Abstract
Boundary elements or insulators subdivide eukaryotic chromosomes into a series of structurally and functionally autonomous domains. They ensure that the action of enhancers and silencers is restricted to the domain in which these regulatory elements reside. Three models, the roadblock, sink/decoy, and topological loop, have been proposed to explain the insulating activity of boundary elements. Strong predictions about how boundaries will function in different experimental contexts can be drawn from these models. In the studies reported here, we have designed assays that test these predictions. The results of our assays are inconsistent with the expectations of the roadblock and sink models. Instead, they support the topological loop model.
EUKARYOTIC chromosomes are subdivided into a series of functionally, biochemically, and structurally autonomous domains. This was first recognized more than a half century ago in cytological studies on amphibian lampbrush chromosomes and insect polytene chromosomes (Alfert 1954; Gall 1956). Over the intervening period, convincing evidence for the organization of chromosomes into discrete domains that have distinct and characteristic chromatin architectures has come from a combination of genetic, molecular, and biochemical experiments (for reviews, see Udvardy 1999; Gaszner and Felsenfeld 2006; Valenzuela and Kamakaka 2006). A classical example of an active domain is the 35-kb β-globin locus in chicken erythrocytes. The chromatin of this locus differs markedly from nearby transcriptionally repressed domains. In addition to an enhanced sensitivity to DNase I digestion compared to sequences in silent domains, β-globin chromatin has other distinctive properties including lower levels of the linker histone H5 and much higher levels of acetylated histones (Bellard et al. 1980; Stalder et al. 1980; Hebbes et al. 1992; Verreault and Thomas 1993). The patterns of methylation of histone H3 at lysine residues 4 and 9 in the β-globin domain also differ from that of silenced domains (Litt et al. 2001). The features that distinguish the active globin domain from the surrounding inactive domains are evident not only elsewhere in the chromosomes of vertebrates, but also in the chromosomes of many other eukaryotes.
The organization of eukaryotic chromosomes into domains having distinct chromatin architectures and genetic activities requires a mechanism for delimiting the domains. Special elements called boundaries or insulators serve this purpose. These elements define the limits of chromosomal domains and function to establish independent units of gene activity. Elements having these properties were first discovered in Drosophila but have since been found in many organisms ranging from yeast to humans (Gaszner and Felsenfeld 2006; Valenzuela and Kamakaka 2006). While mutations in boundary elements can be associated with unusual phenotypic effects (Keppy and Welshons 1977; Gyurkovics et al. 1990), the biological activities of most boundaries have been investigated using two different transgenic assays. The first assay postulates that boundaries function to establish autonomous units of genetic activity and it tests whether these elements can insulate reporter genes against positive and negative chromosomal position effects. It was found that the reporter must be bracketed by boundary elements to guard against position effects, while a single element either upstream or downstream of the reporter was unable to insulate (Kellum and Schedl 1991; Chung et al. 1993). The second assay was based on the fact that enhancers and silencers are not only rather promiscuous in their regulatory interactions but are also able to regulate genes located many kilobases away, independent of orientation and position. This meant that there must be mechanisms in place that constrain the regulatory activities of enhancers/silencers to their designated target genes. It was suggested that this regulatory problem might be solved if boundaries functioned to restrict enhancer activity to the chromosomal domains in which they reside. This idea was tested using a blocking assay in which boundaries were placed between an enhancer or silencer and a target promoter (Holdridge and Dorsett 1991; Geyer and Corces 1992; Kellum and Schedl 1992; Chung et al. 1993). It was found that boundaries can block regulation when interposed between an enhancer or silencer and the promoter, while they have no effect when located distal to the enhancer or silencer.
Like enhancers/silencers, boundaries are now known to be ubiquitous components of the genome in multicellular eukaryotes. For example, recent studies on the chromosomal distribution of the boundary protein CTCF in humans have identified >13,000 in vivo binding sites (Kim et al. 2007; Xie et al. 2007). In addition, >14,000 putative insulators have been reported in the Drosophila genome (Bushey et al. 2009; Negre et al. 2010). As expected for elements that function to prevent adventitious regulatory interactions, the CTCF sites in the human genome are enriched in intergenic regions that separate differentially expressed genes or gene clusters. CTCF-dependent boundaries in Drosophila have also been shown to demarcate the cis-regulatory domains of the homeotic Bithorax and Antennapedia complexes (Moon et al. 2005; Holohan et al. 2007; Negre et al. 2010).
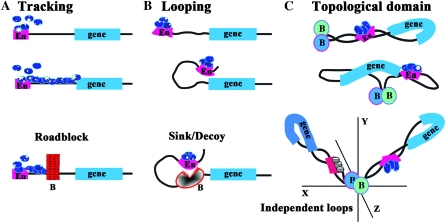
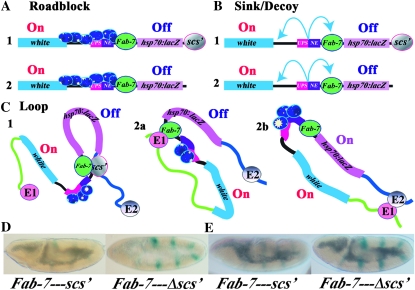
While the importance of boundary elements in delimiting units of independent gene activity has now been well established, how they function to block enhancers (and silencers like the fly Polycomb Response Elements) is not well understood. Three different models have been proposed to explain how boundaries restrict the activity of enhancers. For two of these, it is assumed that the mechanism used by enhancers (or silencers) to act over large distances will dictate how boundaries function and both of these models are based on currently popular ideas for how enhancers contact their target genes (West et al. 2002; Kuhn and Geyer 2003; Schedl and Broach 2003; Gaszner and Felsenfeld 2006). In the first, enhancers (silencers) provide entry points for regulatory machines that track along the chromosome toward the promoter (see Figure 1A). This model has recently come back into favor with the realization that both gene activation and silencing depend upon histone modifications such as methylation and acetylation (Berger 2007). Moreover, as would be expected for complexes that processively alter chromatin structure, many of these histone modifications are not limited to the target promoter, but instead appear to encompass the entire regulatory domain (Noma et al. 2001; Schwartz et al. 2006; Tolhuis et al. 2006). In this model, boundary elements function as roadblocks, blocking the movement of the regulatory complexes along the chromatin fiber (Figure 1A). Roadblock activity could be entirely passive, generated by the assembly of a large nucleoprotein complex at the boundary that disrupts the regular chromatin architecture and prevents the passage of the regulatory machinery (Bi and Broach 1999). The roadblock could also recruit enzymatic activities that locally modify chromatin around the boundary and thus help promote the maintenance of an inactive or active state (depending upon whether the roadblock is blocking an enhancer or silencer, respectively). Evidence for such chromatin-modifying activities has been reported for several yeast and vertebrate boundaries that function to block the spread of repressive heterochromatin (Oki and Kamakaka 2002; West et al. 2004). In this regard, it is interesting to note that in the case of the boundary at the 5′ end of the β-globin locus blocking the spread of heterochromatin is separable from blocking enhancer action. The former appears to involve a “barrier” activity that maintains a local active chromatin environment in the vicinity of the boundary element, while the later depends upon the highly conserved CTCF boundary factor. A similar distinction has been drawn for the gypsy transposon su(Hw) boundary (Kurshakova et al. 2007).
