Abstract
Brassica carinata, an allotetraploid with B and C genomes, has a number of traits that would be valuable to introgress into B. napus. Interspecific hybrids were created between B. carinata (BBCC) and B. napus (AACC), using an advanced backcross approach to identify and introgress traits of agronomic interest from the B. carinata genome and to study the genetic changes that occur during the introgression process. We mapped the B and C genomes of B. carinata with SSR markers and observed their introgression into B. napus through a number of backcross generations, focusing on a BC3 and BC3S1 sibling family. There was close colinearity between the C genomes of B. carinata and B. napus and we provide evidence that B. carinata C chromosomes pair and recombine normally with those of B. napus, suggesting that similar to other Brassica allotetraploids no major chromosomal rearrangements have taken place since the formation of B. carinata. There was no evidence of introgression of the B chromosomes into the A or C chromosomes of B. napus; instead they were inherited as whole linkage groups with the occasional loss of terminal segments and several of the B-genome chromosomes were retained across generations. Several BC3S1 families were analyzed using SSR markers, genomic in situ hybridization (GISH) assays, and chromosome counts to study the inheritance of the B-genome chromosome(s) and their association with morphological traits. Our work provides an analysis of the behavior of chromosomes in an interspecific cross and reinforces the challenges of introgressing novel traits into crop plants.
ONE of the limitations of modern crop breeding is the reduction in available genetic variability in cultivars for most crop species, resulting from successive generations of artificial selection. Wild relatives, on the other hand, maintain a wide range of allelic diversity for important traits such as disease resistance and enhanced stress tolerance. The “advanced backcross” approach allows the transfer of genes controlling useful agronomic traits that are not present in the crops' natural background, while identifying the genomic regions and potentially the genes controlling specific traits (Tanksley and Nelson 1996). To adapt this approach effectively from intra- to interspecific crosses, detailed knowledge of the genetic characteristics of crop plants and their wild relatives is important in predicting the consequences of backcrossing. Interspecific crosses have been widely used to attempt to introgress desired traits into specific genetic backgrounds and increase genetic diversity, but are often unsuccessful. Factors such as limited chromosome homology and low rates of recombination can make it difficult to introgress the target trait and the resultant linkage drag can lead to the simultaneous introduction of undesirable phenotypes (Brown et al. 2003; Desloire et al. 2003).
The genus Brassica, because of extensive genetic mapping and close homology to Arabidopsis, has a number of tools available to assess interspecific hybrids and to detect the transfer of traits between different species. The morphology of leaves, flowers, and pods has been successfully used to distinguish the species of the Brassicaceae (Gomez-Campo 1980), but may be ambiguous for determining true hybrids, which, depending on the trait, are often more similar to one of the parents than to the predicted intermediate phenotype. Cytogenetic methods, such as chromosome banding in wheat (Gill and Kimber 1977) and fluorescent in situ hybridization (Wang et al. 2006), can also be used to visualize the genomic constitution of hybrids and the meiotic behavior of the chromosomes (Attia and Robbelen 1986; Heneen and Jorgensen 2001). These methods have also been used to identify genomic regions in common between closely related species and to detect introgressed segments from wild relatives (Chèvre et al. 2004). Introgressed segments can also be accurately assayed using molecular markers. Currently, a large number of markers are available for the Brassica genomes (A, B, and C) that have been used in recent years to tag simple Mendelian traits and to map quantitative trait loci (Mahmood et al. 2007) and can be used to detect interspecific hybrids and genetic introgression.
Panjabi et al. (2008) used intron polymorphism (IP) markers to identify a high degree of colinearity between the A and B genomes of Brassica juncea with the A genome of B. napus and the B genome of B. nigra, respectively, which suggested low levels of chromosomal change after polyploidization. Cytological observations in digenomic triploids (BBC and CCB) generated from interspecific hybridization between B. carinata and B. nigra and between B. carinata and B. oleracea indicated that the Brassica B genome chromosomes may not form homeologous pairs with chromosomes of the A and C genomes in interspecific crosses (Meng et al. 1998). Likewise on the basis of molecular cytogenetic evidence, the structure of the genome affects the frequency of homologous and homeologous pairing during meiosis, where there were more A–C genome associations observed than A–B or B–C in an interspecific cross of B. napus and B. carinata (Mason et al. 2010a). On the basis of these data and early mapping studies (Lagercrantz and Lydiate 1996), it was recognized that the B genome has significantly diverged from the A and C genomes and was not considered homeologous to any A/C genome chromosomes (Warwick et al. 1992; Axelsson et al. 2000). However, despite the lack of pairing observed, more recent mapping studies have shown that the B genome shares a surprising number of homologous regions with the A and C genomes (Lagercrantz and Lydiate 1996; Panjabi et al. 2008). Comparative mapping of different Brassicaceae lineages indicates the presence of 24 conserved genome blocks in B. napus (Parkin et al. 2005; Schranz et al. 2006). Panjabi et al. (2008) showed intact ancestral block arrangements during evolution, as well as significant homology between three linkage groups of the B genome (B4, B5, and B6) and the A genome (A4, A5, and A6).
One of the goals of Brassica oilseed research programs is the stable introgression of novel traits from wild or closely related species into cultivated canola plants through interspecific crosses (Ky et al. 2000). The Brassica species containing the B genome, B. nigra, B. carinata, and B. juncea, possess valuable agronomic traits including blackleg resistance (caused by Leptosphaeria maculans), heat and drought tolerance (Kumar et al. 1984), aluminum tolerance (Huang et al. 2002), and tolerance to salinity (Malik 1990). Several groups have tried to introgress blackleg resistance and silique shatter resistance from the B genomes of B. juncea and B. nigra into B. napus species; however, these traits have not been successfully transferred into commercial germplasm (Roy 1984; Prakash and Chopra 1988; Gerdemannknorck et al. 1995; Chèvre et al. 1997; Dixelius and Wahlberg 1999; Roussel et al. 1999).
More recently, the introgression of B-genome material into the A/C genome was traced using florescent in situ hybridization (FISH) of B-genome-specific repetitive DNA (Schelfhout et al. 2006). The Brassica B genome appeared to be excluded in favor of homologous and homeologous pairing of A and C genomes in interspecific crosses among Brassica species (Meng et al. 1998); however, in these crosses of B. rapa and B. carinata, several of the B chromosomes were also maintained for several generations.
In this article, we describe the development of an interspecific hybrid population between B. carinata and B. napus with the purpose of using an advanced backcross approach to (1) introgress traits into advanced germplasm, (2) provide information on the inheritance of different chromosomes from a close relative, and (3) map agronomically interesting traits. We demonstrate the production of lines carrying B-genome chromosome segments and track these segments using molecular markers and cytogenetic techniques at each stage of backcrossing. At a more fundamental level, we hypothesized that specific B-genome chromosomes could be maintained and tracked through a backcrossing program. We were particularly interested in whether B-genome material could be introgressed by pairing of homeologous chromosomes. These interspecific hybrid lines provide material for an analysis of the structure of the C genome of B. carinata and represent both foundational germplasm and a tool for the comprehensive analysis of the B genome.
MATERIALS AND METHODS
Development of the plant material:
B. napus PSA12 is an artificially resynthesized B. napus, which was chosen on the basis of the observation that this genotype exhibits reduced pairing control and therefore might undergo nonhomologous pairing more frequently than established commercial cultivars (D. Lydiate, personal communication). BCA-070 is recorded as an Ethiopian B. carinata line, which was derived from an accession that we accessed from the Australian Temperate Field Crops Collection at Horham, Victoria, Australia. This line possessed several desirable agronomic and disease resistance traits (Purwantara et al. 1998) and was kindly donated by Phil Salisbury (University of Melbourne, Australia). One inbred BCA-070 plant was crossed to a single B. napus PSA12 individual through bud pollination and 10 F1 hybrid plants were generated through tissue culture from immature ovules, which were excised from 3-week-old siliques and sterilized in 10% bleach for 1 min. The ovules were then cultured on modified Murashige and Skoog (MS) media containing 300 mg/liter casein hydrolysate, 2.5 g/liter gelrite, and 50 g/liter sucrose. Ovules were transferred to fresh modified-MS media as needed. Surviving ovules were cultured until germination or callus formation and were subsequently transferred to 100 ml of modified-MS medium containing 0.15 mg/ml solution of naphthaleneacetic acid (NAA). Well-rooted plantlets were then transferred to soil-less media (280 MetroMix; Sungro, Vancouver, BC, Canada).
