Abstract
Aging is accompanied by expression changes in multiple genes, and the brain is one of the tissues most vulnerable to aging. Since the α7 nicotinic acetylcholine receptor (nAChR) subunit has been associated with neurodevelopmental disorders and cognitive decline during aging, we hypothesized that its absence might affect gene expression profiles in aged brains. To study whether transcriptional changes occur due to aging, α7 deficiency, or both, we analyzed whole-brain transcriptomes of young (8 weeks) and aged (2 years) α7-deficient and wild-type control mice, using Mouse Genome 430 2.0 microarray. Highly significant expression changes were detected in 47 and 1,543 genes [after Bonferroni and false discovery rate (FDR) correction] in the brains of aged mice compared to young mice, regardless of their genotype. These included genes involved in immune system function and ribosome structure, as well as genes that were previously demonstrated as differentially expressed in aging human brains. Genotype-dependent changes were detected in only three genes, Chrna7 which encodes the α7 nAChR subunit, and two closely linked genes, likely due to a “mouse background effect.” Expression changes dependent on age–genotype interaction were detected in 207 genes (with a low significance threshold). Age-dependent differential expression levels were approved in all nine genes that were chosen for validation by real-time RT-PCR. Our results suggest that the robust effect of aging on brain transcription clearly overcomes the almost negligible effect of α7 nAChR subunit deletion and that germ line deficiency of this subunit has a minor effect on brain expression profile in aged mice.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-010-9155-7) contains supplementary material, which is available to authorized users.
Keywords: Aging, Nicotinic acetylcholine receptor, Microarray, Gene expression, Immune response, Ribosome
Introduction
Aging is associated with structural disorganization, functional decline, decreasing adaptability, and increasing probability of disease and death. The brain is significantly affected by aging as it undergoes widespread changes including in size, vasculature, and cognition. Understanding the molecular mechanisms that characterize the aging-related changes in normal and diseased brains is a constant challenge. Factors that may influence the complex process of brain aging include decline in neurotransmitters (mainly dopamine and serotonin), calcium dysregulation, mitochondrial dysfunction, and production of reactive oxygen species (Burke and Barnes 2006; Mattson et al. 2002; Toescu and Verkhratsky 2007).
Impairment of cholinergic markers is well characterized in normal aging as well as in patients with dementia. These changes include reduction of nicotine and nicotinic agonist binding sites and changes in brain expression of nicotinic acetylcholine receptor (nAChR) subunits (Picciotto and Zoli 2002; Rogers et al. 1998; Sabbagh et al. 2006). The involvement of nAChRs in aging is also supported by multiple epidemiological studies showing the protective effect of smoking against the development of aging-associated diseases such as Alzheimer's and Parkinson's diseases (AD and PD, respectively; Elbaz and Moisan 2008; Fratiglioni and Wang 2000). Substantial evidence further suggests that enhancement of cognitive function in humans suffering from dementia, and in animal models of AD and PD, can be achieved by nicotine and nicotinic agonists. Specifically, agonists of the α7 nAChR have been developed for treatment of cognitive deficits (Briggs et al. 2009; Buccafusco et al. 2005; Sarter et al. 2009; Tietje et al. 2008; Timmermann et al. 2007). Interestingly, progressive degeneration of hippocampal neurons and cognitive deficits were demonstrated in aged mice with deficiency of the nAChR subunit β2 (Zoli et al. 1999).
The nAChRs are homopentameric or heteropentameric membrane proteins that belong to a large family of ligand-gated ion channels and mediate the effects of acetylcholine. Seventeen nAChR subunit encoding genes have been identified. Of them, nine α and three β subunits are expressed in the brain (Taly et al. 2009). The nAChRs have been implicated in complex diseases affecting the nervous system, including epilepsy, schizophrenia, developmental disorders, and aging-associated neurodegenerative diseases such as AD and PD (Freedman et al. 2001; Steinlein et al. 1995). Several recent genome-wide association studies identified genomic alterations in the human α7 nAChR subunit gene locus in patients with schizophrenia, bipolar disorder, autism, developmental delay, and seizures (Ben-Shachar et al. 2009; Consortium 2008; Helbig et al. 2009; Miller et al. 2009; Sharp et al. 2008; Shinawi et al. 2009; Stefansson et al. 2008). α7 subunits can form homopentameric receptor channels, which are activated by acetylcholine (ACh) and blocked by α-bungarotoxin (αBgtx). nAChRs that contain the α7 subunit account for almost all high-affinity αBgtx binding sites in murine brains (Orr-Urtreger et al. 1997). These receptors are highly permeable to Ca2+ and show a rapid desensitization rate. They have been detected in many brain regions, with very high expression in the hippocampus (Gotti et al. 2009).
Aging is not dependent on a single gene. Complex interactions between multiple genes influence age-related changes. Herein, we applied high-throughput microarray technology in order to detect the transcriptional changes that occur in the aging brain and to explore the possibility that an inherited α7 nAChR subunit deficiency affects the brain transcriptome during young adulthood and/or aging.
Methods
Animals
All mice used were from congenic lines that were backcrossed onto a C57BL/6J background for seven generations after germ line transmission. The mice used for expression analyses were deficient for the α7 nAChR subunit (α7−/− mice; Orr-Urtreger et al. 1997) and wild-type (WT) littermate control mice. Each experimental mouse was genotyped twice, once before and once after the experiment. The polymerase chain reaction (PCR) conditions and primer pairs used for genotyping the α7−/− mice were as previously described (Sack et al. 2005).
The study included two age groups: young (6–9 week old) and aged (23–25 month old) mice. Prior to the experiments, the mice were housed in groups of two to five per cage under a 12/12 h light/dark cycle, with food and water ad libitum. All procedures were approved by the institutional animal and care committee, in accordance with the “NIH Guide for the Care and Use of Laboratory Animals.”
Profiling brain expression
Whole-brain (including the cerebrum, olfactory bulbs, thalamus, cerebellum, brain stem, and the proximal tip of the spinal cord) expression profiles were determined in four experimental groups of mice: young WT, young α7−/−, aged WT, and aged α7−/− mice. Each experimental group included five mice. The young mice group included five males and five females (three males in young WT and two in young α7−/−), and the aged mice group included six males and four females (two males in aged WT and four in aged α7−/−). The global expression profiles were analyzed using the GeneChip® Mouse Genome 430 2.0 Array (Affymetrix, Inc., Santa Clara, CA), which includes over 39,000 transcripts (probe sets). Total RNA isolation, target preprocessing, and hybridization to the Mouse Genome 430 2.0 microarray (Affymetrix, Inc.) were conducted according to the manufacturer's instructions and as previously described (Kedmi and Orr-Urtreger 2006). Microarray experiments were designed to comply with minimum information about a microarray experiment (MIAME) guidelines (Brazma et al. 2001). Raw data are available in Supplementary Table 6 and were deposited to the National Center for Biotechnology Information Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO series accession number GSE20411.
Data analysis
The statistical algorithm, implanted in Affymetrix Suite Version 5.0 software (MAS5, Affymetrix, Inc.), generated a signal value (which designates a relative measure of the abundance of the transcript), a detection P value (which indicates the reliability of the transcript's detection call) and a detection call (present, absent, or marginal) for each probe set on the microarray. The detection calls were calculated based on the detection P value, as follows: probe sets with a P value of more than 0.06 were designated as absent, P value of more than 0.04 and less than 0.06 as marginal, and P value of less than 0.04 as present. For interarray comparisons, data from each array were scaled using MAS5 software, and the mean intensity for each array was adjusted by a scaling factor to a set target intensity of 150.