Figure 1.—
Enhancer blocking in the roadblock, sink, and loop domain models. (Top to bottom) (A) In the tracking model, regulatory factors (blue ovals) first assemble at the enhancer (or silencer) (En) and then move processively along the chromatin fiber toward the promoter of the regulatory target. In this model, boundaries (B in section A) act as roadblocks or barriers, preventing the processive movement of the regulatory factors. (B) In the looping model, regulatory factors associate with the enhancer (or silencer) (En). These bound factors then contact the promoter of the regulatory target by the looping over of the chromatin fiber. In this model, boundaries (B in section B) act as sinks, capturing (or repelling) the looping enhancer/DNA complex before it can make contact with the target promoter. Unlike the roadblock model where boundary action is passive, boundaries (B) must actively block regulatory interactions in the looping model. (C) In the topological loop domain model, the chromosome is subdivided by boundaries into a series of topologically independent loops. Enhancers/silencers (En) would contact their regulatory targets by the sliding of the chromatin fiber in the loop against itself (which formally is a combination of both tracking and looping). In this illustration, the enhancer is located 3′ to the transcription unit rather than 5′ as in A and B. Genes are topologically isolated from regulatory elements in adjacent loops, and contacts can only be made by a search in three-dimensional space. In this and subsequent figures, the proximal and distal endpoints of each looped domain are delimited through interactions between boundaries. The endpoints of the looped domain could also be defined by boundary interactions with the nuclear matrix, the chromosome scaffold, or insulator bodies.
The second model for enhancer action postulates that enhancers and silencers contact their target genes by a looping mechanism (see Figure 1B). In this model, the looping of the chromatin fiber brings DNA binding proteins and other factors associated with the enhancer into close proximity with the target promoter (Blackwood and Kadonaga 1998). These factors can then recruit or contact components of the transcriptional apparatus such as Mediator complex, transcription factor II D (TFIID), or PolII or modify the chromatin organization of the promoter and flanking regions to activate or repress transcription. There is now considerable evidence from chromatin conformation capture (3C) experiments that enhancers and their target promoters are in close contact with one other in vivo (Tolhuis et al. 2002; Engel et al. 2008). In this model, the interposed boundary elements must be able to prevent the looping enhancer from making contact with the target gene (Figure 1B). Unlike the roadblock model, preventing the looping enhancer from establishing productive contacts with the target gene demands an active mechanism (Kuhn and Geyer 2003). It has been suggested that the boundary captures and retains the looping enhancer before it can make contact with the target gene by acting as a promoter decoy or sink (Geyer 1997; Blackwood and Kadonaga 1998). The capturing process would likely require the establishment of direct protein:protein contacts between factors associated with the enhancer and the boundary. As boundaries show little specificity in the sorts of enhancers (or silencers) that they can block, this model would require that boundary proteins are quite promiscuous in their potential protein:protein interactions. In other versions of this model, boundaries repel instead of capturing the looping enhancer.
The third mechanism for boundary function is based on the notion that boundaries subdivide the chromosome into a series of topologically independent domains either by interacting with each other or with some nuclear structure(s) (Udvardy and Schedl 1984; Blanton et al. 2003; Bondarenko et al. 2003; Schedl and Broach 2003; Splinter et al. 2006). This idea was first suggested by studies from the Worcel and Laemmli laboratories in the 1970s, which showed that eukaryotic chromosomes are organized into DNA loops of 10–100 kb in length (Paulson and Laemmli 1977; Worcel and Benyajati 1977). In this model, restraining the action of enhancers/silencers and protecting against both positive and negative chromosomal position effects would be a consequence of the subdivision of the chromatin fiber into topologically independent loops. Enhancers or silencers located within a single looped domain could be brought into contact with their target genes by the sliding of the chromatin fiber in the loop against itself (see Figure 1C) or by intraloop contacts. While interactions within a loop are physically constrained, transient contacts between regulatory elements in one loop and a target gene in another loop would require searching in three-dimensional space. In addition, the possibility of independent movement in three-dimensional space could make it more difficult to establish or sustain productive interactions between regulatory elements in one looped domain and potential target genes in another (Rippe 2001; West and Fraser 2005). The idea that boundaries physically restrict enhancer interactions with regulatory targets is supported by chromosome conformation capture (3C) experiments in the mouse H19/Igf2 locus (Engel et al. 2008; Nativio et al. 2009).
The roadblock, sink/decoy, and looped domain models make straightforward predictions about the behavior or functioning boundary elements in different experimental contexts. In the studies reported here, we have used the postulated properties of boundary elements in each model to design assays that test these predictions and point to the likely mechanism of action.
MATERIALS AND METHODS
Fly methods:
Flies were grown on standard cornmeal agar. All crosses were done at 22°. su(Hw)v/TM6, Ubx, su(Hw)f mutants were generously provided by Victor Corces. Transgenic lines were analyzed in a background maternally and zygotically mutant for su(Hw) (Figure 6).
Figure 6.—
Domain definition depends upon boundary identity and relative position. (A) In the starting transgene containing a random segment of Drosophila DNA (from the Bicaudal-D locus) or no DNA (not shown) in between the ftz enhancers and the hsp70:lacZ reporter the ftz enhancers activate both white and hsp70:lacZ. Lines are from Hagstrom et al. (1996). Note that the NE enhancer does not activate white when a competing hsp70:lacZ reporter is accessible. (B) When Fab-7 is placed between the ftz enhancers and the hsp70:lacZ reporter, it subdivides the transgene into two independent regulatory domains. Lines are from Hagstrom et al. (1996). One domain contains white and the ftz enhancers, while the other domain contains the hsp70:lacZ reporter. In this configuration of regulatory domains, the ftz enhancers drive white expression but are blocked from activating hsp70:lacZ expression. (C) The regulatory domains in B are redefined when Fab-8 is interposed between the ftz enhancers and white. In the first example, both ftz enhancers now drive hsp70:lacZ expression, while they no longer activate white. From this pattern of gene activity, we infer that the organization of regulatory domains resembles that in Figure 5, 3b. The ftz enhancers (along with Fab-7) are now in the same regulatory domain as hsp70:lacZ, while white is in a separate regulatory domain. In the second example, the ftz UPS enhancer activates hsp70:lacZ, but not white, while the NE enhancer activates neither of the reporters. From this pattern of gene activity we infer that the organization of regulatory domains at the time when the UPS enhancer is active resembles that in Figure 5, 3b. Later in development, when the NE enhancer is active, the organization of regulatory domains changes and it resembles that shown in Figure 5, 3a. In this case, the ftz NE enhancer is separated from both white and hsp70:lacZ and neither reporter is activated. (D) A similar pattern of domain redefinition is observed when su(Hw) is interposed between the ftz enhancers and white. In 9 of 10 transgenic lines, the ftz UPS enhancer activates hsp70:lacZ, but not white (D, upper and lower panels, Figure 5, 3b), while in the remaining line neither reporter is active (Figure 5, 3a). In midembryogenesis, the Fab-7 boundary is inactive in half of the lines, and the ftz NE enhancer activates hsp70:lacZ but not white (D, top panel). In the remaining lines, Fab-7 boundary activity is reestablished and neither reporter is active (D, bottom panel). (E) Domain redefinition depends upon a functional su(Hw) boundary. In this example, both ftz enhancers drive hsp70:lacZ expression but not white in wild-type flies, while in su(Hw) mutant flies where the su(Hw) boundary is inactive, the ftz enhancers activate white, but not hsp70:lacZ.