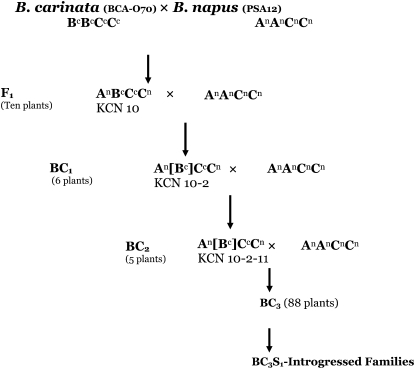
A single female F1 plant (KCN-10) was selected and crossed with the original PSA12 clone to generate 40 BC1 plants, 6 of which were genotyped in detail (Figure 1). These BC1 plants were grown to provide leaf material for genomic DNA extraction and selfed to produce seed (BC1S1) and a single BC1 plant (KCN-10-2) was crossed to the PSA12 clone to generate BC2 seed. Five of the BC2 plants were genotyped in detail. One BC2 plant (KCN-10-2-11) was then chosen to generate a large BC3 mapping population. The resultant BC3 plants were selfed to generate the BC3S1 population. Seventeen BC3S1 families, derived from BC3 plants containing B genome chromosomes (determined by microsatellite markers), were phenotyped and designated as introgressed families (IF). Of these, 5 IFs were genotyped with SSR markers and used in genomic in situ hybridization (GISH) assays (Figure 1). Eleven plants of each of the 17 BC3S1 families and their parents were planted in Metro Mix 290 (Grace Horticultural Products, Ajax, ON, Canada) and grown in a growth cabinet set at 21°/18° (day/night) with a 16-hr photoperiod. Fertilization was done every second week with 200 parts per million liquid solution of Peres 20-20-20 (N-P-K) (Plant Products, Brampton, ON, Canada) to fill up the pots with liquid, which was allowed to drain. Plants were grown in two replicates and evaluated for morphological traits as described below (Figure 2). Each replicate included 11 individuals from the 17 families.
Figure 1.—
Pedigree and crosses used in the development of the BC3 lines and the BC3S1 families used in this study. No selection was performed in any generation, except the F1 plants, which were chosen in part by seed set. At the BC3 generation 88 individuals were selected for SSR genotyping, and 17 individuals were then selected to generate the BC3S1 introgressed families (IFs). These IFs were phenotyped for morphological traits where six families were chosen for SSR genotyping as well as FISH confirmation.
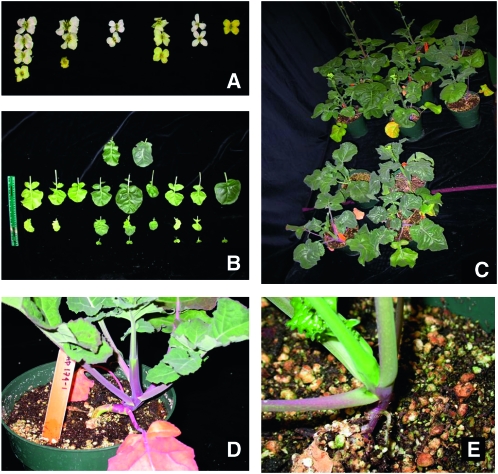
Figure 2.—
Segregation of the morphological traits in the BC3 backcross populations: (A) flower color, (B) leaf shape, (C) leaf color, and (D and E) stem color.
Genomic DNA extractions, Southern hybridization, and microsatellite analysis:
Genomic DNA extractions and Southern hybridizations, confirming the hybrid genotype of KCN-10, were carried out as described in Sharpe et al. (1995). Microsatellite primer pairs, derived from B. nigra, B. juncea, B. rapa, and B. napus, with defined loci in the Brassica A, C, and B genomes were developed by Agriculture and Agri-Food Canada (AAFC), Saskatoon Research Station (http://www.agr.gc.ca). From these, a subset of markers that identified polymorphic loci in the Brassica C and B genomes was used to characterize KCN-10 and the different backcross generations. Amplification and resolution of microsatellite alleles by PCR and capillary electrophoresis were performed as described in Navabi et al. (2010).
Allele identification:
The assignment of the Brassica B and C genomic markers to specific linkage groups was achieved by aligning the new linkage map with published maps through common markers (Parkin et al. 2005; Sun et al. 2007; Panjabi et al. 2008). The universally agreed nomenclature system as introduced at www.brassica.info is adopted. Using the AAFC reference maps (based on two B. napus and one B. juncea reference mapping populations) (Parkin et al. 1995; Sharpe et al. 1995; Lagercrantz and Lydiate 1996; Axelsson et al. 2000), markers were selected that mapped at regular intervals along each B and C genome chromosome and that ideally amplified only one or two loci. The SSR markers were evaluated to compare allele sizes in the parents of KCN-10 and in the experimental population to those characterized in the AAFC reference populations and appropriate representative B. nigra (BB), B. oleracea (CC), and B. rapa (AA) lines. This allowed the assignment of markers to previously mapped loci (on the basis of identical allele size) and confirmed the genome origin for those not previously mapped.
Analysis of specific linkage groups in BC3S1 IF lines:
Leaf tissue samples were collected from 11 plants from each of 17 BC3S1 families at the four- to five-leaf stage, and DNA was extracted using Sigma's GenElute Plant Genomic DNA Miniprep Kit (Sigma, St. Louis).
Six BC3S1 families (IFs) were selected on the basis of SSR analysis and analyzed for the segregation of different morphological traits as described above. From a total of 1242 B-genome microsatellites, 103 markers for the linkage groups B3, B5, B6, and B8 were selected due to the presence of these four B-genome chromosomes in the parental BC3 plants in the six BC3S1 families. The molecular marker analysis was conducted as described in Navabi et al. (2010).
Linkage analysis:
Microsatellite markers that amplified one or more polymorphic loci were scored to generate scoring matrices. The assignment of loci to linkage groups was accomplished using both comparative mapping and linkage analysis. For the comparative mapping, marker loci were assigned to a specific linkage group, on the basis of the published maps as described above. For the initial linkage association, a minimum LOD score of 4.0 and a maximum distance of 50.0 cM were used. A LOD score of 3.0 was used to bridge any larger gaps. Recombination frequencies were converted to Kosambi centimorgan map distances (Kosambi 1944). Following initial assignments, the linkage analysis was completed using a LOD score of 3.0 with the “ripple” and “automap” mapping functions of the MapDisto 1.2.0.3 software package (Lorieux et al. 2000). Segregation distortion for the mapping populations was verified with chi-square tests contrasting expected 1:1 allele segregation with the values observed. Graphical genotypes were constructed by assuming that if two adjacent markers were from the same parent, then the intervening block was also from that parent (Tanksley et al. 1989). Conversely, if adjacent markers differed in parental origin, then the intervening block was assumed to contain a crossover event. The graphical genotype was then constructed to minimize the total number of crossover events required to explain a given genotype. The probability of undetected multiple crossovers between two markers would be low for early generation hybrids and for small marker distance intervals (Rieseberg et al. 2003).
GISH assay on BC3S1 IF lines:
To confirm the presence of B-genome chromosomes, flower buds from the six selected IFs used for marker analysis were used for GISH/FISH analysis using Brassica genomic DNA and a 45S DNA probe. Immature flower buds collected from two to three plants of each family were used for mitotic and meiotic chromosome spreads. Slides were prepared following tissue maceration with pectolyase and cellulase (Kato et al. 2004; Lamb and Birchler 2006). B. nigra and B. oleracea genomic DNA and repeated sequences were labeled fluorescently (Navabi et al. 2010). A 45S DNA clone was also used as a probe to detect nuclear organizing regions (NORs). Fluorescent in situ hybridization was performed following the method of Kato et al. (2004) with slight modifications, as described in Navabi et al. (2010).
Statistical analysis of morphological traits on BC3S1 IF lines:
Morphological traits were measured using predefined descriptors for Brassica (Ibpgr 1990) with minor modifications (supporting information, File S1). Morphological data collected from two trials were used to perform the statistical analysis. A marker regression approach (Kearsey and Hyne 1994) was used to test for an association of the B-genome chromosome(s) with each trait. Analysis of variance was performed using the PROC MIXED command of SAS software, followed by “LSmean” and “boxplot” statements to calculate the statistics of the population (SAS Institute 1989) and distribution of each trait (Figure 3). The null hypothesis was that the presence of the B genome had no effect on the trait under evaluation (Table S1). In the simplest linear model of regression, the phenotypic value of individual j (Zj) is a function of mean value (μ), effects (bi) of different chromosomes (xij) on the phenotype, and residual error (ej) following the model Zj = μ+ ∑bixij + ej. When two or more markers are considered, the effect (bi) corresponds to the multilocus marker genotype; the evidence of a linked QTL is provided by a significant R2, which is the fraction of phenotypic variance accounted for by the marker genotype (Lynch and Walsh 1998). In addition, the segregation of the B-genome chromosomes for fit to a 3:1 ratio was tested by the χ2-test, using the formula χ2 = ∑(Oi − Ei)2/Ei.