The signal values were normalized both per probe set and per the entire microarray by dividing each signal by the median of the probe set and by the median of the microarray. The normalized data were then subjected to filtering, leaving probe sets that were present in at least ten out of the 20 tested arrays. Following filtration, statistical analyses were applied on the data from all 20 microarrays to find genes that significantly distinguish between the different experimental groups' brain expressions. Two-way analysis of variance (ANOVA) with age and genotype parameters identified differentially expressed genes (Fig. 1): (1) Genes differentially expressed between old and young mice were detected by either Bonferroni or FDR corrections for multiple comparisons and a P value cutoff of 0.05. (2) Genes differentially expressed between WT and α7−/− mice were Bonferroni- and FDR-corrected for multiple comparisons with a P value cutoff of 0.05. (3) Genes with interaction between the age parameter and the genotype parameter were detected by applying a P value cutoff of 0.01 (not corrected for multiple comparisons). The normalization, filtration, and statistical analyses procedures were conducted using GeneSpring version 7.3 software (Silicon Genetics, Redwood City, CA).
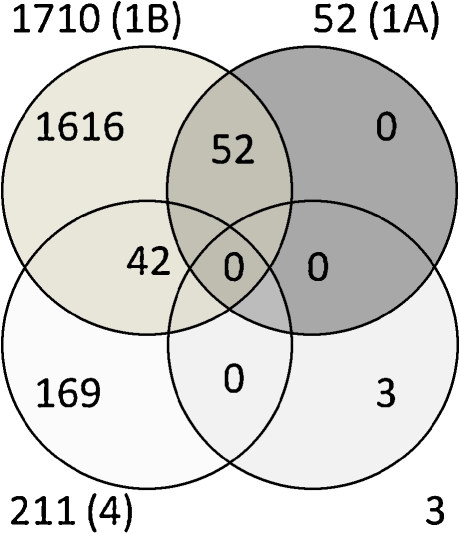
Fig. 1.
Venn diagram of differentially expressed probe sets according to parameters and statistical thresholds of two-way ANOVA analysis. The circles represent the differentially expressed probe sets in each of the following analyses: In dark (upper right), the age-dependent probe sets that passed Bonferroni correction (P < 0.05); in gray (upper left), the age-dependent probe sets that passed FDR correction (P < 0.05); in light gray (lower right), the genotype-dependent probe sets that passed Bonferroni and/or FDR corrections (P < 0.05); and in white (lower left), probe sets resulted from the interaction between age and genotype (P < 0.01 without a correction). Numbers outside the circles represent the total number of probe sets, while numbers in the circles are the unique or shared number of the differentially expressed probe sets. In brackets are the numbers of the specific Supplementary Tables that listed the relevant probe sets and genes
Gene ontology and pathway annotations analyses
The genes that significantly differentiated between the experimental groups were categorized into gene ontologies (GO) according to their molecular functions, associated biological processes, and/or cellular components. Additionally, genes that are included in KEGG pathways were annotated. This was done using DAVID functional annotation online tools (http://david.abcc.ncifcrf.gov/, Dennis et al. 2003; Huang da et al. 2009). EASE score, implemented in DAVID, with a P value threshold of 0.05 was applied in order to detect the significantly overrepresented GO or KEGG pathway annotations among the differentially expressed genes, as compared to all the genes represented on the Mouse Genome 430 2.0 microarray.
Previously described genes with significant expression changes in aging brains
We generated two lists of genes that were previously detected as changed in aging brains in eight mouse or rat expression microarray experiments (Blalock et al. 2003; Burger et al. 2008; Cho et al. 2002; Jiang et al. 2001; Kadish et al. 2009; Prolla 2002; Rowe et al. 2007; Sharman et al. 2007) and in four human experiments (Berchtold et al. 2008; Erraji-Benchekroun et al. 2005; Lu et al. 2004; Tang et al. 2009). These studies profiled different gender compositions and samples from various ages. For example, Berchtold et al. analyzed human brain regions from males and females separately, and Prolla compared 5-month young adults to 30-month aged mice. Additionally, these studies were conducted using different platforms for microarray analysis, as well as different types of arrays. Gene symbols and GenBank ID annotations were updated using NetAffx (http://www.affymetrix.com/analysis/index.affx) or DAVID tools (http://david.abcc.ncifcrf.gov/). The comparisons between these lists and our results were done based on the official gene symbols.
Validation/confirmation by quantitative real-time RT-PCR assay
Quantitative real-time RT-PCR assays were performed to determine the expression levels in whole-brain RNA. The expression analyses of Abca8a (Mm00462472_mH), Avp (Mm00437761_g1), C1qb (Mm00437836_m1), C1qc (Mm00776126_m1), Gstm1 (Mm00833915_g1), Itih3 (Mm00434560_g1), Klf4 (Mm00516105_g1), Pvrl3 (Mm01343006_m1), and Uba6 (Mm00554778_m1) genes were done using TaqMan Universal PCR Master Mix and MGB probes with StepOne Real-Time PCR system (Applied Biosystems, Foster City, CA). For these quantitative RT-PCR analyses, we enlarged the number of samples in each experimental group to ten, including at least five new biological replicates. cDNA was synthesized from 1 μg of brain-derived total RNA using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) in the presence of RNase inhibitor. Quantitative RT-PCR reactions were performed for 2 min at 50°C, 10 min at 95°C, and then 45 cycles of 15 s at 95°C and 1 min at 60°C. The expression levels were determined using the comparative threshold cycle (CT) quantification method (Livak and Schmittgen 2001; Schmittgen and Livak 2008). The CT values of the internal control gene, Sdha (Mm01352360_m1), were subtracted from CT values of each target genes (ΔCT). Then, 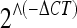 values were compared between the groups and two-way ANOVA followed by one-way ANOVA statistics were applied on the data.
values were compared between the groups and two-way ANOVA followed by one-way ANOVA statistics were applied on the data.
Results
The global expression profiles of whole brains from ten young (8 week old) mice and ten aged (2 year old) mice, with or without deficiency in the α7 nAChR subunit, were compared. After the filtration process described in the “Methods” section, a list of 21,302 probe sets was generated. Further statistical analysis was performed on this list of probe sets to detect genes whose expression levels significantly changed depending on the parameters of age or genotype or the interaction between them (Fig. 1).