Cloning of P-element constructs:
The following boundary fragments were used in the enhancer-blocking assays described in this article:
Fab-7, in the domain definition (ftz-LacZ) assay, a 1.7-kb ApaI–NcoI Fab-7 fragment was used, to allow comparison with the results reported (Hagstrom et al. 1996). In the triple boundary (mini-white) assay, an ∼1.5-kb Fab-7 fragment was used.
Fab-8, an 800-bp Fab-8 fragment was amplified with the following primers:
F8 4620 RI, GGCACAATCAAGAATTCGTTGG
F8 3445 Bam: TGCCTCCGGATCCCGACGGCTGAC.
scs, a 1.7-kb scs fragment from (Vazquez and Schedl 1994) was used.
scs′, a 450-bp BamHI–EcoRI fragment from (Kellum and Schedl 1992) was used.
su(Hw), a 370-bp fragment derived from an XmnI–Bsp1286 fragment of the gypsy retrotransposon was used (Hagstrom et al. 1996). This fragment contains 12 degenerate binding sites for Su(Hw).
The boundary reporter transgenes used in these studies were:
Triple boundary (mini-white) constructs:
XN–XN is described in detail in Hagstrom et al. (1996). The XN vector has unique cloning sites upstream (KpnI) and downstream (XhoI, NotI) of a minimal white enhancer that drives white expression in the eyes and in the testes. For a detailed map of XN, see supporting information, Figure S1).
X8–X8 is a derivative of XN, in which the downstream scs′ boundary has been replaced with an 800-bp Fab-8 fragment (see above). To construct X8, XN was digested with EcoRI, the P-element backbone and white enhancer/mini-white fragments were separately gel purified. The P-element backbone was recircularized and then digested with EcoRI, BamHI to remove scs′. An EcoRI, BamHI Fab-8 fragment was ligated into the P-element backbone. This plasmid was then cut with EcoRI, and the white enhancer/mini-white EcoRI fragment was added back into the plasmid in the proper orientation. Other boundary elements were then cloned into the unique restriction sites upstream (KpnI) and downstream (XhoI, NotI) of the white enhancer.
Domain definition (ftz-LacZ) constructs:
pCfhL–pCfhL is described in detail in Hagstrom et al. (1996). pCfhL contains two fushi-tarazu (ftz) enhancers, the upstream element (UPS), and the neurogenic enhancer (NE). Like XN, pCfhL also has unique cloning sites upstream (KpnI) of the ftz enhancers (between the enhancers and mini-white), and downstream of the ftz enhancers (between the enhancers and LacZ). For a detailed map of pCfhL, see Figure S1).
pCfhL(SceI)–pCfhL(SceI) is identical to pCfhL, except a pair of I-SceI sites have been introduced on either side of scs′. To construct pCfhL(SceI), an I-SceI flanked linker with unique SpeI and NsiI sites was generated. The I-SceI linker also had XhoI and MfeI sites 5′ to the I-SceI sites, and SfiI and XbaI sites 3′ to the I-SceI sites. The sequence of the linker was (I-SceI sites in boldface type):
GTTACTCGAGCAATTGATTACCCTGTTATCCCTACTAGTGATGATGATGATATGCATTACCCTGTTATCCCTAGGCCATATGGCCGCAGCGGCCATATAGGCCTCTAGAATTG.
The I-SceI linker was cloned into the XhoI, XbaI sites of pBluescript to generate pBS-SceI. Next, pCfhL was cut with XhoI, and then partially digested with EcoRI, to isolate an hsp70 promoter–LacZ-hsp70 trailer fragment. This fragment was cloned into the XhoI, MfeI sites of pBS-SceI. The XhoI, SfiI hsp70 promoter–LacZ-hsp70 trailer-I-SceI linker fragment was then liberated and cloned into an XhoI, SfiI cut pCfhL backbone to generate pCfhL-SceI-linker. scs′ was amplified out of pCfhL with the following primers and then cloned into pCRII-TOPO (Invitrogen) in order to flank scs′ with SpeI sites:
scs′ for, CGGGAATTCCAACAAAAACTTTGC;
scs′ rev SpeI, ACTAGTCTGTGAAAATAAAATGCCGT.
The scs′ SpeI fragment was then cloned into SpeI cut pCfhL-SceI-linker plasmid in the same orientation as scs′ in pCfhL to generate pCfhL(SceI).
Other boundary elements were then cloned into the unique restriction sites upstream (KpnI) and downstream (XhoI, NotI) of the ftz enhancers.
P-element–mediated transformation:
A total of 0.5 mg/ml of DNA was co-injected with a P-turbo helper plasmid into w1 embryos, and transformants were selected by the presence of eye color. Individual transformants were then backcrossed to w1 flies for at least two generations and then balanced over Bins, CyO, or TM3Ser, to create a balanced stock and to determine the chromosome of insertion.
Deleting scs′ using the I-SceI nuclease:
To delete scs′ from pCfhl(SceI)(X-Fab7-scs′) and pCfhl(SceI)(Fab-8-Fab7-scs′), flies carrying these transgenes were crossed to w; TM3, Sb, P{4}72C flies, which express the I-SceI nuclease under the control of the Drosophila Ubiquitin (Ubi-p63E) promoter (Preston et al. 2006). Potential deletion events were rebalanced and screened by PCR for deletion of scs′. The following primers were used to confirm the deletion of scs′:
pCfhLSceI-F2, CGATGGATATTCAGGTGCGAA;
pCfhlSceI-R1, GAGTGAGACAGCGATATGATTGT.
Scoring eye color phenotypes in the white enhancer-blocking assay:
Balanced transgenic male flies were crossed to w1 females, and progeny hemizygous for the transgene were scored for levels of mini-white expression. Since age and sex both affect levels of mini-white expression, comparisons were made only between flies of the same sex and age. For X-linked transgenes, only females were scored, as the dosage compensation machinery seems to override enhancer blocking in males (Vazquez and Schedl 1994). To determine whether the transgenic lines had a blocking or nonblocking phenotype and to allow comparison with the data from Vazquez and Schedl (1994), Hagstrom et al. (1996), and Schweinsberg and Schedl (2004), four reference lines were used. Nos. 89.98.1 and 101.93.1 are two lines that have the lightest eye color previously classified as nonblocking (Schweinsberg et al. 2004), while nos. 110.115A and 110.185A have the darkest eye color previously classified as blocking. Eye photos were taken with a Nikon FX-35WA camera and Nikon HFX-IIA photosystem on a Nikon SMZ-2T dissecting microscope.