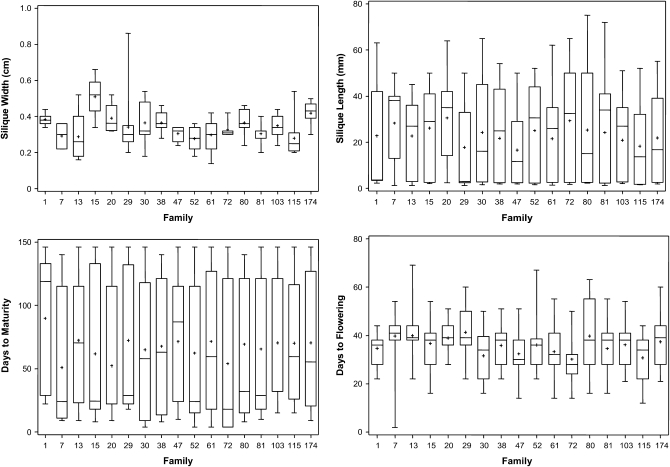
Figure 3.—
Boxplots showing the variation for different morphological traits in 17 BC3S1 introgressed families (IFs). Maximum, minimum, and median values are marked. Each box represents the interquartile range, which contains 50% of the values. The whiskers are lines that extend from the box to the highest and lowest values, excluding outliers. The solid line across the box indicates the median. The mean value is indicated by a “+” sign.
Oi is the observed number of the chromosome of either the B+ or the B− class in each family and Ei is the expected value under a segregation ratio of 3:1, assuming that the chromosomes are segregating in a Mendelian manner.
RESULTS
Generation of B-genome interspecific lines:
The interspecific cross between B. napus (PSA12) and B. carinata (BCA-070) required embryo rescue to generate F1 hybrids. Ten F1 plants were confirmed to be hybrid via RFLP hybridization using four probes (Sharpe et al. 1995; Stead 2009), and the F1 hybrid KCN-10 was crossed with PSA12 to generate the subsequent backcross populations. A single F1 plant (KCN-10) was selected randomly from 10 fertile F1 plants and used to generate a BC1 population of 6 plants. KCN-10-2 was then selected randomly from these 6 plants and used to generate a BC2 population of 5 plants, from which KCN-10-2-11 was chosen. KCN-10-2-11 was then used as the parent of the BC3 mapping population of 88 plants (Figure 1).
Introgression of the C genome of B. carinata with the B. napus C genome:
The marker scoring data for the original F1 through to the BC3 family are presented in Table S2. A number of SSR markers amplified loci from genomes they were not originally derived from. Of 170 B. napus-specific SSR markers 20% (34) also amplified loci in the B genome of B. nigra and B. carinata. Only 2 markers (1.4%) amplified loci in the C genome from a set of 138 B. juncea B-genome-specific markers.
The B. napus C genome has been identified and mapped in a number of Brassica populations by several groups and has been determined to comprise nine linkage groups: N11–N19 (Slocum et al. 1990; Park et al. 1995; Sharpe et al. 1995; Piquemal et al. 2005; Sun et al. 2007). Using the approaches described above, we identified the nine C-genome chromosomes of B. carinata. However, due to the backcrossing approach used, the B. carinata C genome represented only a small percentage of the overall C genome in the BC3 hybrid plants. C-genome-specific microsatellite markers amplified the same or similar loci in B. carinata and B. napus as shown by similar sized alleles, the lack of segregation distortion between the alleles (Table 1), and the pairing and recombination observed for each backcross generation. Most of the alleles from the B. carinata C genome were not inherited in the BC3 population (due to backcrossing to B. napus); however, the KCN-10 BC1 and BC2 families were evaluated to track the B. carinata allele loss from the F1 generation through to the BC3 generation (Table 2). Figure 4A illustrates two of the C-genome linkage groups (N11 and N13), where recombination continued to occur in each backcross generation, resulting in the loss of the B. carinata C-genome alleles as outlined in Table 2. Assuming normal pairing and segregation, the B. carinata C alleles were preferentially lost in the BC1 generation, with on average 38% of the loci retaining an allelic copy from the B. carinata C genome, while in the subsequent backcross generation only 20.5% of loci maintained B. carinata C-genome alleles.
TABLE 1.
Allele segregation χ2-values for the B and C genomes in the BC3 mapping population
| LG | Marker | χ2 1:1 | P | S | LG | Marker | χ2 1:1 | P | S |
|---|---|---|---|---|---|---|---|---|---|
| B3 | sJ1322x | 23.84 | 0 | *****↓ | B7 | sB1871x | 33.05 | 0 | *****↓ |
| B3 | sB3910A | 9.67 | 0.00188 | **↓ | B7 | sJ39119ia | 26.45 | 0 | *****↓ |
| B3 | sJ1071B | 0.02 | 0.87879 | NS | B7 | sB1538BB | 34.77 | 0 | *****↓ |
| B3 | sB2668 | 7.25 | 0.0071 | **↓ | B7 | sJ13133a | 30.6 | 0 | *****↓ |
| B3 | sB0862a | 9.89 | 0.00166 | **↓ | B7 | sJ1536a | 36.47 | 0 | *****↓ |
| B3 | sB02124B | 14.63 | 0.00013 | ***↓ | B7 | sB0570a | 38.22 | 0 | *****↓ |
| B3 | sB1752x | 11.31 | 0.00077 | ***↓ | B7 | sJ4633a | 39.12 | 0 | *****↓ |
| B3 | sB2771a | 8.38 | 0.0038 | **↓ | B8 | sB4727Fa | 51.86 | 0 | *****↓ |
| B3 | sJ0266c | 12.49 | 0.00041 | ***↓ | B8 | sB0860Aa | 30.73 | 0 | *****↓ |
| B5 | sB2334x | 0.11 | 0.74488 | NS | B8 | sB1728a | 34.71 | 0 | *****↓ |
| B5 | sB31111Bx | 0.45 | 0.50233 | NS | B8 | sJ2013B | 27.84 | 0 | *****↓ |
| B6 | sB1839x | 7.53 | 0.00607 | **↓ | B8 | sJ3327Rx | 31.44 | 0 | *****↓ |
| B6 | sB1755B | 8.58 | 0.00341 | **↓ | B8 | sJ0143c | 31.25 | 0 | *****↓ |
| B6 | sB1956a | 8.24 | 0.00409 | **↓ | B8 | sJ0397Ra | 30.73 | 0 | *****↓ |
| B6 | sB4727FB | 88 | 0 | *****↓ | B8 | sB2596a | 30.73 | 0 | *****↓ |
| B6 | sB1772ia | 11.2 | 0.00082 | ***↓ | N17 | sORC76 | 0.05 | NS | |
| B6 | sB0273B | 0.49 | 0.4855 | NS | N17 | sN12508i | 0.1 | NS | |
| B6 | sJ1505a | 3.56 | 0.05935 | NS | N17 | sS1949 | 1.64 | NS | |
| B6 | sJ0502a | 5.73 | 0.0167 | *↓ | N11 | sn1838 | 3.68 | NS | |
| B6 | sB2545a | 27.94 | 0 | *****↓ | N11 | sn11910 | 1.39 | NS | |
| B6 | sJ46102y | 41.02 | 0 | *****↓ | N11 | sN9431 | 2.23 | NS | |
| N13 | sn0744 | 3.68 | NS |
Normal segregation for the C-genome loci on N chromosomes and tendency for distortion against B. carinata alleles in the B genome are shown. LG indicates the linkage group to which core markers were assigned in the KCN-10 mapping population. Chi-square values (χ2) used a 1:1 expected allele ratio. NS, not significant; * to *****, significant (P < 0.05) to highly significant (P < 0.0001). Arrows indicate distortion in favor (↓) of a lower frequency of the B. carinata alleles.
TABLE 2.