Brains of aged vs. young mice
Identification of genes whose expression levels significantly discriminate between brains of aged and young mice
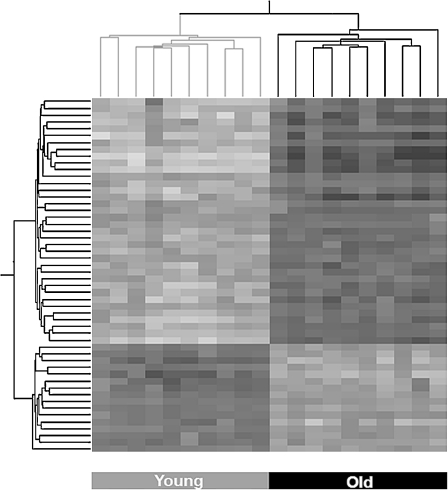
Two-way ANOVA with Bonferroni correction for multiple comparisons on the age parameter yielded a list of 52 differentially expressed probe sets (P < 0.05), while FDR correction resulted in the identification of 1,710 probe sets (P < 0.05), representing 47 and 1,543 genes, respectively. As expected, all Bonferroni-corrected probe sets and genes are included in the FDR-corrected list (Fig. 1 and Supplementary Table 1a and b). Supplementary Table 1a and b detail these probe sets with their expression levels and functional annotations. Among the 52 highly significant probe sets, 16 were downregulated and 36 were upregulated in brains of aged mice. Among the 1,710 probe sets, 767 were downregulated and 958 were upregulated in brains of aged mice. The most downregulated genes in the aged brains were the Col3a1 gene [fold change (FC) = 0.39] among the 52 significantly changed probe sets and 9630013A20Rik (FC = 0.14) among the 1,710 significantly changed probe sets. In both lists, the most upregulated gene in the aged brains was Lyz1 (FC = 7.81). Hierarchical clustering of the 52 differentially expressed probe sets demonstrated that all samples were clearly clustered into two distinct groups according to age (Fig. 2).
Fig. 2.
Hierarchical clustering of the 52 probe sets that significantly distinguished between brains of aged and young mice. The expression signals of the 52 Bonferroni-corrected probe sets, representing 47 genes, were subjected to hierarchical clustering. This clustering clearly divided the samples into the two age groups and the genes into upregulated and downregulated. Lower expression levels are represented in white and high expression in black
Genes involved in immune system function and ribosome structure distinguish between brains of aged and young mice
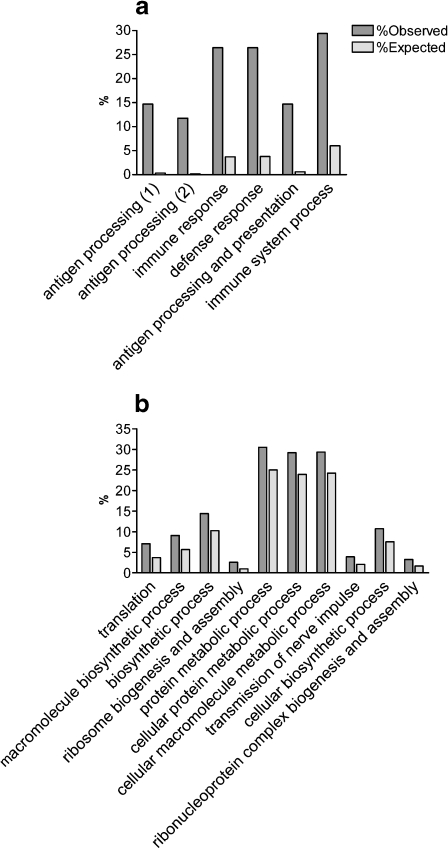
Significant enriched GO annotations were identified among the 47 and 1,543 significantly changed genes (52 and 1,710 probe sets, Fig. 3a, b, Supplementary Table 2a and b, respectively). The most significantly overrepresented GO annotations among the Bonferroni-corrected list were the “antigen processing and presentation of peptide antigen” (five out of 47 genes, P = 2.64 × 10−6, and 9/1,543 genes among the FDR-corrected list, P = 0.0055) and the “major histocompatibility complex (MHC) protein complex” (5/47, P = 4.04 × 10−6 and 11/1,543 P = 0.0026).
Fig. 3.
GO annotation analysis detected significantly enriched biological process among the age-dependent differentially expressed genes. The most significant (P < 0.001) biological processes among the 47 Bonferroni-corrected (a) and 1,543 FDR-corrected (b) differentially expressed genes are presented here. Full descriptions of all significantly overrepresented annotations in all GO categories (biological process, molecular function, and cellular component) are listed in Supplementary Table 2a and b. Dark gray bars represent the observed percentages of genes among the age-dependent differentially expressed genes. Light gray bars depict the expected percentages of genes as represented on the entire array. The annotations on the X-axis are in ascending order of P values. In a, the annotations antigen processing (1) and (2) stand for “antigen processing and presentation of peptide antigen” and “antigen processing and presentation of peptide antigen via MHC class I,” respectively
Importantly, genes belonging to “ribosome” annotations were the most significantly enriched among the 1,543 FDR-corrected differentially expressed genes, including the molecular function “structural constituent of ribosome” (54 genes, P = 6.38 × 10−18) and the cellular component “ribosome” (55 genes, P = 3.12 × 10−17). Two additional highly significant overrepresented groups of genes were the cadherins (27 genes, P = 1.79 × 10−6; InterPro database categories) and lysosomal genes (31 genes, P = 3.61 × 10−4). Interestingly, several neuronal-related annotations were also overrepresented among the 1,543 genes, including transmission of nerve impulse (41 genes, P = 1.20 × 10−4), synaptic transmission (32 genes, P = 0.003), neuron recognition (six genes, P = 0.0045), nervous system development (76 genes, P = 0.006), neurite development (30 genes, P = 0.007), regulation of synapse structure and activity (eight genes, P = 0.007), and neuron differentiation (39 genes, P = 0.0097).
Significantly overrepresented KEGG pathways (Table 1) were also identified among the Bonferroni- and FDR-corrected differentially expressed genes. The most significant were pathways related to the immune system and ribosome function. Of interest, Alzheimer's disease (ten genes, P = 4.92 × 10−4) and Parkinson's disease (seven genes, P = 0.013) pathways were both overrepresented among the 1,543 differentially expressed genes.
Table 1.
Significantly enriched KEGG pathways among the age-dependent differentially expressed genes
| KEGG pathway | No. | P value | Fold enrichment |
|---|---|---|---|
| Among the 47 Bonferroni-corrected genes | |||
| Antigen processing and presentation | 5 | 1.06 × 10−04 | 17.27 |
| Natural killer cell mediated cytotoxicity | 5 | 6.14 × 10−04 | 10.96 |
| Cell adhesion molecules (CAMs) | 5 | 8.72 × 10−04 | 9.99 |
| Type I diabetes mellitus | 4 | 8.79 × 10−04 | 18.64 |
| Among the 1,543 FDR-corrected genes | |||
| Ribosome | 47 | 4.94 × 10−26 | 5.85 |
| Porphyrin and chlorophyll metabolism | 12 | 2.79 × 10−04 | 3.57 |
| Alzheimer's disease | 10 | 4.92 × 10−04 | 3.97 |
| Pentose and glucuronate interconversions | 8 | 0.002855 | 3.89 |
| Glycosphingolipid biosynthesis—lactoseries | 5 | 0.006394 | 5.95 |
| Neurodegenerative Diseases | 10 | 0.009659 | 2.68 |
| Parkinson's disease | 7 | 0.012904 | 3.41 |
| Long-term potentiation | 13 | 0.026452 | 1.99 |
| Axon guidance | 20 | 0.040278 | 1.60 |
| Antigen processing and presentation | 14 | 0.049101 | 1.76 |
P value calculated using EASE score with DAVID online tools. Fold enrichment is the ratio between observed and expected number of genes in each pathway.