β-Galactosidase stainings:
To determine levels of β-galactosidase expression, transgenic males were crossed to w1 females. Embryos 0–15 hr old were collected from yeasted apple juice plates and stained as previously described (Bellen et al. 1989; Hagstrom et al. 1996), with the following modifications. Heptane was saturated with gluteraldehyde by mixing 10 ml heptane, 7.5 ml PBS, and 2.5 ml gluteraldehyde and vortexing vigorously. The phases were then allowed to separate and the top layer, containing gluteraldehyde-saturated heptane was added to a multi-well dish containing the embryos. Embryos were fixed for 20 min at room temperature, shaking. Embryos were stained for 20–24 hr, then transferred to 30% glycerol and left overnight. Embryos were then mounted on glass slides and pictures were taken using a Nikon DXM200F digital camera on a Nikon Microphot-SA light microscope. Comparisons were made between embryos stained in parallel, in the same dish. Positive and negative control lines were also used in each staining. Lines containing either no insert, or a random DNA insert in the blocking position, 56.33.1 and 25.129.1, respectively, were used as positive controls for β-galactosidase staining, and as standards for a nonblocking phenotype (Hagstrom et al. 1996). Lines containing a 1.2-kb Fab-7 insert, 117.17B, 117.31B, and 117.3B, were used as negative controls for β-galactosidase staining, and as standards for an enhancer-blocking phenotype (Schweinsberg and Schedl 2004).
In situ hybridizations:
In situ hybridizations were done as previously described (Tautz and Pfeifle 1989). Briefly, a probe for white was prepared by in vitro transcription using T7 polymerase in the presence of digoxigenin (DIG)-labeled dNTPs (Roche). The white-containing plasmid was obtained from Jumin Zhou. Embryos 0–15 hr old were collected on apple juice plates. Embryos were washed onto 40-μm mesh cell strainers (BD Falcon) with distilled water (dH20) and then dechorionated by submerging in bleach (Na hypochlorite 7%) for two minutes. Embryos were washed several times with dH20 and then fixed in 4% paraformaldehyde in 1:1 PBS:heptane in a glass scintillation vial for 20 min at room temperature, shaking. After shaking, the bottom (aqueous) layer was removed, and 5 ml of 100% methanol was added. The vials were then shaken and vortexed vigorously, and embryos that settled on the bottom of the vial were transferred to a microfuge tube. Embryos were then rehydrated in a stepwise manner in 25, 50, 75, and 100% PBST in methanol. Following rehydration, embryos were washed five times with PBST and then treated with 10 μg/ml proteinase K (in PBST) for 2 min. Embryos were then refixed in 4% formaldehyde in PBS for 20 min at room temperature, while rocking. The embryos were then washed thoroughly with PBST, washed once with 1:1 PBST:hybridization buffer (50% formamide, 5× SSC, 50 μg/ml heparin, 0.1% Tween 20, 100 μg/ml sonicated salmon sperm DNA), and allowed to prehybridize in hybridization buffer for 2 hr at 55°. The DIG-labeled white probe was diluted 1:100, heated to 80°, added to the embryos, and incubated at 55° overnight to hybridize. The probe was then removed and the sample was washed extensively with hybridization buffer, followed by 1:1 PBST:hybridization buffer, and then five times with PBST. The embryos were then probed with 1:2000 HRP-conjugated anti-DIG antibody (Roche) for 1.5 hr. Upon removal of the antibody, the embryos were washed extensively with PBST and then washed twice with developing solution (0.1 m NaCl, 0.1 m Tris-HCl pH 9.0, 0.05 m MgCl2, 0.1% Tween 20). The embryos in developing solution were transferred to a glass dish and 20 μl of NBT/BCIP solution was added. In situs were developed for between 30 and 120 min. Comparisons were made between groups of embryos that were developed together for the same amount of time. The reaction was stopped by washing twice with PBST. The stained embryos were dehydrated in a series of 25, 50, 75, 90, 95, and 100% ethanol (in PBS) washes, and then washed twice with 100% ethanol. All traces of the ethanol were removed, and the embryos were incubated in 1 ml of methyl salicylate overnight. Embryos were then mounted on glass slides in 100 μl of Permount (Fischer) and pictures were taken using a Nikon DXM200F digital camera on a Nikon Microphot-SA light microscope.
RESULTS
Triple boundary:
Experimental design:
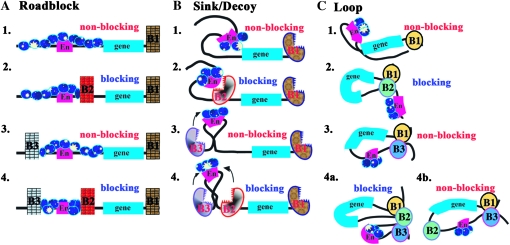
The enhancer-blocking assay tests the ability of a boundary element to prevent regulatory interactions. As illustrated in Figure 2, the starting reporter gene is typically flanked by a downstream boundary (B1) to protect against potential position effects at the site of insertion (1). In all three models, regulatory interactions are blocked when a boundary (B2) is interposed between the enhancer (2 in Figure 2, A–C) and the reporter. Both the roadblock and loop models predict that placing a boundary (B3) upstream of the enhancer instead of in between will have no influence on regulatory interactions (3 in Figure 2, A and C). In the sink/decoy model, boundaries capture or otherwise actively interfere with the regulatory element and, thus, an upstream boundary might be expected to attenuate, at least modestly, enhancer action on the reporter (3 in Figure 2B). Since this has not been observed, it is assumed either that enhancer activity is in excess or that boundaries might repel rather than capture the looping enhancer.
Figure 2.—
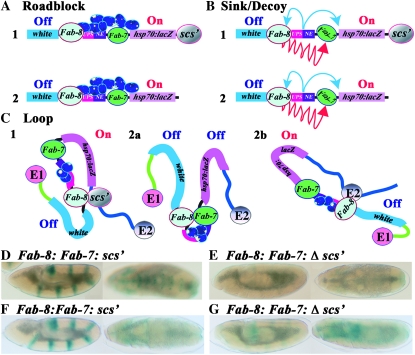
Triple boundary assay. The starting transgene used for the triple boundary assay has a white reporter (“gene” indicated by blue line) and the white enhancer (En, red line). As shown in A–C1, a boundary (B1), either scs′ or Fab-8, is placed downstream of the reporter. Additional boundaries are then introduced into the transgene in between the enhancer and the reporter and upstream of the enhancer as indicated in 2, 3, and 4. As shown in A–C1, the white enhancer is expected to activate white (nonblocking) in all three models when the transgene contains only a single boundary downstream of the reporter. Likewise in A–C2, a boundary placed between the white enhancer and reporter (B2) will block activation (blocking) in all three models. As shown in 3, no blocking (nonblocking) will be observed when a boundary is placed upstream (B3) of the white enhancer in the roadblock (A3) and loop (C3) models. In the sink (B3) model, the upstream boundary should have the potential to capture (arrow) the looping enhancer before it can contact the promoter and thus some attenuation of enhancer activity could be observed. In 4, boundaries are placed upstream of the enhancer (B3) and in between the enhancer and the reporter (B2). In the roadblock model (A4), the addition of an upstream boundary (B3) should have no effect on the blocking activity (blocking) of the interposed (B2) boundary. In the sink/decoy model (B4), flanking the enhancer with boundaries is predicted to have no effect or increase blocking activity (see arrows in 4). In the loop model, blocking activity can be maintained or lost depending on how the loops are defined. As illustrated in C4a, blocking activity will be maintained if the enhancer and gene are in separate loops (via boundary interactions as illustrated, or by interactions with the nuclear matrix/chromosome scaffold). In C4b, blocking will be lost (nonblocking) if the loop is defined by the upstream (B3) and downstream boundaries (B1) (Note that the 3′ end of the white reporter also contains the recently described wari insulator (Chetverina et al. 2008), though it is not indicated here. The presence of wari does not alter the predictions of each model. Moreover, this boundary does not appear to insulate in combination with scs or Fab-7).