Loss of B. carinata C alleles in the KCN-10 mapping populations: the percentage of loci on each linkage group having one C-genome allele from the B. carinata parent, at each backcross generation
| Generation (% expected) | N11 | N12 | N13 | N14 | N15 | N16 | N17 | N18 | N19 | Mean |
|---|---|---|---|---|---|---|---|---|---|---|
| KCN-10-F1, 1 individual, no. of markers used | 15 | 8 | 15 | 14 | 5 | 4 | 11 | 8 | 7 | 10.3 |
| KCN-10-BC1, 6 individuals, % loci with B. carinata alleles (50%) | 28 | 27 | 43 | 40 | 40 | 37.5 | 51 | 35 | 42 | 38 |
| KCN-10-BC2, 5 individuals, % loci with B. carinata alleles (25%) | 16 | 5 | 24 | 18 | 40 | 20 | 25 | 20 | 22 | 20.5 |
| KCN-10-BC3, 88 individuals, no. of alleles present | 4 | 0 | 2 | 1 | 0 | 1 | 3 | 2 | 2 | 2.3 |
The percentages of lines that had inherited B. carinata C alleles in the BC1, BC2, and BC3 generations of the KCN-10 population are shown. Percentages are calculated on the basis of the number of loci where B. carinata C-genome alleles were detected for each C-genome linkage group, present at each generation, F1 plants behaved normally, and the polymorphic SSR markers had one B. napus and one B. carinata allele.
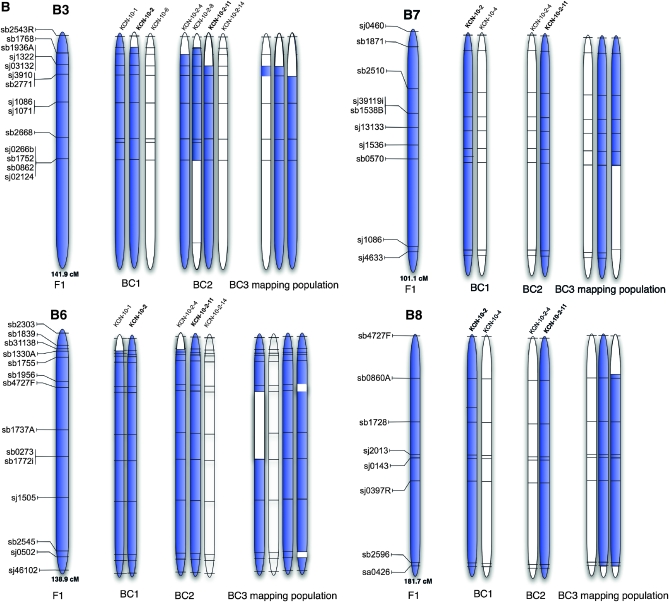
Figure 4.—
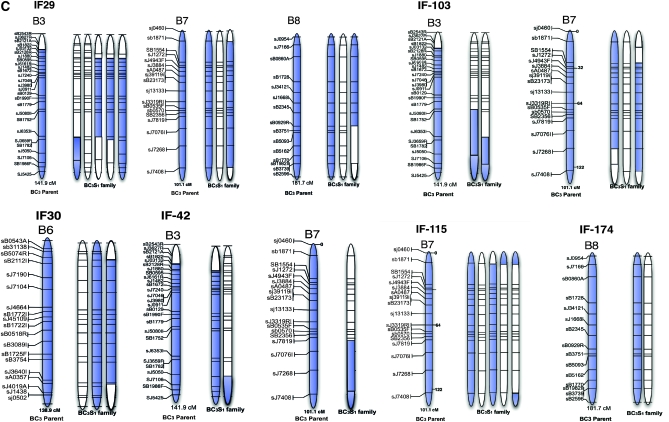
Physical representation of a selection of linkage groups in the genetic material used in this study. (A) Segregation pattern of N11 and N13 chromosomes of the C genome from F1 to BC3 generation; (B) segregation pattern of B3, B6, B7, and B8 chromosomes of the B genome from F1 to BC3 generation; (C) segregation pattern of B-genome chromosomes observed in the 11 individuals of BC3S1 families. The location of the markers selected on the linkage groups is based on the AAFC reference maps. Blue parts represent the presence of the segment and white segments represent its absence. The parent of the BC3 mapping population is indicated in boldface type.
Mapping of the B-genome linkage groups in the backcross populations:
The B-genome linkage groups of the KCN-10 mapping population were identified by aligning putative linkage groups with the AAFC reference maps via common marker alleles (Parkin and Lydiate 1997; Axelsson et al. 2000). Table 3 and Figure 4B illustrate the B-genome content of the KCN-10 population during each successive generation of backcrossing, by analyzing a subset of lines from the BC1 and BC2 populations. In contrast to the normal recombination behavior between the C genomes of B. napus and B. carinata, the chromosomes from the B genome were either missing or present largely as complete chromosomes, although there were a number of cases where small terminal deletions had occurred. Linkage groups B2 and B3 from the B genome were, at least partially, retained in most of the BC1 population while the remaining B linkage groups were maintained in at least one of the BC1 individuals (Table 3). At the BC2 stage B5 had been almost completely lost in all of the lines. There was one interesting region of B5 (identified by the markers sB2334x and SB31111Bx) that could be tracked through to the BC3 generation. It appeared that this segment of B5 may have been translocated to either N1 or N11, due to cosegregation of these markers with a marker that can be mapped to either of these two homeologous linkage groups. The mapping parent for the third backcross, KCN-10-2-11, carried loci from only four B-genome linkage groups (B3, B6, B7, and B8), in addition to the two markers from B5. A selection of markers specific to B1, B2, and B4 were assayed to confirm that the remaining B-genome linkage groups were absent from the BC2 and BC3 lines (Table S2).
TABLE 3.
B-genome content of the KCN-10 BC1 and BC2 families
| Individual | B1, % | B2, % | B3, % | B4, % | B5, % | B6, % | B7, % | B8, % |
|---|---|---|---|---|---|---|---|---|
| F1 | ||||||||
| KCN-10 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| BC1 | ||||||||
| KCN-10-1 | 0 | 94 | 100 | 0 | 100 | 81 | 62 | 100 |
| KCN-10-2 | 100 | 100 | 93 | 88 | 36 | 100 | 100 | 100 |
| KCN-10-3 | 20 | 78 | 100 | 75 | 21 | 100 | 100 | 100 |
| KCN-10-4 | 0 | 44 | 100 | 0 | 28 | 27 | 0 | 0 |
| KCN-10-5 | 100 | 94 | 100 | 88 | 100 | 100 | 100 | 100 |
| KCN-10-6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BC2 | ||||||||
| KCN-10-2-4 | 0 | 85 | 81 | 71 | 15 | 88 | 0 | 0 |
| KCN-10-2-8 | 0 | 0 | 88 | 0 | 0 | 0 | 100 | 100 |
| KCN-10-2-11 | 0 | 0 | 73 | 0 | 20 | 100 | 100 | 100 |
| KCN-10-2-13 | 100 | 85 | 88 | 0 | 16 | 100 | 13 | 100 |
| KCN-10-2-14 | 85 | 0 | 0 | 0 | 8 | 100 | 20 | 0 |
Individuals comprising the KCN-10 sibling families are aligned on the vertical axis, and the B-genome chromosomes are aligned along the top. The percentages of markers present on each LG are calculated on the basis of the portion of markers that showed amplification.
Inheritance of B-genome linkage groups in the BC3S1 families:
Most of the B-genome chromosomes in BC3S1 plants tended to be inherited as intact linkage groups; however, loss of terminal segments or translocations was detected in several cases (Figure 4C). For example, the family IF-42 carried only the terminal segment of B7, while it was present as a whole chromosome in IF-29, IF-103, and IF-115 (Figure 4C). Similarly, the terminal segment of B3 was present in IF-103, while B3 was mostly conserved in IF-29 and IF-42 (Figure 4C). B8 was found to exist as a whole chromosome in IF-174; no deletions of this chromosome appeared to have occurred in this family (Figure 4C).
In the BC3S1 families where B-genome chromosome(s) were detected, the BC3 female plant would have been carrying one copy of these chromosomes. Assuming Mendelian segregation in these BC3 plants, it is expected that the chromosomes would segregate in the BC3S1 plants in a 1:2:1 ratio. On the basis of the SSR marker data it was not possible to differentiate between the BC3S1 plants carrying one or two copies of a given B-genome chromosome; therefore a 3:1 segregation ratio for the presence or absence of the B-genome chromosome was tested in the BC3S1 families. The transmission rate of B-genome chromosomes from the BC3 to the BC3S1 generation was found to vary depending on the chromosome. For example, 8 of the 11 plants of the family IF-30 inherited B6, and 6 of the 11 plants of the family IF-29 inherited the chromosome B8. We observed that B3 and B6 segregated in a Mendelian fashion, whereas B7 and B8 are clearly selected against.