No. number of differentially expressed genes that belong to this pathway
Comparison to multiple age-related murine and human microarray databases
We compiled a list of genes that were previously reported as changed in brains of aged mice or rats (Blalock et al. 2003; Burger et al. 2008; Cho et al. 2002; Jiang et al. 2001; Kadish et al. 2009; Prolla 2002; Rowe et al. 2007; Sharman et al. 2007; Supplementary Table 3a). Of these, 2,032 genes are represented on the Mouse Genome 430 2.0 array (by 3,942 probe sets). Of the 47 highly significant differentially expressed genes defined herein, 14 were previously reported (Supplementary Table 3b). In addition, 235 of the previously reported genes (Supplementary Table 3c) are included in our list of 1,543 differentially expressed genes. Of note, among our lists of Bonferroni- and FDR-corrected genes (Supplementary Table 1a and b), the proportion of genes that were previously reported as changed in aged murine brains was significantly higher than expected by chance (P < 0.0001, χ2 test with Yates correction, χ2 = 19.97 for the 47 changed genes and χ2 = 51.83 for the 1,543 changed genes). Of note, some of the previously published studies analyzed distinct brain regions, compared to our study that used whole-brain tissues. For example, Prolla profiled mouse's cerebellum and cortex. While genes such as Avp, Bdnf, and Ifit1 that were differentially expressed in our study changed only in one of these regions, other genes, such as B2m, C4b, and Gfap changed in both regions (Prolla 2002; Supplementary Table 3b and c).
We further generated a list of 8,564 genes whose expression was changed in human aged brains (Berchtold et al. 2008; Erraji-Benchekroun et al. 2005; Lu et al. 2004; Tang et al. 2009; Supplementary Table 3d). This list included 5,411 genes that are represented on Mouse Genome 430 2.0 array. Of these, 14 and 563 differentially expressed genes (Supplementary Table 3e and f) were included in our lists of Bonferroni-corrected (47) and FDR-corrected (1,543) genes. The human brain area in which these genes changed included the superior-frontal gyrus, postcentral gyrus, entorhinal cortex and hippocampus (Berchtold et al. 2008), prefrontal cortex (Erraji-Benchekroun et al. 2005; Tang et al. 2009), and frontal cortex (Lu et al. 2004).
The genes previously described as changed in old human brains were significantly overrepresented among our list of 1,543 genes (P < 0.0001, χ2-test with Yates correction, χ2 = 91.62). Of note, 127 genes (Supplementary Table 3g) were included in our current and previously published murine and human databases (Supplementary Tables 1b, 3a and d).
Functional in silico analysis was performed to detect overrepresented GO annotations and KEGG pathways. Among the 235 age-related murine brain genes (Supplementary Table 3c), the most significantly enriched GO annotations were the “cytoplasm” (132 genes, P = 8.61 × 10−11) and “lysosome” (17 genes, P = 2.69 × 10−8) cellular components. These two annotations were also overrepresented among the combined human and murine gene list (127 genes in Supplementary Table 3g, 74 genes, P = 6.44 × 10−7 and nine genes P = 1.78 × 10−4, respectively). Additionally, ribosomal genes, and specifically genes that are part of the ribosome KEGG pathway, were highly enriched (17 out of 235 genes, P = 1.46 × 10−10). Other enriched annotations that are relevant to the aging process were the “response to oxidative stress” (5/127 genes, P = 0.002) and “immunoglobulin-mediated immune response” (7/235 genes, P = 9.19 × 10−4). Interestingly, the most significant GO annotation among the human age-related genes (Supplementary Table 3f) was the “transmission of nerve impulse” (29/563 genes, P = 1.77× 10−7). The full list of significantly overrepresented annotations is available in Supplementary Table 3h.
Comparing the expression of brains from α7 nAChR null and control mice
Only three genes were detected as significantly changed when comparing brains of young and aged α7−/− to WT mice (two-way ANOVA with either Bonferroni or FDR corrections for multiple comparisons on the genotype parameter, P < 0.05): Chrna7 itself and two closely linked genes, Ipw and Gabrb3, that are located 3.32 and 5.39 Mbp upstream to Chrna7.
As we previously described (Kedmi and Orr-Urtreger 2006), the expression of genes that flank the “knocked-out gene” are likely affected by the background strain. Briefly, this phenomenon results from using two mouse strains to generate genetically manipulated mice: 129SvEv to introduce the mutation in embryonic stem cells and C57BL/6J for blastocysts. Therefore, even after backcrossing, the null mice with C57BL/6J for multiple generations, the expression of genes that are closely linked to the mutant Chrna7 allele potentially represent the pattern of expression of the 129SvEv-driven alleles and not the wild-type control C57BL/6J allele. Thus, the very close locations of Ipw and Gabrb3 to Chrna7 suggest that their altered expression resulted from the “background flanking genes effect.”
Aging–genotype interaction affected the expression of genes involved in neuronal development, cellular organization, and MAPK inactivation
Interaction analysis to detect genes whose expression was affected by both aging and genotype parameters was performed (two-way ANOVA). None of the genes passed statistical correction for multiple comparisons; however, 211 probe sets with P < 0.001 were detected (Supplementary Table 4), representing 207 genes. Forty-two probe sets were also included in the list of 1,710 FDR-corrected age-dependent differentially expressed probe sets (Fig. 1, Supplementary Table 1b).
Significantly enriched GO annotations were identified among the 207 changed genes (Supplementary Table 5). The two most significantly enriched GO annotations were the “cellular component organization and biogenesis” (44 genes, P = 8.16 × 10−4) and the “inactivation of MAPK activity” (three genes, P = 0.006) ontologies. Several enriched GO annotations involved in neuronal development were also identified including: neurite development (eight genes, P = 0.008), neurite morphogenesis (seven genes, P = 0.014), and neuron differentiation (nine genes, P = 0.017). Additionally, among these age–genotype interacting changed genes, a group of mitochondrial inner membrane genes was also overrepresented (eight genes, P = 0.015).
Validation/confirmation of expression by quantitative RT-PCR
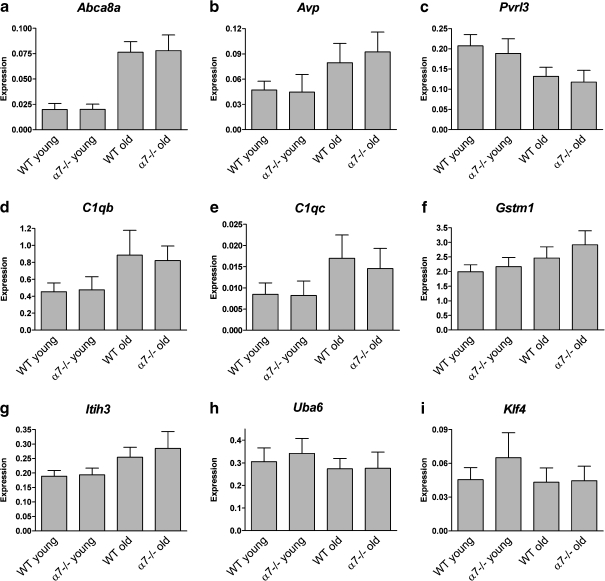
The expression levels of nine significantly changed genes were validated in a larger sample size (40 samples) by real-time RT-PCR (Fig. 4). The genes for validation were selected from the different lists described above (Supplementary Tables 1a, b and 4). The Abca8a, Avp, and Pvrl3 were among the 47 Bonferroni-corrected genes whose expression significantly changed between brains of aged and young mice (Fig. 4a–c). The C1qb, C1qc, and Gstm1 were included in the list of 1,543 FDR-corrected age-dependent genes (Fig. 4d–f). The Itih3 and Uba6 were included both in the list of 1,543 age-changed genes and in the list of 207 age–genotype interaction genes (Fig. 4g–h), and Klf4 (Fig. 4i) was one of the 207 genes.