It is possible to distinguish between the three models by determining what happens when a boundary located in between the enhancer and promoter is “challenged” by placing a boundary upstream of the enhancer. In the roadblock model, the ability to stop the processive “tracking” of regulatory factors from the enhancer toward the promoter is an intrinsic feature of each boundary and does not depend upon the activity of other nearby boundaries. Thus, the addition of an upstream boundary (B3) should have no effect on the blocking activity of the interposed boundary (B2), and full blocking activity should be retained (4 in Figure 2A). In the sink model, blocking activity is also an intrinsic property of the boundary and depends only upon the ability of the interposed boundary to capture or repel the looping enhancer before it can contact the promoter (4 in Figure 2B). However, since the enhancer would be bracketed by elements that act as sinks or repellents, the addition of an upstream boundary (B3) could accentuate the blocking activity of the interposed boundary (B2).
The loop model differs from the other models in that blocking activity does not depend solely on the intrinsic properties of the boundary element. Instead, because two boundaries are required to define the endpoints of a looped domain, blocking activity will be determined by how a boundary functions in relation to or in combination with the other boundaries in the transgene and with boundaries in the surrounding chromosomal environment. Since the boundaries (and their DNA sequence contexts) are different, some boundary combinations will be better matches than others and for this reason the outcome of the triple boundary assay will depend upon boundary competition. Additionally as the competition between the boundaries included in the transgene will be influenced by endogenous boundaries near the site of insertion, position effects are also to be expected. If the favored boundary combination generates one loop containing the enhancer and a second loop containing the reporter, blocking activity in the triplet (4a in Figure 2C) will be retained. In contrast, if the favored boundary combination generates a loop containing both the enhancer and the reporter (4b in Figure 2C), blocking activity will be lost. The factors that determine which boundary combinations are used to define the regulatory domains will ultimately depend upon how loops are generated. As suggested by Mirkovitch et al. (1984), Yusufzai et al. (2004), and others (Razin et al. 1981; Cockerill and Garrard 1986; Loc and Stratling 1988), two boundaries could generate a loop by interacting with sites in the nuclear matrix or chromosomal scaffold. Alternatively, as proposed by Udvardy et al. (1985), loop formation could be dictated by direct physical interactions between boundary elements. Yet another model for loop formation would be the recruitment of boundaries into specialized, boundary-specific structures, such as the “insulator bodies” (Gerasimova et al. 2000, 2007). For simplicity, loop formation is illustrated in this and subsequent figures (Figures 2, 5, 7, and 8) by physical interactions between the boundaries that define the endpoints of the looped domain.
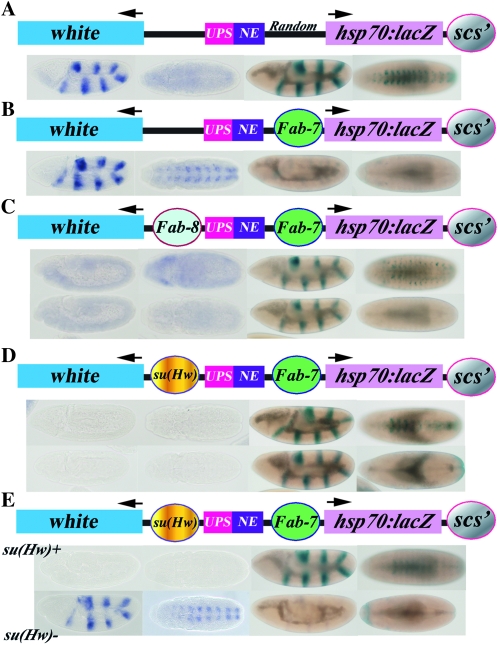
Figure 5.—
Domain definition assay. The transgene in the domain definition assay has white (blue) and hsp70:lacZ (mauve) reporters flanking two enhancers, the UPS stripe enhancer (red) and the NE neurogenic (purple) enhancer, from the ftz gene, which are active during early and midembryogenesis. scs′ is located downstream of the hsp70:lacZ reporter. Additional boundaries are then introduced into the transgene between the ftz enhancers and the hsp70:lacZ reporter (B2) and/or between the ftz enhancers and the white reporter (B3). As shown in A–C1, the ftz enhancers are expected to activate both the white and hsp70:lacZ reporters in all three models when there are no intervening boundaries (On). In (A–C2), a boundary (B2) placed between the ftz enhancers and the hsp70:lacZ reporter is expected to generate two regulatory domains, one containing the hsp70:lacZ reporter and the other containing the ftz enhancers and the white reporter. In this domain configuration, the ftz enhancers will activate the lacZ (Off) but not the white (On) reporter in all three models. Similarly, a boundary (B3) placed between the ftz enhancers and white is expected to block activation of the white but not the hsp70:lacZ reporter in all three models (not shown). As indicated in 3 for the roadblock (A) and sink (B) models, flanking the ftz enhancers with boundaries B2 and B3 will subdivide the transgene into three independent domains, containing, respectively, white, the ftz enhancers, and hsp70:lacZ. In the loop model, domain definition depends on the identity and relative position of the boundary elements in the transgene and in the neighboring chromosomal DNA segments. Among the many possible domain configurations, two different examples are shown for the loop model in C3a and C3b. In C3a, a loop domain is defined by B2 and B3 [interacting with each other as shown, or with some nuclear structure(s)]. This subdivides the transgene into three separate regulatory domains are formed containing, respectively, white, the ftz enhancers and hsp70:lacZ. In this case both reporters are protected from the ftz enhancers. In C3b, a loop domain is defined by B3 and scs′. This subdivides the transgene into a domain containing white, and a domain containing the ftz enhancers, boundary B2 and the hsp70:lacZ reporter. In this case, the ftz enhancers are blocked from activating white, but are not prevented from activating hsp70:lacZ.