Cytological tracking of the B-genome chromosomes:
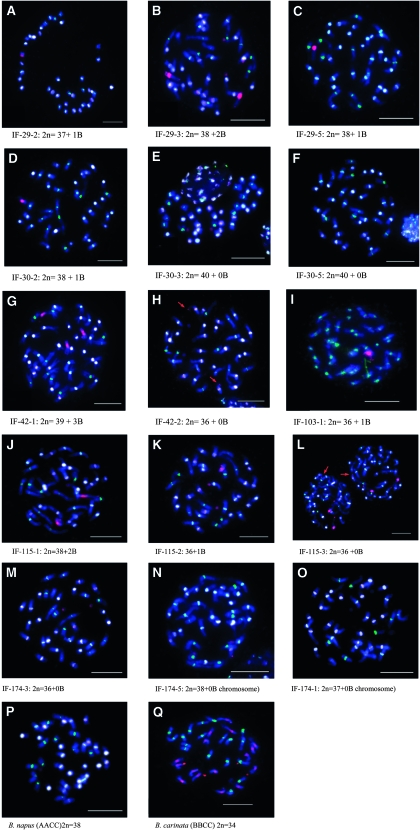
Using in situ hybridization, we determined which BC3 plants containing the different B-genome chromosomes were chromosomal addition lines (Figure 5). Genomic DNA from B. nigra was used to detect the B-genome chromosome content in the BC3S1 families (Figure 5). On the basis of the molecular marker data, it was possible to predict the number of B-genome chromosomes introgressed into the BC3S1 segregating families. The GISH technique allowed visualization of the B chromosomes in these materials. On the basis of the presence of B-genome-specific alleles detected using molecular markers, it was determined that the BC3S1 family IF-29, as well as its corresponding BC3 parent, carried the chromosomes B3, B7, and B8. GISH analysis of three plants from this family showed one or two signals from B-genome chromosomes (Figure 5, A–C), with chromosome counts of 2n = 37 + 1B, 2n = 38 + 2B, or 2n = 38 + 1B (Table 3). The line IF-29-2 was found to carry 37 AC chromosomes (2n = 37 + 1B), apparently due to the loss of an A- or C-genome chromosome and substitution with a B-genome chromosome, while the other lines tested appeared to be addition lines. In the case of the IF-30, signals from B6 were expected to be observed. Although GISH analysis supported this in one of the three plants tested (2n = 38 + 1B) (Figure 5D), the other two plants had 2n = 40 chromosomes with no B genome signals observed. These data suggested that these plants have gained an A- and/or C-genome chromosome due to abnormal chromosome segregation (Table 3 and Figure 5, D and F). The family IF-42 segregated for both B3 and B7 (Table 4). One of two plants from this family produced three signals specific to B-genome chromosomes (2n = 39 + 3B; Figure 5G). On the basis of SSR marker data for this line, either B3 or B7 has fragmented and been introgressed fully in the AACC genomic background (Figure 5G). The other plant from this family (IF-42-2) carried 2n = 36 chromosomes and produced only faint signals from small segments of the B genome (Figure 5H and Table 4). These signals appear to result from the translocation of a small segment of the B7 onto the terminal segment of either an A or a C chromosome (Figure 5H and Table 4). The two plants of IF-103 carried B7, and a single B-genome signal was observed. These plants had 37 AC-genome chromosomes, suggesting the loss of an A or a C-genome chromosome pair and the gain of one B-genome chromosome (Figure 5I and Table 4). In the case of the family IF-115, two B-genome chromosomes, B3 and B7, were expected to be observed. The GISH assay identified two signals in one of the three plants tested, presumably from two B-genome chromosomes, and that plant had a chromosome count of 2n = 38 + 2B (Figure 5J). The second plant carried 37 chromosomes (Figure 5K) with a single signal from either B3 or B7. The third plant of this family had 2n = 36 chromosomes with no observable B genome, indicating the loss of two AC chromosomes (Table 4). On the basis of molecular marker data, it was expected that the chromosome B8 would be present in plants of IF-174. However, no B-genome chromosome signal was detected in any of the three plants studied (Figure 5, M–O). These plants had chromosome numbers of 2n = 36, 37, and 38 (Table 4), indicating that two of these plants had lost AC chromosomes.
Figure 5.—
(A–Q) Chromosome painting at late prophase II with 60× magnification, using the GISH technique. B. nigra genomic DNA is fluorescently labeled in red, B. oleracea genomic DNA is labeled green, and the 45S DNA is labeled white. Arrows show chromosomal fragments. Bars: 10 μm.
TABLE 4.
Summary of GISH assay for individual plants of the five selected BC3S1 families
| BC3S1 plant | Expected B chromosome | No. of red signals | Total chromosome count |
|---|---|---|---|
| IF−29–2 | B3/B7/B8 | 1 | 38 |
| IF−29–3 | B3/B7/B8 | 2 | 40 |
| IF−29–5 | B3/B7/B8 | 1 | 39 |
| IF−30–2 | B6 | 1 | 39 |
| IF−30–3 | B6 | 0 | 40 |
| IF−30–5 | B6 | 0 | 40 |
| IF−42–1 | B3/tip of B7 | 3 | 42 |
| IF−42–2 | B3/tip of B7 | Fragments | 36 |
| IF−103–1 | B7 | 1 | 37 |
| IF−103–2 | B7 | 1 | 37 |
| IF−115–1 | B3/B7 | 2 | 40 |
| IF−115–2 | B7 | 1 | 37 |
| IF−115–3 | B7 | 0 | 36 |
| IF−174–1 | B8 | 0 | 37 |
| IF−174–3 | B8 | 0 | 36 |
| IF−174–5 | B8 | 0 | 38 |
| Westar | None | 0 | 38 |
| Carinata | All | 8 | 34 |
Expected B-genome chromosome on the basis of SSR marker analysis, total chromosome number, and observed number of B. nigra genomic signals are presented.
Distribution of morphological traits in backcross populations:
Morphological traits specific to the B. carinata parent were observed to segregate in all backcross generations (Figure 2). Significant differences (P <0.01), revealed by the ANOVA test, occurred between the introgressed families (IFs) for the following traits: cotyledon retention, leaf incision, blade blistering, days to flowering, stem color, stem length, number of branches, silique length, silique width, days to maturity, and stem color (Table S1). For days to flowering (DTF), the family IF-72 with no B-genome chromosome content had the earliest flowering, with a mean value of 28.9 DTF; while the family IF-29, segregating for three B-genome chromosomes, had the greatest number of DTF with a mean of 41.1. For days to maturity, the family IF-1, in which no B-genome chromosome could be detected, was the latest to mature with a mean value of 79.4 days to maturity; while the family IF-30, carrying B6, was the earliest maturing line, requiring an average 65.0 days (Table S1). For stem color, the family IF-1, with no B-genome chromosomes, had the greenest color whereas the families IF-29 and IF-30 had the highest qualitative value of 3.3 with green–purple stem color (Table S1). The family IF-81, carrying B3, was the tallest, with a mean height of 82.2 cm, while the family IF-72 had the lowest height (58.8 cm) among the introgressed families (Table S1).
The distributions of the BC3S1 plants in each of the 17 families for different morphological traits are presented as boxplots in Figure 3. These boxplots compare the distribution between several sets of data and show the mean, median, maximum, and minimum values in each IF line. For qualitative traits such as leaf division and seed color, there was little diversity within the families (data not presented).
Among the two ripening-related traits, the number of days to flowering showed much less variability, with 50% of the data points within the families being closer to the median, compared to days to maturity, where significantly greater variation was observed (Figure 3). Similarly silique width generally showed less variability within the families compared to silique length (Figure 3). The families IF-29 and IF-30 had a much darker purple stem color, while IF-115 and IF-7 were significantly different in the number of primary branches (data not shown).
The alien B-genome chromosome(s) in the AC-genome background were found to have significant effects on cotyledon retention, leaf margin, leaf incision, days to flowering, stem color, flower color, stem length, beak length, and days to maturity. The R2 values, although small due to the limitations of this study, showing the amount of variation explained by these B-genome chromosomes are presented in Table 5.
TABLE 5.