Fig. 4.
a–i Validation/conformation of differentially expressed genes. Quantitative real-time RT-PCR analyses were performed to validate and replicate the expression changes of nine genes. Each gene's expression level was normalized to Sdha gene expression level and tested in the four experimental groups (young and old mice, wild-type, and α7 nAChR-subunit-deficient mice). Each group included ten samples. Bars represent mean ± SD of expression levels, measured as 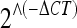 (see text for details). Expression levels in all genes tested were significantly changed in brains of aged compared to young mice (two-way ANOVA, *P < 0.0005 and #P < 0.05). In addition, genotype-dependent significant expression changes were detected for the Gstm1 and Klf4 genes (two-way ANOVA, P = 0.011 and P = 0.04, respectively)
(see text for details). Expression levels in all genes tested were significantly changed in brains of aged compared to young mice (two-way ANOVA, *P < 0.0005 and #P < 0.05). In addition, genotype-dependent significant expression changes were detected for the Gstm1 and Klf4 genes (two-way ANOVA, P = 0.011 and P = 0.04, respectively)
The age-dependent expression changes in all nine tested genes were validated (two-way ANOVA, age parameter, P < 0.0005 except for Uba6 and Klf4 P < 0.05). In contrast, the age–genotype interaction of Itih3, Uba6, and Klf4 did not reach significant level. Interestingly, genotype-related significant changes were seen in Gstm1 and Klf4 (two-way ANOVA, genotype parameter, P = 0.011 and P = 0.04, respectively).
Discussion
Our study aimed to explore the transcriptional changes occurring in the aged-mouse brain and the effect of α7 nAChR subunit deficiency on expression patterns. The transcriptomes compared here included brains from 2-month old mice and aged (2 year old) mice. This work presents the first expression brain profiling of young and old α7 knockout mice. Since the parameters of age (aged vs. young mice) and genotype (α7−/− vs. wild-type mice) had equal statistical power in our study, we can conclude that the effect of aging on brain transcription overcomes the effect of deleting the α7 nAChR subunit gene. The age-dependent expression changes were much more robust and profound than those dependent on the genotype. Moreover, since the interaction between age and genotype detected only a small number of differentially expressed genes, with low statistical significance, our data suggest that germ line α7 deficiency had a relatively minor effect on the differential transcriptome of the aging mouse brain.
α7-containing nAChRs have unique characteristics such as rapid desensitization rate and high permeability to Ca2+. This subunit has important roles in brain development and neuron differentiation and migration (Gotti et al. 2009; Taly et al. 2009), and its chromosomal locus was associated in humans with developmental and neurological disorders (Ben-Shachar et al. 2009; Consortium 2008; Helbig et al. 2009; Miller et al. 2009; Sharp et al. 2008; Shinawi et al. 2009; Stefansson et al. 2008). Despite the importance of the α7 nAChR subunit, its deficiency results in viable, anatomically normal α7-null mice (Orr-Urtreger et al. 1997; Paylor et al. 1998). The only abnormal CNS-related phenotypes reported in these mice were mild cognitive deficits (Young et al. 2004, 2007) and reduced nicotine withdrawal symptoms (Salas et al. 2007). It has been suggested that overexpression of other types of nAChRs compensates for the depletion of α7-nAChRs, and in fact, the upregulation of α3 and α4 nAChR subunits was demonstrated in brains of postnatal α7−/− mice (Yu et al. 2007). Although we did not detect any expression changes in other nAChR subunit genes in young and aged brains, we cannot rule out the possibility that other compensatory mechanisms may be responsible for the lack of α7-associated transcriptional changes in the knockout brains. Such mechanisms may involve bypass of the α7 subunit functions by proteins that interact with α7-containing receptors. Such interactions were described with proteins involved in cellular structural support, basic metabolism, scaffolding, trafficking, and targeting, as well as with kinases, chaperones, and other signal transduction proteins (Baer et al. 2007; Berg and Conroy 2002; Paulo et al. 2009; Shoop et al. 2000). It is therefore possible that regulation of these protein levels may explain the minor mRNA transcriptional changes in brains of α7-deficient mice. Finally, the limited effect of α7 deletion on brain expression in young and aged mice is in agreement with the minor phenotypic changes described in these mice; however, this is still somewhat surprising in light of the important roles attributed to α7-containing nAChRs.
The major functional group of genes whose expression changes were associated with aging was the immune response genes. The significantly enriched categories included complement component genes and MHC class I and class II genes, as well as genes involved in regulation of T and B cell proliferation and signaling. Upregulation of immune response genes was previously demonstrated in murine and human brains (Lee et al. 2000; Terao et al. 2002). Specifically, the expression of the complement component genes C1qa, C1qb, C1qc, and C4b was increased in aged murine's hippocampus, neocortex, cerebellum, and whole brain (Blalock et al. 2003; Burger et al. 2008; Prolla 2002; Rowe et al. 2007; Sharman et al. 2007), and C1qa and C1qb expression levels were also upregulated in normal-aged human brains (Berchtold et al. 2008). Interestingly, glial cells of AD patients secreted more C1q than nondemented elderly controls (Lue et al. 2001). Moreover, the levels of various complement components increase during the course of neurodegenerative disorders such as AD, PD, Huntington, and prion diseases (reviewed by Lucin and Wyss-Coray 2009). In other aging tissues, such as muscle, lung, thymocytes, and kidney, an increase in immune response genes was also demonstrated (Aoshiba and Nagai 2007; Lustig et al. 2009; Rodwell et al. 2004; Zahn et al. 2006). Recently, Swindell analyzed published expression microarray data derived from many different mouse tissues of aged mice and detected common patterns of age-related expression changes, such as upregulation of genes associated with immune response (Swindell 2009). Taken together, our results further suggest that the involvement of the immune system is a common aging-associated phenomenon, seen across different species and tissue types.
The age-dependent transcriptional changes detected here were also highly and significantly enriched in ribosomal genes, including genes encoding for large and small cytosolic ribosomal subunits, mitochondrial ribosomal proteins, and proteins involved in ribosome biogenesis. Of note, the expression levels of almost all of these ribosomal genes were upregulated in aged brains. Similar expression changes were also demonstrated in other studies that profiled the transcriptional changes in aging brain, muscle, thymocytes, and kidney in murine, human, or both (Blalock et al. 2003; Lustig et al. 2009; Oh et al. 2010; Rodwell et al. 2004; Zahn et al. 2006). In addition to the increased aging-related ribosomal gene expression described in multiple studies, there is a known decrease in protein synthesis during aging (reviewed by Tavernarakis 2008). It is therefore possible that upregulation of ribosome-associated gene expression during aging may be a compensatory mechanism. Of interest, an impairment of ribosomal function, including decreased ribosomal RNA and decreased protein synthesis, was observed in cortical areas of AD patients (Ding et al. 2005). Additionally, deletion of several ribosomal proteins in yeast and Caenorhabditis elegans led to increased lifespan (Hansen et al. 2007; Steffen et al. 2008; reviewed by Lempiainen and Shore 2009), and administration of rapamycin, which can inhibit mRNA translation, promoted longevity in mice (reviewed by Kennedy and Kaeberlein 2009). Our data demonstrate that numerous ribosome-associated genes are involved in the aging process. The detailed information presented here can help understand the spectrum of ribosomal proteins and ribosomal regulatory systems and their roles in aging.