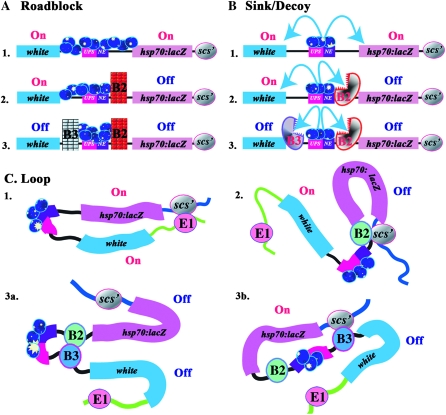
Figure 7.—
Single or in combination? Predictions of the barrier, sink/decoy, and loop models when the downstream scs′ boundary is present or absent are shown in A–C. When scs′ is present (A–C, 1) all three models predict that the Fab-7 boundary will block the ftz enhancer from activating hsp70:lacZ. In the barrier and sink/decoy models blocking activity is an intrinsic property of a boundary. Consequently, as shown in A2 and B2, removing the downstream scs′ boundary should have no effect on Fab-7, and it should still block the ftz enhancers from activating hsp70:lacZ. In contrast, in the loop model, the regulatory domains will be redefined whenever a boundary is removed or added. In the redefined domains, Fab-7 could still block the ftz enhancers from activating hsp70:lacZ (C2a), or Fab-7 blocking activity could be lost (C2b). In the former case, a looped domain would be defined by Fab-7 and an endogenous boundary upstream of the transgene insertion site (E1). In the later case, a looped domain is formed between boundaries upstream (E1) and downstream (E2) of the transgene insertion site. (D and E) UPS-dependent stripe expression (Fab-7–scs′) before and after (Fab-7–Δscs′) for two different transgenic lines. In both lines, Fab-7 blocks the UPS enhancer in the Fab-7–scs′ transgene, and there is only a low level of hsp70:lacZ stripe expression as seen previously for Fab-7 (Hagstrom et al. 1996). However, when scs′ is deleted, UPS-dependent stripe expression is activated.
Figure 8.—
Boundary resurrection. In the roadblock and sink/decoy novel mechanisms must be postulated to account for the loss of Fab-7 blocking activity in the domain definition assay. One mechanism, illustrated for the roadblock model in A1, postulates that when enhancers are tightly confined by two flanking boundaries, excess enhancer activity accumulates and eventually overcomes the weaker boundary. A second mechanism, illustrated for the sink/decoy model in B1, postulates that certain boundaries have a novel ability to neutralize or inactivate (red arrows) other nearby boundaries. A2 and B2 show that the special mechanisms postulated to cause the loss of Fab-7 blocking activity in the barrier and sink/decoy models (confining the ftz enhancers or boundary inactivation) should be completely indifferent to the presence or absence of a downstream scs′ element. In contrast, the loop model predicts that the configuration of regulatory domains will be redefined whenever a boundary is removed. In C2a, Fab-7 and Fab-8 define a new domain that contains the ftz enhancers. In this domain configuration, Fab-7 boundary activity will be restored. In C2b, the new domain is formed by Fab-8 and an endogenous boundary (E2) downstream of the transgene insertion site. In this configuration, the ftz enhancers, Fab-7 and hsp70:lacZ, are in the same regulatory domain, and the reporter will be activated. (D–G) UPS- and NE-dependent hsp70:lacZ activity before (D and F) and after (E and G) the removal of scs′ for two different transgenic lines.
To test these predictions, we used a white reporter and enhancer to monitor regulatory interactions (Vazquez and Schedl 1994). For boundaries, we selected scs and scs′ from the 87A7 heat-shock locus (Udvardy and Schedl 1984; Kellum and Schedl 1991), Fab-7 (Hagstrom et al. 1996) and Fab-8 (Barges et al. 2000) from the Bithorax complex (BX-C), and su(Hw) from the gypsy transposon (Geyer and Corces 1992). These boundaries differ in the DNA binding proteins thought to be important for their activity: Zw5 contributes to scs activity (Gaszner et al. 1999), while scs′ requires boundary element-associated factor 32 (BEAF) (Zhao et al. 1995). Fab-7 is unusual in that different factors confer boundary activity at different stages of development (Schweinsberg and Schedl 2004). In the early embryo, the Elba factor seems to be especially important, while boundary activity later in embryogenesis and in the adult depends upon distinct, but as yet unknown factors (Aoki et al. 2008). The other Bithorax boundary Fab-8 is a CTCF-dependent boundary (Mohan et al. 2007). Finally, the su(Hw) element requires the DNA binding protein su(Hw) as well as a collection of other proteins [Mod(mdg4), CP190, and dTopors] (Pai et al. 2004; Capelson and Corces 2006; Gaszner and Felsenfeld 2006).
An interposed scs retains blocking activity when challenged with upstream boundary:
In the first experiment, scs′ was placed downstream (B1) of white, while scs was interposed (B2) between the 1-kb white enhancer and reporter (Figure 3A; also see detailed map in Figure S1). In this configuration, scs effectively blocks the white enhancer, and in all transgenic lines examined the flies had a yellow/orange eye color phenotype (Vazquez and Schedl 1994). As the upstream (B3) boundary, we used either su(Hw) or Fab-8. In the roadblock and sink models, placing either of these boundaries upstream of the white enhancer should have no effect on blocking activity or might even strengthen it. As for the loop model, 3C experiments indicate that the endogenous scs and scs′ are physically associated in vivo (Blanton et al. 2003). If these scs–scs′ interactions are recapitulated in the triple boundary transgene or if the two elements generate loops by interacting with sites in the nuclear matrix or by targeting to insulator bodies, the blocking activity of scs should be unaffected by the upstream su(Hw) or Fab-8 boundaries (Figure 2, C4a). However, it is also possible that scs′ will preferentially combine with su(Hw) or Fab-8, looping out scs. In this case, the enhancer and reporter would be in the same loop (Figure 2, C4b) and blocking activity would be lost.
Figure 3.—
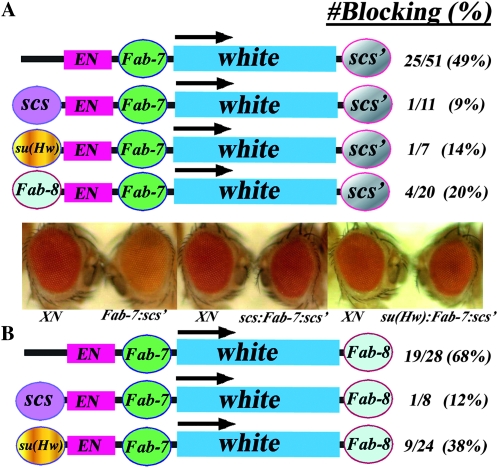
Triple boundary with scs or Fab-8 in the middle. (A) scs in between the white enhancer and the white reporter. When scs is interposed between the enhancer and reporter, blocking is observed in all of the lines (3 X-scs-scs′ lines were reported in Vazquez and Schedl (1994). scs is then challenged by placing su(Hw) or Fab-8 upstream of the enhancer. In each case, blocking is retained in the majority of the transgenic lines. The photographs show eyes of flies transformed with the starting transgene XN (Hagstrom et al. 1996), which has scs′ downstream of the white reporter but no other boundaries and flies transformed with either the su(Hw):scs:scs′ transgene or the Fab-8:scs:scs′ transgene as indicated. Flies transgenic for the starting transgene, XN, have red eyes, while flies transgenic for either su(Hw):scs:scs′ (8/9) or Fab-8:scs:scs′ (20/21) have yellow to orange eyes as illustrated. (B) Fab-8 in between the white enhancer and the white reporter. When Fab-8 is interposed between the enhancer and the reporter, blocking is observed in ∼80% of the lines and the eye color ranges from light yellow to orange (like that illustrated for su(Hw)scs:scs′ and Fab-8:scs:scs′ in A). Half of the lines retain blocking when Fab-8 is challenged by an upstream scs (6/12) and have yellow to orange eye color. None of the lines (0/6) show blocking when Fab-8 is challenged with su(Hw). Photographs in B show examples of two of the nonblocking lines for scs:Fab-8:scs′.