Effect of the B-genome chromosomes on different morphological traits
| Trait |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LG | Value | CR | MLD | ILD | DTF | STC | FC | STL | NB | SW | BL | DTM | SC |
| B3 | P | NS | * | NS | NS | * | NS | * | NS | NS | NS | NS | — |
| R2 | 0.04 | 0.16 | 0.01 | 0 | 0.05 | 0 | 0.05 | 0 | 0 | 0.03 | 0.02 | 0 | |
| B6 | P | ** | NS | NS | NS | * | * | NS | NS | NS | NS | NS | NS |
| R2 | 0.22 | 0 | 0.02 | 0.03 | 0.06 | 0.08 | 0.01 | 0 | 0.01 | 0.01 | 0.01 | 0 | |
| B7 | P | NS | * | * | NS | NS | * | NS | NS | NS | * | NS | NS |
| R2 | 0.03 | 0.23 | 0.08 | 0 | 0.03 | 0.06 | 0.02 | 0.02 | 0.02 | 0.05 | 0.02 | 0.03 | |
| B8 | P | NS | — | NS | * | NS | * | NS | NS | NS | NS | * | — |
| R2 | 0 | 0 | 0.02 | 0.07 | 0.01 | 0.06 | 0 | 0.03 | 0.02 | 0.03 | 0.05 | 0 | |
R2 values explain the amount of phenotypic variation due to the B-genome chromosome. CR, cotyledon retention; MLD, margin leaf division; ILD, incision of leaf division; DTF, days to flowering; STC, stem color; FC, flower color; STL, stem length; NB, number of primary branches; SW, silique width; BL, beak length; DTM, days to maturity; SC, seed color. *significant at p < 0.01; **significant at p < 0.05.
The B-genome chromosome B3 was found to have a significant effect on leaf margin, explaining 16% of the phenotypic variation. This linkage group (LG) also had a minor but significant effect on stem color and stem length, explaining 5% of the total phenotypic variation for these traits (Table 5). The chromosome B6 explained 22% of the total phenotypic variation for cotyledon retention and had a minor but significant effect on stem color and flower color, explaining 6 and 8% of the phenotypic variation, respectively (Table 5). While B7 explained 23% of the variation for leaf margin, this chromosome also had significant effect on leaf incision, flower color, and beak length, explaining 8, 6, and 5% of the variation, respectively (Table 5). The only chromosome having a significant effect on days to flowering was B8, which explained 8% of the total phenotypic variation. This chromosome also explained 6 and 8% of the variation for flower color and days to maturity, respectively (Table 5).
DISCUSSION
An understanding of the effect of whole chromosome, chromosome segment, and gene introgression has been essential to the analysis of key agronomic traits. The Mi gene that was introgressed from Lycopersicon peruvianum into L. esculentum is the key gene providing nematode resistance in tomato and its value has been estimated at $3 billion US per year (Milligan et al. 1998; Rossi et al. 1998; Seah et al. 2004). In the genus Brassica, the Rfo restorer gene from radish (Raphanus sativus) has proven to be the key restorer gene used in the commercial production of hybrid canola (B. napus) (Brown et al. 2003). Although there has been great interest in introgressing genes of agronomic traits from the B genome, there has not been a systematic analysis of B-genome introgression into B. napus until now. This has been largely due to lack of tools available to study the chromosome composition of interspecific crosses. In addition, there has been a limit to the extent that B-genome chromosomes will pair with their A- and C-genome homeologous counterparts, leading to a total loss of chromosomes or introgression of entire chromosomes.
This study is part of our efforts to construct B-genome addition lines, to facilitate the use of Brassica species as a model system for studying allopolyploidy, and to introgress traits from distant relatives into crops. Analysis of the BC3S1 segregating families with a combination of molecular markers and GISH assays provided an opportunity to study the inheritance of the alien B-genome chromosomes. We observed that B-genome linkage groups were largely maintained and inherited as intact segments in the BC3 plants and BC3S1 families (Figures 4 and 5 and Table 4). There was limited evidence for intergenomic recombination between the B-genome chromosomes and the A- or C-genome chromosomes (Figure 4, A–C). Different B-genome chromosomes were detected in the BC3S1 families that appeared to be associated with significant differences for a number of morphological and agronomic traits. High levels of segregation distortion for the B-genome chromosomes were observed in this study at the BC3 level. This is due to the aneuploid nature of the BC3 plants, which can be explained by the interspecific origin of the material and the fact that homeologous chromosomes from the B and A genomes do not pair very frequently (Parkin and Lydiate 1997; Ky et al. 2000; Lorieux et al. 2000; Mason et al. 2010a). This was previously reported in interspecific Brassica hybrids that contained distantly related genomes (Chèvre et al. 1998, 2007).
Certain B-genome linkage groups appeared to segregate normally in the BC3S1 families, which suggests that they must be forming pairing structures during meiosis, presumably through homeologous interactions. Ten of 16 of the lines analyzed by GISH had even chromosome numbers, a slight bias toward maintaining a balanced nucleus, which can be explained by diploid-like meiotic behavior in allopolyploid species, which is genetically controlled (Cifuentes et al. 2010). It has been shown in resynthesized B. napus that the first meiosis promotes a lot of meiotic driven genetic changes and genome rearrangements, which are transmitted to the progeny (Szadkowski et al. 2010). The progeny are then naturally selected in favor of the establishment and maintenance of fertile natural allopolyploids to eliminate further aberrant meiotic behavior that would reduce fertility (Gaeta and Pires 2010). However, meiosis in interspecific hybrids of B. napus × B. carinata is disturbed and results in homologous and homeologous chromosome exchange, as revealed in microspore-derived plants derived from unreduced gametes from the hybrids (Nelson et al. 2009).
Recently, FISH based on a repetitive DNA marker from the B genome was used to show B-genome introgressions in backcross progeny of B. napus × B. juncea (Schelfhout et al. 2006). However, the absence of locus-specific molecular markers prevented the identification of B-genome linkage groups that were retained. In the current study, although the GISH assay allowed the B-genome chromosomes to be distinguished from those of the A and C genome, it could not differentiate the linkage groups or detect translocations. Conversely, although SSRs have been used to determine allele copy number (Mason et al. 2010b), they do not normally provide information on the exact copy number of chromosomes or indicate whether they are addition or substitution lines. However, a combination of these two approaches allows both linkage group differentiation and chromosomal copy number to be determined. It is not possible to identify specific B-genome linkage groups using GISH with B. nigra genomic DNA, since the probe paints the centromeric area of any B-genome chromosome (Navabi et al. 2010). To distinguish and visualize different B-genome chromosomes, one needs to use probes of BAC clones specific for a particular B-genome chromosome (Hasterok et al. 2005; Howell et al. 2008; Xiong et al. 2010).
Due to the nature of the cross, we could recognize lines with B chromosomal segments and those without any. In some cases, lines without B chromosomes had either lost or gained an A or a C chromosome, which might not be surprising, considering the use of a newly resynthesized B. napus line, the number of backcross generations, and the fact that the A and C chromosomes can pair readily (Attia et al. 1987; Parkin and Lydiate 1997). Future studies are needed to identify which chromosomes pair in these backcrosses, similar to the approach taken by Howell et al. (2008) to detect the A7/C6 translocation in B. napus var. “Westar.” Hence for future work, we would fully characterize the A and C component of the Brassica genome in these B-genome addition, substitution, and introgression lines.
Using the allele size amplified by the primers for a specific SSR marker and the available reference maps, we were able to associate almost all of the SSR markers used in this study to specific A-, B-, or C-genome LGs. While the BC1 and BC2 families showed some evidence of preferential retention of B-genome chromosomes, the BC3 B-genome chromosomes were inherited with an average frequency of 27%. This observation was reflected in the allelic segregation distortion observed for the KCN-10 mapping population. The loci associated with the B genome in the mapping population were inherited at frequencies that were significantly different from normal Mendelian segregation with the B-genome alleles being preferentially lost during the BC2 meioses. We also observed that there are differences in the frequency with which certain chromosomes were retained; for instance, B3 and B6 are present in a higher percentage of lines than B1 and B4 (Table 3).
Recombination frequency of B-genome chromosomes:
Recombination between B-genome chromosomes and chromosomes from the A or C genomes was not observed, with the exception of the terminal tip of B5. Rather, B-genome loci were generally inherited as coincident blocks. Figure 4B illustrates the initial maintenance of whole B-genome chromosomes and the progressive loss of terminal segments through the three generations of backcrossing. In some instances such as for B3 in IF-29 and IF-42 (Figure 4C), the small retained segment of the chromosome may have translocated onto another LG; however, identifying which LG will require more extensive studies. We reported a similar pattern of chromosome inheritance in our previous work in a smaller population (Navabi et al. 2010).