Although the signature of expression changes in immune- and ribosomal-associated genes is shared by multiple organs during aging, transcriptional changes in genes and genetic pathways specific to the aging brain were also detected here. For example, the expression levels of genes that are involved in transmission of nerve impulse and in nervous system development and genes that belong to the AD and PD pathways were changed in the 2-year-old mouse brains (listed in Supplementary 2b). These include Apoe, App, Nr4a2, Bdnf, and Gstm1 and other previously published genes that were shown as differentially expressed in aging brains, as well as novel genes that were not previously associated with aging. Notably, our list of genes whose expression changed in brains of aged mice was significantly enriched for genes that were previously demonstrated as differentially expressed in the aging human brains (Berchtold et al. 2008; Erraji-Benchekroun et al. 2005; Lu et al. 2004; Tang et al. 2009). Our results suggest that mouse and human brains undergo similar molecular changes during aging and further emphasize the importance of murine models in the study of the aging processes of the brain.
In summary, in contrast to the robust effect of aging on gene expression in the brain, the effect of α7 nAChR subunit deletion on brain transcription was almost negligible. Data presented here regarding the age-dependent expression alterations of both common and unique, known, and novel genes and genetic pathways help to enhance our understanding of the complex molecular mechanisms of the aging brain.
Electronic supplementary material
Below is the link to the electronic supplementary material.
a The 52 probe sets whose expression significantly differentiated brains of aged mice from brains of young mice (two-way ANOVA, Bonferroni-corrected, P < 0.05). b The 1,710 probe sets whose expression significantly differentiated brains of aged mice from brains of young mice (two-way ANOVA, FDR-corrected, P < 0.05). (XLS 1099 kb)
a Overrepresented GO annotations among the 47 significantly changed age-dependent genes (Bonferroni-corrected 52 probe sets). b Overrepresented GO annotations among the 1,543 significantly changed age-dependent genes (FDR-corrected 1,710 probe sets). (XLS 254 kb)
a A compiled list of genes that were previously published in eight different microarray studies as changed in brains of aged mice or rats (Blalock et al. 2003; Burger et al. 2008; Cho et al. 2002; Jiang et al. 2001; Kadish et al. 2009; Prolla 2002; Rowe et al. 2007; Sharman et al. 2007). b Genes that appear in both Supplementary Tables 1a and 3a. c Genes that appear in both Supplementary Tables 1b and 3a. d A compiled list of genes that were previously published in four different microarray studies as changed in brains of aged humans (Berchtold et al. 2008; Erraji-Benchekroun et al. 2005; Lu et al. 2004; Tang et al. 2009). e Genes that appear in both Supplementary Tables 1a and 3d. f Genes that appear in both Supplementary Tables 1b and 3d. g Genes that appear in all three Supplementary Tables 1b, 3a and 3d. h Overrepresented GO annotations among the age-dependent differentially expressed genes that were previously described in human and murine (genes that are listed in Supplementary Tables 3c, f, and g). (XLS 3721 kb)
The 211 probe sets whose expression is dependent on the interaction between age and genotype parameters (two-way ANOVA, P < 0.001). (XLS 162 kb)
Overrepresented GO annotations among the 207 age–genotype interaction genes (211 probe sets in Supplementary Table 4). (XLS 29 kb)
Pivot table of the raw expression signals data. (XLS 10180 kb)
Acknowledgments
This work was supported by Kahn Foundation. We would like to thank Alona Gochberg-Sarver for technical assistance and Dr Uri Rozovski for statistical advice at the beginning of the study.
References
- Aoshiba K, Nagai A. Chronic lung inflammation in aging mice. FEBS Lett. 2007;581:3512–3516. doi: 10.1016/j.febslet.2007.06.075. [DOI] [PubMed] [Google Scholar]
- Baer K, Burli T, Huh KH, Wiesner A, Erb-Vogtli S, Gockeritz-Dujmovic D, Moransard M, Nishimune A, Rees MI, Henley JM, et al. PICK1 interacts with alpha7 neuronal nicotinic acetylcholine receptors and controls their clustering. Mol Cell Neurosci. 2007;35:339–355. doi: 10.1016/j.mcn.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar S, Lanpher B, German JR, Qasaymeh M, Potocki L, Nagamani SC, Franco LM, Malphrus A, Bottenfield GW, Spence JE, et al. Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet. 2009;46:382–388. doi: 10.1136/jmg.2008.064378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, Beach T, Miller C, Troncoso J, Trojanowski JQ, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg DK, Conroy WG. Nicotinic alpha 7 receptors: synaptic options and downstream signaling in neurons. J Neurobiol. 2002;53:512–523. doi: 10.1002/neu.10116. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Chen KC, Sharrow K, Herman JP, Porter NM, Foster TC, Landfield PW. Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci. 2003;23:3807–3819. doi: 10.1523/JNEUROSCI.23-09-03807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Briggs CA, Gronlien JH, Curzon P, Timmermann DB, Ween H, Thorin-Hagene K, Kerr P, Anderson DJ, Malysz J, Dyhring T, et al. Role of channel activation in cognitive enhancement mediated by alpha7 nicotinic acetylcholine receptors. Br J Pharmacol. 2009;158:1486–1494. doi: 10.1111/j.1476-5381.2009.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccafusco JJ, Letchworth SR, Bencherif M, Lippiello PM. Long-lasting cognitive improvement with nicotinic receptor agonists: mechanisms of pharmacokinetic–pharmacodynamic discordance. Trends Pharmacol Sci. 2005;26:352–360. doi: 10.1016/j.tips.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Burger C, Lopez MC, Baker HV, Mandel RJ, Muzyczka N. Genome-wide analysis of aging and learning-related genes in the hippocampal dentate gyrus. Neurobiol Learn Mem. 2008;89:379–396. doi: 10.1016/j.nlm.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke SN, Barnes CA. Neural plasticity in the ageing brain. Nat Rev Neurosci. 2006;7:30–40. doi: 10.1038/nrn1809. [DOI] [PubMed] [Google Scholar]
- Cho KS, Choi J, Ha CM, Son YJ, Choi WS, Lee BJ. Comparison of gene expression in old versus young rat hippocampus by cDNA array. NeuroReport. 2002;13:285–289. doi: 10.1097/00001756-200203040-00008. [DOI] [PubMed] [Google Scholar]
- Consortium IS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Ding Q, Markesbery WR, Chen Q, Li F, Keller JN. Ribosome dysfunction is an early event in Alzheimer's disease. J Neurosci. 2005;25:9171–9175. doi: 10.1523/JNEUROSCI.3040-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz A, Moisan F. Update in the epidemiology of Parkinson's disease. Curr Opin Neurol. 2008;21:454–460. doi: 10.1097/WCO.0b013e3283050461. [DOI] [PubMed] [Google Scholar]
