx | |||
| Residual | 0.0151 | 0.1675 | >0.05 |
| Residual + FIC | 0.0012 | −0.0391 | >0.05 |
| Heart Grx | |||
| Residual | 0.1194 | −0.5137 | >0.05 |
| Residual + FIC | 0.0299 | −0.213 | >0.05 |
| Brain Grx | |||
| Residual | 0.2752 | −1.8917 | <0.05 |
| Residual + FIC | 0.0353 | −1.0398 | >0.05 |
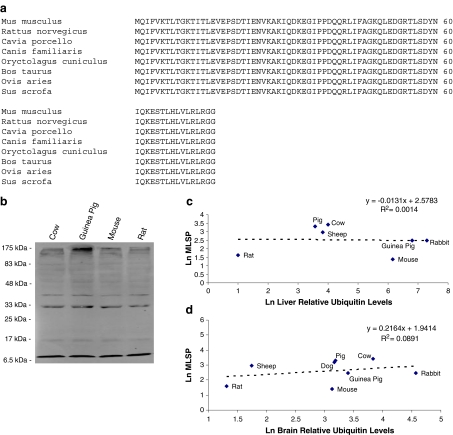
In vivo activity of the 26S proteasome will be affected by the concentration of its substrates (i.e. ubiquitylated proteins). Steady-state levels of protein ubiquitylation were measured in liver and brain tissues using quantitative immunodetection. Surprisingly, the amino acid sequence of ubiquitin is 100% conserved amongst eight species for which we could obtain complete sequence data (Fig. 3a). This allowed the quantitative cross-species western quantification of ubiquitylated protein levels (Fig. 3b shows a representative blot). The total ubiquitin signal (bound and free) per lane was quantified and plotted against MLSP for these species (Fig. 3c, d). The analysis revealed no correlation between ubiquitylated protein levels and MLSP or body mass (data not shown) in either liver or brain tissue. The results were unaffected when the signal from free ubiquitin (8.5 kDa) was excluded from the analysis (data not shown).
Fig. 3.
Ubiquitin protein levels do not correlate with MLSP. a The amino acid sequence of ubiquitin is conserved in the seven species analysed. b A representative western detection of ubiquitin in electrophoresed rabbit liver homogenate proteins of four individuals. Sizes in kilodalton represent protein marker sizes. Lanes represent ubiquitin protein levels in liver of different species. c, d No correlation between extent of protein ubiquitylation and MLSP in c liver and d brain tissue. All values are natural log-transformed and each data point represents the mean from duplicate measurement from three to eight individuals of a given species
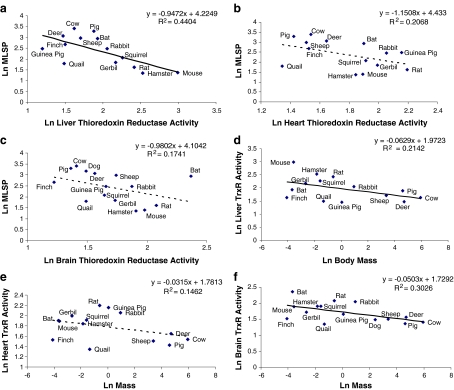
TrxR activity appeared to be negatively correlated with MLSP in all three tissues; however, this reached statistical significance only in liver (Fig. 4a–c). Again, the liver relationship was driven by the negative correlation of TrxR with body mass (Fig. 4d, Table 2). Interestingly, while brain TrxR activity did not correlate significantly with MLSP (Fig. 4e) or with mass-specific metabolic rate (Table S1), the correlation of brain TrxR activity with body mass was significant (Fig. 4f, Table 3).
Fig. 4.
Correlation between thioredoxin reductase (TrxR) activity and MLSP or body mass. TrxR activity is negatively correlated with MLSP a in liver (p < 0.025), but not in b heart or c brain tissue. d–f Correlation of TrxR activity with the species’ body mass. TrxR is negatively correlated with body mass in d liver (p < 0.05), and f brain tissue, but not in e heart tissue. All values are natural log-transformed and each data point represents the mean from duplicate measurement from three to eight individuals of a given species
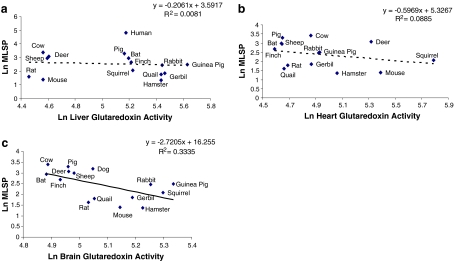
Grx activity was not correlated with either MLSP (Fig. 5a, b) or body mass (not shown) for either liver or heart. However, brain Grx activity was significantly negatively correlated with MLSP (Fig. 5c). In this instance, the correlation remained following residual analysis (Table 2), though was lost following FIC analysis (Table 2). Correlations between Grx activity and either species’ body mass (Table 3) or mass-specific metabolic rate (Table S1) were also not statistically significant.