As can be seen in Figure 3A, placing either su(Hw) or Fab-8 in the upstream position has little effect on scs boundary activity, and in both cases, blocking is still observed in the vast majority of the lines. These results are consistent with all three models.
Blocking activity of an interposed Fab-8 is compromised by the upstream boundary:
We next interposed the BX-C CTCF-dependent boundary Fab-8 between the white enhancer and reporter. When coupled with scs′ at the downstream position, Fab-8 is a somewhat less effective enhancer blocker than scs and the white enhancer is blocked in ∼80% of lines. We then challenged Fab-8 with either scs or su(Hw). In the roadblock and sink models, the outcome is expected to be the same as that predicted when scs is challenged by an upstream Fab-8 and su(Hw) boundary—blocking activity should be retained. In the loop model, blocking the intervening Fab-8 boundary will be bypassed if the favored boundary combination is between scs and scs′ as would be suggested by the results described in the previous section. Similarly, since Yamaguchi et al. (2001) reported that there are genetic interactions between BEAF and the gene encoding the Su(Hw) protein, it is possible that scs′ will preferentially combine with su(Hw) rather than Fab-8, and Fab-8 blocking activity will be lost.
The results of these experiments are shown in Figure 3B. When Fab-8 is challenged by scs, its blocking activity is not detected in half of the lines and the eye color is comparable to that in control constructs lacking an interposed boundary. In addition, as shown in Figure S2, in the lines that still show blocking, there is a shift in the phenotypic spectrum toward a darker eye color. This would suggest that even in these lines the upstream scs element competes with Fab-8 and weakens its blocking activity. The effects of an upstream su(Hw) element are equivalent if not more dramatic than scs. As indicated in Figure 3B, blocking activity is absent in all six of the su(Hw):wen:Fab8:white:scs′ lines that were recovered. These findings do not fit with the predicted behavior of boundary elements in either the roadblock or sink models. They would, however, be compatible with the predictions of a model that invokes loop formation as the mechanism of boundary function.
Fab-7 blocking activity is compromised by an upstream boundary:
To further test the predictions of the roadblock, sink, and loop models, we interposed the CTCF-independent BX-C boundary, Fab-7, between the white enhancer and reporter and then challenged with an upstream boundary (see Figure 4A). When coupled with scs′ downstream of the white reporter, Fab-7 boundary activity is subject to chromosomal position effects and only about half of the lines have blocking activity. As was seen for the interposed Fab-8 element, the introduction of a boundary at the upstream position (B3 in Figure 2) weakens Fab-7 blocking activity. When scs is placed upstream, enhancer-blocking activity is only maintained in ∼10% of the transgenic lines. This is close to the background frequency of wen:white:scs′ transgenic lines that have a lighter eye color phenotype. An equivalent reduction in Fab-7 blocking activity is observed when su(Hw) is placed in the upstream position.
Figure 4.—
Triple boundary with Fab-7 in the middle. (A) Fab-7 in between the white enhancer and the white reporter and scs′ downstream. When Fab-7 is interposed between the enhancer and reporter, blocking is observed in 50% of the transgenic lines (Hagstrom et al. 1996); compare XN to Fab-7:scs′ on the left. Fab-7 is then challenged by placing scs (see scs:Fab-7:scs′ in the middle), su(Hw) (see su(Hw):Fab-7:scs′ on the right) or Fab-8 (not shown) upstream of the enhancer. In all three cases, Fab-7 blocking is compromised, and the number of transgenic lines showing blocking activity is reduced. (B) Fab-7 in between the white enhancer and the white reporter with Fab-8 rather scs′ downstream of the reporter. Replacing the downstream scs′ with Fab-8 improves Fab-7 blocking activity and instead of 50%, ∼70% of the transgenic lines have blocking activity. When scs is placed upstream of the white enhancer, Fab-7 blocking activity is again absent in most of the transgenic lines. While Fab-7 blocking activity in this transgene is also reduced when su(Hw) is placed upstream of the white enhancer, the effects of su(Hw) are not as strong as those observed in the transgene that has scs′ downstream of the reporter.
The upstream scs and su(Hw) boundaries and the downstream scs′ boundary are derived from chromosomal DNA segments (or transposons) that have no apparent functional relationship to the homeotic genes or cis-regulatory domains of the BX-C. Thus, it seemed possible that the deleterious effects of the upstream boundaries on Fab-7 and Fab-8 blocking activity might be an anomaly arising from some sort of incompatibility between BX-C boundaries and boundaries derived from elsewhere in the fly genome. To explore this possibility, we placed the Fab-8 boundary in the upstream position. As shown in Figure 4A, the Fab-8 boundary functions in this context much like an upstream scs or su(Hw) element and substantially reduces the enhancer-blocking activity of the interposed Fab-7 boundary.
To examine the compatibility question further, we replaced the scs′ element downstream of the white reporter with Fab-8. As shown in Figure 4B, the frequency of blocking Fab-7 transgenic lines increases to slightly more than two-thirds when Fab-8 is downstream of the reporter. We then introduced scs and su(Hw) upstream of the w enhancer. While a downstream Fab-8 afforded little protection against scs, the effects of an upstream su(Hw) element are partially ameliorated and the frequency of lines retaining blocking activity when Fab-8 is in the downstream position is increased compared to that observed when scs′ was located downstream. Taken together with results described above, these findings indicate the blocking activity of a boundary element depends upon the configuration and identity of the flanking boundaries.
Domain definition:
Experimental design:
The behavior of boundaries in the triple boundary assay does not conform to the predictions of either the roadblock or sink models; instead, the changes in blocking activity induced by altering the boundary configuration would be most consistent with a mechanism involving topologically independent looped domains. To further distinguish between the three models, we devised a domain definition assay. In this assay, two reporters, white and hsp70:LacZ, flank the 4-kb upstream (UPS) and neurogenic (NE) enhancers from the fushi tarazu (ftz) gene. The UPS enhancer drives expression in seven stripes during early embryogenesis, while the NE enhancer drives expression in the embryonic CNS during midembryogenesis (Hagstrom et al. 1996). As shown in Figure 5 (see also detailed map in Figure S1), the BEAF-dependent boundary scs′ was placed downstream of the hsp70:LacZ reporter. When there are no other boundaries present in the transgene, the two reporters and the two ftz enhancers lie in the same regulatory domain. In this configuration, the ftz enhancers can activate both reporters; however, the NE enhancer preferentially turns on the LacZ reporter, presumably because the hsp70 promoter is more compatible with the NE enhancer than the white promoter (see Figure 6A).