In contrast, the B. carinata C genome was inherited in small dispersed fragments, as would be expected via homologous recombination. The observation of normal homologous pairing between the C genome of B. carinata and effectively the C genome of B. napus indicates that the C genome of B. carinata has undergone limited or no major chromosomal rearrangement since the fusion of the B and C genomes. We observed selection against the B. carinata C-genome chromosomes during backcrossing to B. napus (Table 2); the C-genome alleles were underrepresented, relative to the C-genome alleles from the recurrent B. napus parent at the BC2 and BC3 generations. As studied before, this can be highly influenced by the genotype of either parent of the hybrid (Mason et al. 2010a,b). Homeologous pairing between the three genomes of Brassica in interspecific crosses is complexly influenced by genome structure and allelic composition (Mason et al. 2010a).
Applications and future insights:
The steps involved in QTL mapping include identifying the significant QTL, positioning them in the genome, and exploring the effect of different QTL combinations (Darvasi 1998). However, mapping of quantitative traits in populations derived from interspecific crosses can be very complex, due to segregation distortion (Lorieux et al. 2000). Although interval mapping could not be applied to this data set, as recombination between the B- and AC-genome chromosomes was highly restricted, we used a marker regression method for QTL mapping that detects the association of the trait value and the genotype for a single locus (in this case a full chromosome). From a plant breeding perspective, the material generated and characterized in this study is an excellent source of diverse genotypes, which possess valuable traits such as early flowering, early maturity, and number of seeds per silique, plus other traits. Although it should be noted that this material also carries some negative traits such as high glucosinolate content in the seed (data not shown). Such linkage drag, if associated with a desired trait, will need to be addressed in future research using this germplasm. Recurrent crossing with selection for the desired trait(s) often leads to breaking of the large chromosomal segments and can be used to generate desirable lines.
In summary, the current genetic material allowed us to track the B-genome content in a collection of B. napus introgression lines. Perhaps not surprisingly, the C genome of B. carinata was able to recombine normally with the C genome of B. napus. However, the B-genome linkage groups were, in almost all cases, inherited as single chromosomes. The one exception was a small terminal region of B5, which we believe may have been translocated onto one of the A- or C-genome linkage groups. The material generated in this program represents a large publicly available resource of interspecific germplasm with known B/C-genome content and we are currently focusing on specific lines with known B-genome content, with a view to introgressing specific regions of the different chromosomes.
Acknowledgments
We thank Reinhold Mayerhofer, Erica Wheeler, and Andreas Madlung for their valuable comments on the manuscript and also Derek Lydiate, Erin Higgins, and Christine Sidebottom for their assistance with the AAFC reference maps. This work was funded in part by a National Sciences and Engineering Research Council Discovery Grant (to A.G.G.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.124925/DC1.
References
- Attia, T., and G. Robbelen, 1986. Cytogenetic relationship within cultivated Brassica analyzed in amphihaploids from the 3 diploid ancestors. Can. J. Genet. Cytol. 28 323–329. [Google Scholar]
- Attia, T., C. Busso and G. Robbelen, 1987. Digenomic triploids for an assessment of chromosome relationships in the cultivated diploid Brassica species. Genome 29 326–330. [Google Scholar]
- Axelsson, T., C. M. Bowman, A. G. Sharpe, D. J. Lydiate and U. Lagercrantz, 2000. Amphidiploid Brassica juncea contains conserved progenitor genomes. Genome 43 679–688. [PubMed] [Google Scholar]
- Brown, G. G., N. Formanova, H. Jin, R. Wargachuk, C. Dendy et al., 2003. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 35 262–272. [DOI] [PubMed] [Google Scholar]
- Chèvre, A. M., P. Barret, F. Eber, P. Dupuy, H. Brun et al., 1997. Selection of stable Brassica napus B-juncea recombinant lines resistant to blackleg (Leptosphaeria maculans). 1. Identification of molecular markers, chromosomal and genomic origin of the introgression. Theor. Appl. Genet. 95 1104–1111. [Google Scholar]
- Chèvre, A. M., F. Eber, A. Baranger, G. Hureau, P. Barret et al., 1998. Characterization of backcross generations obtained under field conditions from oilseed rape wild radish F-1 interspecific hybrids: an assessment of transgene dispersal. Theor. Appl. Genet. 97 90–98. [Google Scholar]
- Chèvre, A. M., H. Ammitzboll, B. Brekling, A. Dietz-Pfeilstetter, F. Eber et al., 2004. A review on interspecific gene flow from oilseed rape to wild relatives, pp. 235–251 in Introgression From Genetically Modified Plants Into Wild Relatives, edited by H. C. M. Den Nijs, D. Bratsch and J. Sweet. CABI Publishing, Cambridge, MA.
- Chèvre, A. M., K. Adamczyk, F. Eber, V. Huteau, O. Coriton et al., 2007. Modelling gene flow between oilseed rape and wild radish. I. Evolution of chromosome structure. Theor. Appl. Genet. 114 209–221. [DOI] [PubMed] [Google Scholar]
- Cifuentes, M., L. Grandont, G. Moore, A. M. Chèvre and E. Jenczewski, 2010. Genetic regulation of meiosis in polyploid species: new insights into an old question. New Phytol. 186 29–36. [DOI] [PubMed] [Google Scholar]
- Darvasi, A., 1998. Experimental strategies for the genetic dissection of complex traits in animal models. Nat. Genet. 18 19–24. [DOI] [PubMed] [Google Scholar]
- Desloire, S., H. Gherbi, W. Laloui, S. Marhadour, V. Clouet et al., 2003. Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 4 588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixelius, C., and S. Wahlberg, 1999. Resistance to Leptosphaeria maculans is conserved in a specific region of the Brassica B genome. Theor. Appl. Genet. 99 368–372. [Google Scholar]
- Gaeta, R. T., and C. J. Pires, 2010. Homoeologous recombination in allopolyploids: the polyploid ratchet. New Phytol. 186 18–28. [DOI] [PubMed] [Google Scholar]
- Gerdemannknorck, M., S. Nielen, C. Tzscheetzsch, J. Iglisch and O. Schieder, 1995. Transfer of disease resistance within the genus Brassica through asymmtric somatic hybridization. Euphytica 85(1-3): 247–253. [Google Scholar]
- Gill, B. S., and G. Kimber, 1977. Recognition of translocations and alien chromosome transfers in wheat by Giemsa C-banding technique. Crop Sci. 17 264–266. [Google Scholar]
- Gomez-Campo, C., 1980. Morphology and morpho-taxonomy in the tribe Brassiceae, pp. 3–31 in Brassica Crops and Wild Allies, edited by S. Tsunoda, K. Hinata and C. Gomez-Campo. Japan Scientific Societies Press, Tokyo.
- Hasterok, R., T. Ksiazczyk, E. Wolny and J. Maluszynska, 2005. FISH and GISH analysis of Brassica genomes. Acta Biol. Cracoviensia Ser. Bot. 47 185–192. [Google Scholar]
- Heneen, W. K., and R. B. Jorgensen, 2001. Cytology, RAPD, and seed colour of progeny plants from Brassica rapa-alboglabra aneuploids and development of monosomic addition lines. Genome 44 1007–1021. [DOI] [PubMed] [Google Scholar]
- Howell, E. C., M. J. Kearsey, G. H. Jones, G. J. King and S. J. Armstrong, 2008. A and C genome distinction and chromosome identification in Brassica napus by sequential fluorescence in situ hybridization and genomic in situ hybridization. Genetics 180 1849–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B., Y. Liu, X. Xue and L. Chang, 2002. Comparison of aluminium tolerance in the brassicas and related species. Plant Breed. 121 360–362. [Google Scholar]
- Ibpgr, 1990. Descriptors for Brassica and Raphanus. International Board for Plant Genetic Resources (IBPGR), Rome.