Fig. 5.
Correlation between glutaredoxin (Grx) activity and MLSP. Grx activity in a liver and b heart does not correlate with MLSP. Brain Grx does correlate negatively with MLSP (c) (p < 0.025). All values are natural log-transformed and each data point represents the mean from duplicate measurement from three to eight individuals of a given species
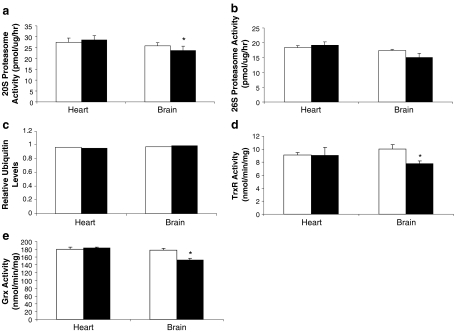
We measured the same protein repair and degradation activities in Snell normal and dwarf mice, an intra-specific model of increased longevity. There were no significant differences between normal and dwarf mice in 20S or 26S proteasome activities, TrxR or Grx in heart tissue. However, brain 20S proteasome activity was significantly lower in Snell dwarf compared to normal mice (Fig. 6). The same trend was observed for brain 26S proteasome activity, though this did not reach statistical significance. Protein ubiquitination levels did not differ between dwarf and normal heart or brain tissues. However, both TrxR and Grx activities were also lower in the brain of Snell dwarf compared to normal mice (Fig. 6).
Fig. 6.
Protein degradation and protein repair enzymes are not upregulated in long-lived Snell dwarf mice compared to normal controls. a 20S proteasome activity is significantly lower in Snell normal mice in brain tissue but no significance is found in heart tissue. b 26S proteasome activity and c ubiquitin protein levels are not different between Snell dwarf and Snell normal mice in brain tissue. d Thioredoxin reductase and e glutaredoxin activity levels are significantly higher in Snell normal mice compared to Snell dwarf mice in brain tissue but not in heart tissue. White bars represent Snell normal mice and black bars represent Snell dwarf mice. Values are means ± SEM of duplicate measurements made in each of five individuals per genotype. *p < 0.05, significantly different using a two-tailed t test
Discussion
The aim of this study was to test the hypothesis that longer-lived species or mouse strains have greater protein repair and/or recycling capacity. The hypothesis stems from the disposable soma theory of aging, which suggests that increases in energy invested in cellular maintenance will translate into increases in lifespan. This theory is supported by observations, in a wide variety of experimental models, of enhanced cellular stress resistance concomitant with greater longevity (reviewed in Robb et al. 2009). The 20S/26S proteasome appears to be a good candidate for positive selection by species evolving increased lifespan. It is required for the efficient removal of damaged proteins following oxidative stress (Divald and Powell 2006) and is stress-inducible (Grune et al. 1998). In C. elegans, fully functional 26S proteasome activity is required for normal stress resistance and lifespan (Ghazi et al. 2007). Its inhibition induces mitochondrial dysfunction, ROS overproduction and oxidative stress (Torres and Perez 2008). It is therefore interesting that we found no evidence for positive selection of either 20S or 26S proteasome activity during the evolution of longevity in vertebrate endotherms. Indeed, in this study, longer-lived species generally possessed lower 20S/26S proteasome activities in liver, though this trend was driven by the negative correlation of proteasome activity with adult body mass. It was surprising that another negative correlate of body mass, mass-specific metabolic rate, was found also not to explain this negative correlation. While we do not have an explanation for the correlation between body mass and proteasome activity, we can in any case reject our hypothesis that the latter is related to MLSP.