- Erraji-Benchekroun L, Underwood MD, Arango V, Galfalvy H, Pavlidis P, Smyrniotopoulos P, Mann JJ, Sibille E. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry. 2005;57:549–558. doi: 10.1016/j.biopsych.2004.10.034. [DOI] [PubMed] [Google Scholar]
- Fratiglioni L, Wang HX. Smoking and Parkinson's and Alzheimer's disease: review of the epidemiological studies. Behav Brain Res. 2000;113:117–120. doi: 10.1016/S0166-4328(00)00206-0. [DOI] [PubMed] [Google Scholar]
- Freedman R, Leonard S, Gault JM, Hopkins J, Cloninger CR, Kaufmann CA, Tsuang MT, Farone SV, Malaspina D, Svrakic DM, et al. Linkage disequilibrium for schizophrenia at the chromosome 15q13-14 locus of the alpha7-nicotinic acetylcholine receptor subunit gene (CHRNA7) Am J Med Genet. 2001;105:20–22. doi: 10.1002/1096-8628(20010108)105:1<20::AID-AJMG1047>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Gotti C, Clementi F, Fornari A, Gaimarri A, Guiducci S, Manfredi I, Moretti M, Pedrazzi P, Pucci L, Zoli M. Structural and functional diversity of native brain neuronal nicotinic receptors. Biochem Pharmacol. 2009;78:703–711. doi: 10.1016/j.bcp.2009.05.024. [DOI] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, Muhle H, Kovel C, Baker C, Spiczak S, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jiang CH, Tsien JZ, Schultz PG, Hu Y. The effects of aging on gene expression in the hypothalamus and cortex of mice. Proc Natl Acad Sci U S A. 2001;98:1930–1934. doi: 10.1073/pnas.98.4.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadish I, Thibault O, Blalock EM, Chen KC, Gant JC, Porter NM, Landfield PW. Hippocampal and cognitive aging across the lifespan: a bioenergetic shift precedes and increased cholesterol trafficking parallels memory impairment. J Neurosci. 2009;29:1805–1816. doi: 10.1523/JNEUROSCI.4599-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedmi M, Orr-Urtreger A. The differential brain transcriptome of {beta}4 nAChR subunit deficient mice: is it the effect of the null mutation or the background strain? Physiol Genomics. 2006;28:213–222. doi: 10.1152/physiolgenomics.00155.2006. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Kaeberlein M. Hot topics in aging research: protein translation. Aging Cell. 2009;8:617–623. doi: 10.1111/j.1474-9726.2009.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- Lempiainen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Lucin KM, Wyss-Coray T. Immune activation in brain aging and neurodegeneration: too much or too little? Neuron. 2009;64:110–122. doi: 10.1016/j.neuron.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue LF, Rydel R, Brigham EF, Yang LB, Hampel H, Murphy GM, Jr, Brachova L, Yan SD, Walker DG, Shen Y, Rogers J. Inflammatory repertoire of Alzheimer's disease and nondemented elderly microglia in vitro. Glia. 2001;35:72–79. doi: 10.1002/glia.1072. [DOI] [PubMed] [Google Scholar]
- Lustig A, Carter A, Bertak D, Enika D, Vandanmagsar B, Wood W, Becker KG, Weeraratna AT, Taub DD. Transcriptome analysis of murine thymocytes reveals age-associated changes in thymic gene expression. Int J Med Sci. 2009;6:51–64. doi: 10.7150/ijms.6.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Chan SL, Duan W. Modification of brain aging and neurodegenerative disorders by genes, diet, and behavior. Physiol Rev. 2002;82:637–672. doi: 10.1152/physrev.00004.2002. [DOI] [PubMed] [Google Scholar]
- Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, Cox GF, Dickinson H, Gentile J, Harris DJ, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Tseng GC, Sibille E (2010) Reciprocal phylogenetic conservation of molecular aging in mouse and human brain. Neurobiol Aging (in press) [DOI] [PMC free article] [PubMed]
- Orr-Urtreger A, Goldner FM, Saeki M, Lorenzo I, Goldberg L, Biasi M, Dani JA, Patrick JW, Beaudet AL. Mice deficient in the alpha7 neuronal nicotinic acetylcholine receptor lack alpha-bungarotoxin binding sites and hippocampal fast nicotinic currents. J Neurosci. 1997;17:9165–9171. doi: 10.1523/JNEUROSCI.17-23-09165.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo J, Brucker W, Hawrot E. Proteomic analysis of an 7 nicotinic acetylcholine receptor interactome. J Proteome Res. 2009;4:1849–1858. doi: 10.1021/pr800731z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paylor R, Nguyen M, Crawley JN, Patrick J, Beaudet A, Orr-Urtreger A. Alpha7 nicotinic receptor subunits are not necessary for hippocampal-dependent learning or sensorimotor gating: a behavioral characterization of Acra7-deficient mice. Learn Mem. 1998;5:302–316. [PMC free article] [PubMed] [Google Scholar]
- Picciotto MR, Zoli M. Nicotinic receptors in aging and dementia. J Neurobiol. 2002;53:641–655. doi: 10.1002/neu.10102. [DOI] [PubMed] [Google Scholar]
- Prolla TA. DNA microarray analysis of the aging brain. Chem Senses. 2002;27:299–306. doi: 10.1093/chemse/27.3.299. [DOI] [PubMed] [Google Scholar]
- Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, Xiao W, Mindrinos M, Crane E, Segal E, et al. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2:e427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SW, Gahring LC, Collins AC, Marks M. Age-related changes in neuronal nicotinic acetylcholine receptor subunit alpha4 expression are modified by long-term nicotine administration. J Neurosci. 1998;18:4825–4832. doi: 10.1523/JNEUROSCI.18-13-04825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe WB, Blalock EM, Chen KC, Kadish I, Wang D, Barrett JE, Thibault O, Porter NM, Rose GM, Landfield PW. Hippocampal expression analyses reveal selective association of immediate-early, neuroenergetic, and myelinogenic pathways with cognitive impairment in aged rats. J Neurosci. 2007;27:3098–3110. doi: 10.1523/JNEUROSCI.4163-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbagh MN, Shah F, Reid RT, Sue L, Connor DJ, Peterson LK, Beach TG. Pathologic and nicotinic receptor binding differences between mild cognitive impairment, Alzheimer disease, and normal aging. Arch Neurol. 2006;63:1771–1776. doi: 10.1001/archneur.63.12.1771. [DOI] [PubMed] [Google Scholar]