- Kato, A., J. C. Lamb and J. A. Birchler, 2004. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey, M. J., and V. Hyne, 1994. QTL analysis—a simple marker-regression approach. Theor. Appl. Genet. 89 698–702. [DOI] [PubMed] [Google Scholar]
- Kosambi, D. D., 1944. The estimation of map distance from recombination values. Ann. Eugen. 12 172–175. [Google Scholar]
- Kumar, A., P. Singh, D. P. Singh, H. Singh and H. C. Sharma, 1984. Differences in osmoregulation in Brassica species. Ann. Bot. 54 537–541. [Google Scholar]
- Ky, C. L., P. Barre, M. Lorieux, P. Trouslot, S. Akaffou et al., 2000. Interspecific genetic linkage map, segregation distortion and genetic conversion in coffee (Coffea sp.). Theor. Appl. Genet. 101 669–676. [Google Scholar]
- Lagercrantz, U., and D. Lydiate, 1996. Comparative genome mapping in Brassica. Genetics 144 1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J. C., and J. A. Birchler, 2006. Retroelement genome painting: cytological visualization of retroelement expansions in the genera Zea and Tripsacum. Genetics 173 1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux, M., M. N. Ndjiondjop and A. Ghesquiere, 2000. A first interspecific Oryza sativa×Oryza glaberrima microsatellite-based genetic linkage map. Theor. Appl. Genet. 100 593–601. [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Mapping and characterizing of QTLs: inbred line crosses, pp. 431–489 in Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Mahmood, T., M. H. Rahman, G. R. Stringam, F. Yeh and A. G. Good, 2007. Quantitative trait loci for early maturity and their potential in breeding for earliness in Brassica juncea. Euphytica 154 101–111. [Google Scholar]
- Malik, R. S., 1990. Prospects for Brassica carinata as an oilseed crop in India. Exp. Agric. 26 125–129. [Google Scholar]
- Mason, A. S., V. Huteau, F. Eber, O. Coriton, G. Yan et al., 2010. a Genome structure affects the rate of autosyndesis and allosyndesis in AABC, BBAC and CCAB Brassica interspecific hybrids. Chromosome Res. 18 655–666. [DOI] [PubMed] [Google Scholar]
- Mason, A. S., M. N. Nelson, M. C. Castello, G. Yan and W. A. Cowling, 2010. b Genotypic effects on the frequency of homoeologous and homologous recombination in Brassica napus × B. carinata hybrids. Theor. Appl. Genet. 22(3): 543–553. [DOI] [PubMed] [Google Scholar]
- Meng, J. L., S. W. Shi, L. Gan, Z. Y. Li and X. S. Qu, 1998. The production of yellow-seeded Brassica napus (AACC) through crossing interspecific hybrids of B-campestris (AA) and B-carinata (BBCC) with B-napus. Euphytica 103 329–333. [Google Scholar]
- Milligan, S. B., J. Bodeau, J. Yaghoobi, I. Kaloshian, P. Zabel et al., 1998. The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10 1307–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navabi, Z. K., I. A. P. Parkin, J. C. Pires, Z. Xiong, M. R. Thiagarajah et al., 2010. Introgression of B-genome chromosomes in a doubled haploid population of Brassica napus × B. carinata. Genome 53 619–629. [DOI] [PubMed] [Google Scholar]
- Nelson, M. N., A. S. Mason, M. C. Castello, L. Thomson, G. Yan et al., 2009. Microspore culture preferentially selects unreduced (2n) gametes from an interspecific hybrid of Brassica napus L. × Brassica carinata Braun. Theor. Appl. Genet. 119 497–505. [DOI] [PubMed] [Google Scholar]
- Panjabi, P., A. Jagannath, N. C. Bisht, K. L. Padmaja, S. Sharma et al., 2008. Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics 9 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, Y. S., O. K. Song, S. W. Hong, J. M. Kwak, M. J. Cho et al., 1995. Frequent in-frame length variations are found in the diverged simple repeat sequences of the protein-coding regions of 2 putative protein-kinase genes of Brassica-napus. Plant Mol. Biol. 27 829–833. [DOI] [PubMed] [Google Scholar]
- Parkin, I., and D. J. Lydiate, 1997. Conserved patterns of chromosome pairing and recombination in Brassica napus crosses. Genome 40 496–504. [DOI] [PubMed] [Google Scholar]
- Parkin, I., S. M. Gulden, A. Sharp, L. Lukens, M. Trick et al., 2005. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171 765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin, I. A. P., A. G. Sharpe, D. J. Keith and D. J. Lydiate, 1995. Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38 1122–1131. [DOI] [PubMed] [Google Scholar]
- Piquemal, J., E. Cinquin, F. Couton, C. Rondeau, E. Seignoret et al., 2005. Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theor. Appl. Genet. 111 1514–1523. [DOI] [PubMed] [Google Scholar]
- Prakash, S., and V. L. Chopra, 1988. Introgression of resistance to shattering in Brassica napus from Brassica juncea through non-homologous recombination. Plant Breed. 101 167–168. [Google Scholar]
- Purwantara, A., P. A. Salisbury, W. A. Burton and B. J. Howlett, 1998. Reaction of Brassica juncea (Indian mustard) lines to Australian isolates of Leptosphaeria maculans under glasshouse and field conditions. Eur. J. Plant Pathol. 104 895–902. [Google Scholar]
- Rieseberg, L. H., O. Raymond, D. M. Rosenthal, Z. Lai, K. Livingstone et al., 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301 1211–1216. [DOI] [PubMed] [Google Scholar]
- Rossi, M., F. L. Goggin, S. B. Milligan, I. Kaloshian, D. E. Ullman et al., 1998. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. USA 95 9750–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel, S., M. Nicole, F. Lopez, P. Ricci, J. P. Geiger et al., 1999. Leptosphaeria maculans and cryptogein induce similar vascular responses in tissues undergoing the hypersensitive reaction in Brassica napus. Plant Sci. 144 17–28. [Google Scholar]
- Roy, N. N., 1984. Interspecific transfer of Brassica juncea-type high blackleg resistance to Brassica napus. Euphytica 33 295–303. [Google Scholar]
- SAS Institute, 1989. SAS/STAT. SAS Institute, Cary, NC.
- Schelfhout, C. J., R. Snowdon, W. A. Cowling and J. M. Wroth, 2006. Tracing B-genome chromatin in Brassica napus x B-juncea interspecific progeny. Genome 49 1490–1497. [DOI] [PubMed] [Google Scholar]
- Schranz, M. E., M. A. Lysak and T. Mitchell-Olds, 2006. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11 535–542. [DOI] [PubMed] [Google Scholar]
- Seah, S., J. Yaghoobi, M. Rossi, C. A. Gleason and V. M. Williamson, 2004. The nematode-resistance gene, Mi-1, is associated with an inverted chromosomal segment in susceptible compared to resistant tomato. Theor. Appl. Genet. 108 1635–1642. [DOI] [PubMed] [Google Scholar]
- Sharpe, A. G., I. A. P. Parkin, D. J. Keith and D. J. Lydiate, 1995. Frequent nonreciprocal translocations in the amphidiploid genome of oilseed rape (Brassica napus). Genome 38 1112–1121. [DOI] [PubMed] [Google Scholar]
- Slocum, M. K., S. S. Figdore, W. C. Kennard, J. Y. Suzuki and T. C. Osborn, 1990. Linkage arrangement of restriction fragment length polymorphism loci in Brassica oleracea. Theor. Appl. Genet. 80 57–64. [DOI] [PubMed] [Google Scholar]
- Stead, K., 2009. Mapping the introgression of the Brassica carinata C and B genomes into Brassica napus. Ph.D. Thesis. University of Alberta, Edmonton, Alberta, Canada.
- Sun, Z., Z. Wang, J. Tu, J. Zhang, F. Yu et al., 2007. An ultradense genetic recombination map for Brassica napus, consisting of 13551 SRAP markers. Theor. Appl. Genet. 114 1305–1317. [DOI] [PubMed] [Google Scholar]
- Szadkowski, E., F. Eber, V. Huteau, M. Lodé, C. Huneau et al., 2010. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 186 102–112. [DOI] [PubMed] [Google Scholar]
- Tanksley, S. D., and J. C. Nelson, 1996. Advanced backcross QTL analysis: a method for simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor. Appl. Genet. 92 191–203. [DOI] [PubMed] [Google Scholar]
- Tanksley, S. D., N. D. Young, A. H. Paterson and M. W. Bonierbale, 1989. RFLP mapping in plant breeding: new tools for an old science. Nat. Biotechnol. 7 257–264. [Google Scholar]
- Wang, Y. P., K. Sonntag, E. Rudloff, P. Wehling and R. J. Snowdon, 2006. GISH analysis of disomic Brassica napus-Crambe abyssinica chromosome addition lines produced by microspore culture from monosomic addition lines. Plant Cell Rep. 25 35–40. [DOI] [PubMed] [Google Scholar]
- Warwick, S. I., L. D. Black and I. Aguinagalde, 1992. Molecular systematics of Brassica and allied genera (subtribe Brassicinae, Brassiceae): chloroplast DNA variation in the genus Diplotaxis. Theor. Appl. Genet. 83 839–850. [DOI] [PubMed] [Google Scholar]
- Xiong, Z., J. S. Kim and J. C. Pires, 2010. Integration of genetic, physical, and cytogenetic maps for Brassica rapa chromosome A7. Cytogenet. Genome Res. 129 190–198. [DOI] [PubMed] [Google Scholar]