A similar result from our intra-specific model of increased longevity, i.e. the Snell dwarf mouse, further strengthens our conclusion that the longevity of vertebrate endotherms is not associated with enhanced proteasome activity. This conclusion is consistent with two recent reports of proteasome activity in the context of mammalian species lifespan. Salmon et al. (2009) also reported significantly lower 20S proteasome activity in livers of two long-lived bat species, Tadarida brasiliensis and Myotis velifer, compared to mice. Interestingly, we did not find that liver 20S and 26S proteasome activities in our bat species, Eptesicus fuscus, were lower than those in mice. Indeed, bat liver 20S/26S proteasome activities were in the mid-range of all species values, whereas heart and brain proteasome activities were actually in the high end of the species range. There are numerous possible reasons for the differences between the present study and Salmon et al. (2009), including the use of different bat species. However, these contrasting results highlight the importance of using as many species as possible in such studies to avoid biasing conclusions.
One factor that will affect 26S proteasome activity in vivo is the concentration of its substrates, i.e. ubiquitinylated proteins. We measured the relative levels of protein ubiquitylation in liver and brain tissue from seven/eight species for which we were able to verify 100% conservation of the ubiquitin amino acid sequence. It is imperative to restrict this analysis to ubiquitin sequences for which 100% conservation can be verified as any between-species differences in sequence may affect the antigen–antibody interaction and thus render the quantification inaccurate. The absence of a correlation between ubiquitylation levels and MLSP, combined with the 26S proteasome results, indicate that the overall rate of protein degradation via the ubiquitylation route in vivo is unlikely to be elevated in the longer-lived species. This suggests that degradative protein recycling via the proteasome does not contribute to any differences in protein homeostasis that may exist either between species or in long-lived Snell dwarf mice.
Protein homeostasis in stressed cells may also be compromised by oxidation of redox-sensitive thiols, such as cysteine. Reversal of this oxidative damage is catalyzed by a number of enzymes using NADPH and glutathione to supply reducing equivalents. Here we tested, and reject, the hypothesis that longevity would be associated with greater activities of two such enzymes, TrxR and Grx, thus providing a greater ability to rapidly respond to oxidative damage to vulnerable cysteines. TrxR and Grx were either not correlated or negatively correlated with MLSP (depending upon tissue). In the latter instance, the relationship was found to be driven either by body mass (TrxR) or perhaps directly by MLSP (Grx). Combined with marginally lower TrxR and Grx activities in brain tissue from Snell dwarf, relative to normal mice, it would appear that maintenance of constitutively high protein repair activities does not contribute to longevity. These results are similar to those reported by Perez et al. (2009a), showing that neither glutaredoxin nor methionine sulfoxide reductase A protein levels are higher in naked mole rat liver compared to mouse (though this conclusion is limited by potential between-species differences in amino acid sequences, which were not known, in this study). Thus, longevity is not associated with enhanced protein repair activities in the major oxidative tissues.
Our results indicate that protein repair and degradative recycling activities are not elevated in longer-lived animals. Nonetheless, longevity is broadly associated with enhanced cellular stress resistance, including resistance to oxidative stressors (e.g. Kapahi et al. 1999; Ogburn et al. 2001; Robb et al. 2009; Miller et al. 2010). Perhaps protein homeostasis is maintained by preventing the occurrence of damage, and research should focus on these mechanisms. However, some possible mechanisms of achieving this can also be eliminated. There is little evidence for greater antioxidant enzyme capacity in longer-lived species (reviewed in Perez et al. 2009b; also see Page et al. 2010a). Protein oxidative damage could also be prevented in longer-lived species by the systematic evolutionary replacement of cysteine residues with amino acids less susceptible to oxidative modification (as Moosmann and Behl (2008) demonstrated for mitochondrial DNA-encoded proteins); however, there is no evidence that this has occurred in nuclear DNA-encoded proteins that function either in the mitochondria or cytosol (Moosmann and Behl 2008). Similarly, proteins of naked mole rats actually have higher cysteine content than shorter-lived mice (Perez et al. 2009a). On the other hand, proteins in crude liver homogenates of some long-lived species, such as bats or naked mole rats, have been shown to resist urea-induced denaturation, which suggests some inherent stabilizing property (Perez et al. 2009a; Salmon et al. 2009). Perez et al. (2009a) report that this is not attributable to differences in heat shock protein-70 levels. While this between-species immunodetection approach requires validation for conservation of amino acid sequence, it is nonetheless in agreement with the observation that HSP70 levels are also lower in stress-resistant fibroblasts of long-lived Snell dwarf mice compared to those from normal littermates (Maynard and Miller 2006). A further study examining several different heat shock protein mitochondrial RNAs (mRNAs) in Snell dwarf mice found no coordinated elevation of heat shock proteins (HSPs) in Snell dwarf tissues or fibroblasts (Swindell et al. 2009). These latter mRNA data may not reflect protein levels, as changes in gene transcript levels often do not parallel changes in corresponding protein levels, even for transcriptionally regulated proteins (see for example Ideker et al. 2001; Le Naour et al. 2001). Investigation into HSP protein levels in this context is therefore probably still warranted. In addition, the possibility that specific post-translational protein modifications impart stability should be explored.