- Sack R, Gochberg-Sarver A, Rozovsky U, Kedmi M, Rosner S, Orr-Urtreger A. Lower core body temperature and attenuated nicotine-induced hypothermic response in mice lacking the beta4 neuronal nicotinic acetylcholine receptor subunit. Brain Res Bull. 2005;66:30–36. doi: 10.1016/j.brainresbull.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Salas R, Main A, Gangitano D, Biasi M. Decreased withdrawal symptoms but normal tolerance to nicotine in mice null for the alpha7 nicotinic acetylcholine receptor subunit. Neuropharmacology. 2007;53:863–869. doi: 10.1016/j.neuropharm.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarter M, Parikh V, Howe WM. nAChR agonist-induced cognition enhancement: integration of cognitive and neuronal mechanisms. Biochem Pharmacol. 2009;78:658–667. doi: 10.1016/j.bcp.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sharman EH, Bondy SC, Sharman KG, Lahiri D, Cotman CW, Perreau VM. Effects of melatonin and age on gene expression in mouse CNS using microarray analysis. Neurochem Int. 2007;50:336–344. doi: 10.1016/j.neuint.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, Schroer RJ, Novara F, Gregori M, Ciccone R, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinawi M, Schaaf CP, Bhatt SS, Xia Z, Patel A, Cheung SW, Lanpher B, Nagl S, Herding HS, Nevinny-Stickel C, et al. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat Genet. 2009;41:1269–1271. doi: 10.1038/ng.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoop RD, Yamada N, Berg DK. Cytoskeletal links of neuronal acetylcholine receptors containing alpha 7 subunits. J Neurosci. 2000;20:4021–4029. doi: 10.1523/JNEUROSCI.20-11-04021.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. Yeast life span extension by depletion of 60 s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinlein OK, Mulley JC, Propping P, Wallace RH, Phillips HA, Sutherland GR, Scheffer IE, Berkovic SF. A missense mutation in the neuronal nicotinic acetylcholine receptor alpha 4 subunit is associated with autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet. 1995;11:201–203. doi: 10.1038/ng1095-201. [DOI] [PubMed] [Google Scholar]
- Swindell WR. Genes and gene expression modules associated with caloric restriction and aging in the laboratory mouse. BMC Genomics. 2009;10:585. doi: 10.1186/1471-2164-10-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taly A, Corringer PJ, Guedin D, Lestage P, Changeux JP. Nicotinic receptors: allosteric transitions and therapeutic targets in the nervous system. Nat Rev Drug Discov. 2009;8:733–750. doi: 10.1038/nrd2927. [DOI] [PubMed] [Google Scholar]
- Tang B, Chang WL, Lanigan CM, Dean B, Sutcliffe JG, Thomas EA. Normal human aging and early-stage schizophrenia share common molecular profiles. Aging Cell. 2009;8:339–342. doi: 10.1111/j.1474-9726.2009.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernarakis N. Ageing and the regulation of protein synthesis: a balancing act? Trends Cell Biol. 2008;18:228–235. doi: 10.1016/j.tcb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Terao A, Apte-Deshpande A, Dousman L, Morairty S, Eynon BP, Kilduff TS, Freund YR. Immune response gene expression increases in the aging murine hippocampus. J Neuroimmunol. 2002;132:99–112. doi: 10.1016/S0165-5728(02)00317-X. [DOI] [PubMed] [Google Scholar]
- Tietje KR, Anderson DJ, Bitner RS, Blomme EA, Brackemeyer PJ, Briggs CA, Browman KE, Bury D, Curzon P, Drescher KU, et al. Preclinical characterization of A-582941: a novel alpha7 neuronal nicotinic receptor agonist with broad spectrum cognition-enhancing properties. CNS Neurosci Ther. 2008;14:65–82. doi: 10.1111/j.1755-5949.2008.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermann DB, Gronlien JH, Kohlhaas KL, Nielsen EO, Dam E, Jorgensen TD, Ahring PK, Peters D, Holst D, Chrsitensen JK, et al. An allosteric modulator of the alpha7 nicotinic acetylcholine receptor possessing cognition-enhancing properties in vivo. J Pharmacol Exp Ther. 2007;323:294–307. doi: 10.1124/jpet.107.120436. [DOI] [PubMed] [Google Scholar]
- Toescu EC, Verkhratsky A. The importance of being subtle: small changes in calcium homeostasis control cognitive decline in normal aging. Aging Cell. 2007;6:267–273. doi: 10.1111/j.1474-9726.2007.00296.x. [DOI] [PubMed] [Google Scholar]
- Young JW, Finlayson K, Spratt C, Marston HM, Crawford N, Kelly JS, Sharkey J. Nicotine improves sustained attention in mice: evidence for involvement of the alpha7 nicotinic acetylcholine receptor. Neuropsychopharmacology. 2004;29:891–900. doi: 10.1038/sj.npp.1300393. [DOI] [PubMed] [Google Scholar]
- Young JW, Crawford N, Kelly JS, Kerr LE, Marston HM, Spratt C, Finlayson K, Sharkey J. Impaired attention is central to the cognitive deficits observed in alpha 7 deficient mice. Eur Neuropsychopharmacol. 2007;17:145–155. doi: 10.1016/j.euroneuro.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Yu WF, Guan ZZ, Nordberg A. Postnatal upregulation of alpha4 and alpha3 nicotinic receptor subunits in the brain of alpha7 nicotinic receptor-deficient mice. Neuroscience. 2007;146:1618–1628. doi: 10.1016/j.neuroscience.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Zahn JM, Sonu R, Vogel H, Crane E, Mazan-Mamczarz K, Rabkin R, Davis RW, Becker KG, Owen AB, Kim SK. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:e115. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoli M, Picciotto MR, Ferrari R, Cocchi D, Changeux JP. Increased neurodegeneration during ageing in mice lacking high-affinity nicotine receptors. EMBO J. 1999;18:1235–1244. doi: 10.1093/emboj/18.5.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
a The 52 probe sets whose expression significantly differentiated brains of aged mice from brains of young mice (two-way ANOVA, Bonferroni-corrected, P < 0.05). b The 1,710 probe sets whose expression significantly differentiated brains of aged mice from brains of young mice (two-way ANOVA, FDR-corrected, P < 0.05). (XLS 1099 kb)
a Overrepresented GO annotations among the 47 significantly changed age-dependent genes (Bonferroni-corrected 52 probe sets). b Overrepresented GO annotations among the 1,543 significantly changed age-dependent genes (FDR-corrected 1,710 probe sets). (XLS 254 kb)
a A compiled list of genes that were previously published in eight different microarray studies as changed in brains of aged mice or rats (Blalock et al. 2003; Burger et al. 2008; Cho et al. 2002; Jiang et al. 2001; Kadish et al. 2009; Prolla 2002; Rowe et al. 2007; Sharman et al. 2007). b Genes that appear in both Supplementary Tables 1a and 3a. c Genes that appear in both Supplementary Tables 1b and 3a. d A compiled list of genes that were previously published in four different microarray studies as changed in brains of aged humans (Berchtold et al. 2008; Erraji-Benchekroun et al. 2005; Lu et al. 2004; Tang et al. 2009). e Genes that appear in both Supplementary Tables 1a and 3d. f Genes that appear in both Supplementary Tables 1b and 3d. g Genes that appear in all three Supplementary Tables 1b, 3a and 3d. h Overrepresented GO annotations among the age-dependent differentially expressed genes that were previously described in human and murine (genes that are listed in Supplementary Tables 3c, f, and g). (XLS 3721 kb)
The 211 probe sets whose expression is dependent on the interaction between age and genotype parameters (two-way ANOVA, P < 0.001). (XLS 162 kb)
Overrepresented GO annotations among the 207 age–genotype interaction genes (211 probe sets in Supplementary Table 4). (XLS 29 kb)
Pivot table of the raw expression signals data. (XLS 10180 kb)