An important caveat to our study is that we measured total activities of proteins of interest in whole tissue homogenates, which precluded us from identifying between-species or between-strain differences that might be present within a specific cellular compartment. For example, aspects of mitochondrial function have been strongly implicated in aging and longevity (Barja and Herrero 2000; Boffoli et al. 1994; Navarro and Boveris 2007). All of the protein activities measured in this study have mitochondrial counterparts. Specific thioredoxin, TrxR and Grx isoforms localize to mitochondria in mammalian, and presumably avian, cells (see Page et al. 2010b). Similarly, mitochondria contain a Lon protease with similar stress-inducible protein degradation functions to the 20S/26S proteasome (Ngo and Davies 2009). These specific mitochondrial activities may be upregulated concomitantly with increased longevity, a possibility that should be explored in future studies.
In conclusion, we show that neither protein repair nor proteasomal degradation is positively correlated with longevity in either a multi-species or an intra-species comparison. These results support the idea that enhanced cellular stress resistance associated with longevity does not arise from increased ability to repair or remove stress-induced protein damage.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Statistical analysis of enzyme activities as correlates of mass-specific metabolic rate. (PDF 11 kb)
Phylogeny used for FIC analyses (PPT 61 kb)
Acknowledgements
KDS was supported by a Faculty of Mathematics and Science Dean’s Graduate Scholarship. MMP was supported by an Ontario Graduate Scholarship. This research was supported by infrastructure grants to PAF and JAS from the Canada Foundation for Innovation (CFI) and the Ontario Innovation Trust (OIT) and by Discovery Grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada (JAS, PAF, GB). JAS is also supported by an Early Researcher Award from the Ontario Ministry of Research and Innovation (OMRI).
References
- Arner ESJ. Focus on mammalian thioredoxin reductases—important selenoproteins with versatile functions. Biochim Biophys Acta. 2009;1790:495–526. doi: 10.1016/j.bbagen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Arner ES, Zhong L, Holmgren A. Preparation and assay of mammalian thioredoxin and thioredoxin reductase. Meth Enzymol. 1999;300:226–239. doi: 10.1016/S0076-6879(99)00129-9. [DOI] [PubMed] [Google Scholar]
- Bader M, Steller H. Regulation of cell death by the ubiquitin–proteasome system. Curr Opin Cell Biol. 2009;21:878–884. doi: 10.1016/j.ceb.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barja G, Herrero A. Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB J. 2000;14:312–318. doi: 10.1096/fasebj.14.2.312. [DOI] [PubMed] [Google Scholar]
- Boffoli D, Scacco SC, Vergari R, Solarino G, Santacroce G, Papa S. Decline with age of the respiratory chain activity in human skeletal muscle. Biochim Biophys Acta. 1994;1226:73–82. doi: 10.1016/0925-4439(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Bokov A, Lindsey M, Khodr C, Sabia M, Richardson A. Long-lived Ames dwarf mice are resistant to chemical stressors. J Gerontol A Biol Sci Med Sci. 2009;64A:819–827. doi: 10.1093/gerona/glp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daulney A, Tansey WP. Damage control: DNA repair, transcription, and the ubiquitin–proteasome system. DNA Repair. 2009;8:444–448. doi: 10.1016/j.dnarep.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Davies KJ. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/S0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- Magalhães JP, Costa J, Toussaint O. HAGR: the human ageing genomic resources. Nucleic Acids Res. 2005;33:D537–D543. doi: 10.1093/nar/gki017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divald A, Powell SR. Proteasome mediates removal of proteins oxidized during myocardial ischemia. Free Radic Biol Med. 2006;40:156–164. doi: 10.1016/j.freeradbiomed.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. doi: 10.1086/284325. [DOI] [Google Scholar]
- Ghazi A, Henis-Korenblit S, Kenyon C. Regulation of Caenorhabditis elegans lifespan by a proteasomal E3 ligase complex. PNAS. 2007;104:5947–5952. doi: 10.1073/pnas.0700638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Bozzella MJ, Seluanov A. Rodents for comparative aging studies: from mice to beavers. Age. 2008;30:111–119. doi: 10.1007/s11357-008-9053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune T, Blasig IE, Sitte N, Roloff B, Haseloff R, Davies KJ. Peroxynitrite increases the degradation of aconitase and other cellular proteins by proteasome. J Biol Chem. 1998;273:10857–10862. doi: 10.1074/jbc.273.18.10857. [DOI] [PubMed] [Google Scholar]
- Grune T, Jung T, Merker K, Davies KJ. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int J Biochem Cell Biol. 2004;36:2519–2530. doi: 10.1016/j.biocel.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Hackett SJ, Kimball RT, Reddy S, Bowie RCK, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han K-L, Harshman J, Huddleston CJ, Marks BD, Miglia KJ, Moore WS, Sheldon FH, Steadman DW, Witt CC, Yuri T. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish J, Christmas R, Buhler J, Eng J, Bumgarner R, Goodlett D, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- Jung T, Catalgol B, Grune T. The proteasomal system. Mol Aspects Med. 2009;30:191–296. doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Boulton ME, Kirkwood TBL. Positive correlation between mammalian life span and cellular resistance to stress. Free Radic Biol Med. 1999;26:495–500. doi: 10.1016/S0891-5849(98)00323-2. [DOI] [PubMed] [Google Scholar]
- Kirkwood T, Kapahi P, Shanley D. Evolution, stress and longevity. J Anat. 2000;197:587–590. doi: 10.1046/j.1469-7580.2000.19740587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Holmgren A. Induction of thioredoxin, thioredoxin reductase and glutaredoxin activity in mouse skin by TPA, a calcium ionophore and other tumor promoters. Carcinogenesis. 1999;22:1761–1767. doi: 10.1093/carcin/20.9.1761. [DOI] [PubMed] [Google Scholar]
- Naour F, Hohenkirk L, Grolleau A, Misek D, Lescure P, Geiger J, Hanash S, Beretta L. Profiling changes in gene expression during differentiation and maturation of monocyte-derived dendritic cells using both oligonucleotide microarrays and proteomics. J Biol Chem. 2001;276:17920–17931. doi: 10.1074/jbc.M100156200. [DOI] [PubMed] [Google Scholar]
- Lofgren S, Fernando MR, Xing KY, Wang Y, Kuszynski CA, Ho YS, Lou MF. Effect of thioltransferase (glutaredoxin) deletion on cellular sensitivity to oxidative stress and cell proliferation in lens epithelial cells of thioltransferase knockout mice. IOVS. 2008;49:4497–4505. doi: 10.1167/iovs.07-1404. [DOI] [PubMed] [Google Scholar]
- Martinez A, Portero-Otin M, Pamplona R, Ferrer I. Protein targets of oxidative damage in human neurodegenerative diseases with abnormal protein aggregates. Brain Pathol. 2008;20:281–297. doi: 10.1111/j.1750-3639.2009.00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard S, Miller R. Fibroblasts from long-lived Snell dwarf mice are resistant to oxygen-induced in vitro growth arrest. Aging Cell. 2006;5:89–96. doi: 10.1111/j.1474-9726.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- Miller RA, Williams J, Kiklevich JV, Austad S, Harper JM (2010) Comparative cellular biogerontology: primer and prospectus. Ageing Res Rev (in press). doi:10.1016/j.arr.2010.01.002 [DOI] [PMC free article] [PubMed]
- Miranda-Vizuete A, Gonzalez J, Gahmon G, Burghoorn J, Navas P, Swoboda P. Lifespan decrease in Caenorhabditis elegans mutant lacking TRX-1, a thioredoxin expressed in ASJ sensory neurons. FEBS Lett. 2005;580:484–490. doi: 10.1016/j.febslet.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Moosmann B, Behl C. Mitochondrially encoded cysteine predicts animal lifespan. Aging Cell. 2008;7:32–46. doi: 10.1111/j.1474-9726.2007.00349.x. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Boveris A. The mitochondrial energy transduction system and the aging process. Am J Physiol Cell Physiol. 2007;292:C670–C686. doi: 10.1152/ajpcell.00213.2006. [DOI] [PubMed] [Google Scholar]
- Ngo J, Davies K. Mitochondrial Lon protease is a human stress protein. Free Radic Biol Med. 2009;46:1042–1048. doi: 10.1016/j.freeradbiomed.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogburn C, Carlberg K, Ottinger MA, Holmes D, Martin GM, Austad SN. Exceptional cellular resistance to oxidative damage in long-lived birds requires active gene expression. J Gerontol (Bio Sci) 2001;56A:B468–B474. doi: 10.1093/gerona/56.11.b468. [DOI] [PubMed] [Google Scholar]
- Page MM, Peters CW, Staples JF, Stuart JA (2009) Intracellular superoxide dismutases are not altered during hibernation in the 13-lined ground squirrel Spermophilus tridecemlineatus. Comp Biochem Physiol A Mol Integr Physiol 152:115–122 [DOI] [PubMed]
- Page M, Richardson J, Wiens B, Tiedtke E, Peters C, Faure P, Burness G, Stuart JA. Antioxidant enzyme activities are not broadly correlated with longevity in 14 vertebrate endotherm species. Age (Dordr) 2010;32(2):255–270. doi: 10.1007/s11357-010-9131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M, Robb E, Salway K, Stuart J. Mitochondrial redox metabolism: aging, longevity and dietary effects. Mech Ageing Dev. 2010;131:242–252. doi: 10.1016/j.mad.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Perez V, Buffenstein R, Masamsetti V, Leonard S, Salmon AB, Mele J, Andziak B, Yang T, Edrey Y, Friguet B, Ward W, Richardson A, Chaudhuri A. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, naked mole-rat. Proc Natl Acad Sci U S A. 2009;106:3059–3064. doi: 10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez V, Bokov A, Remmen H, Mele J, Ran Q, Ikeno Y, Richardson A. Is the oxidative stress theory of aging dead? Biochim Biophys Acta. 2009;1790:1005–1014. doi: 10.1016/j.bbagen.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad AB, Allard MW, Green ED, NISC Comparative Sequencing Program Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol. 2008;25:1795–1808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price T. Correlated evolution and independent contrasts. Philos Trans R Soc Lond B Bio Sci. 1997;352:519–529. doi: 10.1098/rstb.1997.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJ, Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem J. 1998;335:637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb E, Page M, Stuart J. Mitochondria, cellular stress resistance, somatic cell depletion and lifespan. Curr Ageing Sci. 2009;16:12–27. doi: 10.2174/1874609810902010012. [DOI] [PubMed] [Google Scholar]
- Rodgers KJ, Dean RT. Assessment of proteasome activity in cell lysates and tissue homogenates using peptide substrates. Int J Biochem Cell Biol. 2003;35:716–727. doi: 10.1016/S1357-2725(02)00391-6. [DOI] [PubMed] [Google Scholar]
- Salmon AB, Murakami S, Bartke A, Kopchick J, Yasumura K, Miller RA. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Physiol Endocrinol Metab. 2005;289:E23–E29. doi: 10.1152/ajpendo.00575.2004. [DOI] [PubMed] [Google Scholar]
- Salmon AB, Leonard S, Masamsetti V, Pierce A, Podlutsky A, Podlutskaya N, Richardson A, Austad S, Chaudhuri A. The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein oxidation. FASEB J. 2009;23:2317–2326. doi: 10.1096/fj.08-122523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR. Correlations between physiology and lifespan—two widely ignored problems with comparative studies. Aging Cell. 2005;4:167–175. doi: 10.1111/j.1474-9726.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- Springer MS, Murphy WJ. Mammalian evolution and biomedicine: new views from phylogeny. Biol Rev. 2007;82:375–392. doi: 10.1111/j.1469-185X.2007.00016.x. [DOI] [PubMed] [Google Scholar]
- Stuart GW, Moffett K, Leader JJ. A comprehensive vertebrate phylogeny using vector representations of protein sequences from whole genomes. Mol Biol Evol. 2002;19:554–562. doi: 10.1093/oxfordjournals.molbev.a004111. [DOI] [PubMed] [Google Scholar]
- Swindell W, Masternak M, Kopchick J, Conover C, Bartke A, Miller R. Endocrine regulation of heat shock protein mRNA levels in long-lived dwarf mice. Mech Ageing Dev. 2009;130:393–400. doi: 10.1016/j.mad.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres CA, Perez VI. Proteasome modulates mitochondrial function during cellular senescence. Free Radic Biol Med. 2008;44:403–414. doi: 10.1016/j.freeradbiomed.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Statistical analysis of enzyme activities as correlates of mass-specific metabolic rate. (PDF 11 kb)
Phylogeny used for FIC analyses (PPT 61 kb)