The regulatory domain is redefined when a boundary element, such as Fab-7, is interposed between the ftz enhancers and the hsp70:LacZ reporter (2 in Figure 5, A–C). In both the roadblock and sink models, the presence of the intervening Fab-7 boundary places the ftz enhancers and the hsp70:LacZ reporters in different regulatory domains and blocking should be observed (see Figure 5, A2 and B2). In the loop-domain model, domain definition could be influenced by endogenous boundaries near the site of insertion; however, if the loops are defined by a combination of the B2 boundary, Fab-7, with scs′ (see Figure 5, C2), blocking will be observed since the ftz enhancers and the hsp70:LacZ reporter will be in different domains. As is illustrated in Figure 6B, the robust stripe and CNS expression of β-galactosidase seen in the absence of the boundary disappears when Fab-7 is inserted between the enhancers and the hsp70:LacZ reporters. On the other hand, this configuration of boundary elements places the ftz enhancers and the white reporter in the same regulatory domain. As would be predicted by all three models, white is expressed in a stripe pattern during early embryogenesis and in a segmentally repeating pattern in the CNS during midembryogenesis (Figure 6B). [The regulatory domain should also be redefined when a boundary is placed between white and the ftz enhancers (B3). In this case, the ftz enhancers will drive expression of the hsp70:LacZ reporter, while activation of the white reporter should be blocked.]
The three models for boundary function make different predictions about how regulatory domains will be defined when boundaries are interposed simultaneously between the two reporters and the ftz enhancers. In both the roadblock and sink models, boundaries B2 and B3 will generate three independent regulatory domains—the white reporter, the two ftz enhancers, and the hsp70:LacZ reporter—and neither reporter will be active [Figure 5 roadblock (A3) and sink/decoy, (B3)] In the loop model, reporter activity will be determined by which combination of boundaries is used to generate the loops. If three separate loops are formed—the first containing white, the second the two ftz enhancers, and the third the hsp70:LacZ reporter—then both reporters will be off (Figure 5, C3a). On the other hand, if the loop is defined by combining boundaries B3 and scs′ (Figure 5, C3b), the ftz enhancers and the hsp70:LacZ reporter will reside in the same looped domain (together with the unpaired boundary B2) and β-galactosidase expression will be activated. Other sorts of loop configurations and regulatory interactions are possible, depending upon the identity of boundaries B2 and B3 and the potential effects of endogenous boundaries flanking the transgene insertion site. Moreover, if the factors conferring blocking activity in early and midembryogenesis are distinct, as is the case for Fab-7, the domain definition pattern could change from one stage to the other. (In the roadblock and sink models, this would only happen if one of the boundaries is inactive at a specific stage or tissue.)
Challenging Fab-7 with Fab-8:
In the first experiment, a Fab-7 boundary located between the ftz enhancers and the hsp70:lacZ reporter was challenged by placing Fab-8 in between the ftz enhancers and white. Figure 6C shows that the interposed Fab-8 boundary blocks both the UPS and NE enhancers from activating white and in all of the lines examined both white stripe and CNS expression are substantially reduced. While this observation would be consistent with the expectations of the roadblock and sink models, the pattern of β-galactosidase expression is not. We found that Fab-7 blocking of the UPS enhancer is compromised in seven out of eight transgenic lines containing the upstream Fab-8 boundary, and as illustrated for two of the lines in Figure 6C, the UPS enhancer activates robust stripe expression. In addition, the effects of the upstream Fab-8 boundary on Fab-7 blocking activity are tissue and/or enhancer specific. While there is little or no blocking of the stripe enhancer in early embryos, Fab-7 boundary activity seems to be reestablished in midembryogenesis and it is able to block the NE enhancer from activating β-galactosidase expression in the CNS in five out of the seven Fab-8:UPS/NE:Fab-7 lines we tested. In the two remaining lines, blocking activity is compromised and the NE enhancer activates β-galactosidase expression.
Challenging Fab-7 with su(Hw):
In the second experiment, Fab-7 was challenged with the su(Hw) boundary. Consistent with the expectations of all three models, we found that the interposed su(Hw) boundary blocks the ftz enhancers from activating white in all of the transgenic lines that were isolated. On the other hand, as was observed for Fab-8, the effects of incorporating su(Hw) into the transgene are inconsistent with the predictions of both the roadblock and sink models. We found that the Fab-7 boundary is bypassed when it is challenged by the su(Hw) boundary and the UPS enhancer drives β-galactosidase stripe expression in nine out of ten transgenic lines. This is illustrated for two of the lines in Figure 6D. Thus, placing su(Hw) between ftz enhancers and the white reporter effectively abrogates the UPS enhancer-blocking activity of the Fab-7 boundary. The results obtained for the NE enhancer are also inconsistent with the predictions of both the roadblock and sink models. At this point in development, the boundary activity of the Fab-7 element is insertion-site dependent. In half the lines, Fab-7 boundary activity is disrupted by the upstream su(Hw) element and the NE enhancer drives β-galactosidase expression in the CNS (Figure 6D, top). In the other half, Fab-7 boundary activity is apparently unperturbed, and there is a clear reduction in NE-dependent β-galactosidase expression (Figure 6D, bottom).
Fab-7 blocking activity is restored by mutations in su(Hw):
The patterns of domain definition observed when Fab-7 is combined with Fab-8 or su(Hw) are inconsistent with the predictions of both the roadblock and sink models and to account for these findings, some sort of “special” explanation has to be invoked. For example, it is possible that the three-boundary combination targets the transgene to chromosomal sites that are unfavorable for Fab-7 activity especially in early embryos. If this were correct, then the inactivation of Fab-7 in the domain definition reporter would not depend upon the enhancer-blocking function of the Fab-8 or su(Hw) boundary. To test this possibility, we introduced three of the su(Hw)–Fab-7 domain definition inserts into a su(Hw) mutant background. The enhancer-blocking activity of the su(Hw) boundary requires the Su(Hw) protein, and, as would be expected from many previous studies, we found that the UPS and NE enhancers drive white expression in the su(Hw) mutant embryos in all three lines (see example in Figure 6E). In addition to eliminating blocking by the su(Hw) boundary, the su(Hw) mutation also restores Fab-7 boundary activity. The reestablishment of Fab-7 blocking activity was observed in all three transgenic lines and is shown for one of the lines (in which both UPS and NE blocking activity of the Fab-7 element is lost when su(Hw) boundary is functional) in Figure 6E. These findings indicate that special chromosomal position effects are unlikely to be responsible for the loss of Fab-7 blocking activity in the domain definition experiment. They also argue that the Fab-7 boundary in these transgenes is fully “functional” in early and midstage wild-type embryos even in the lines in which no blocking is observed.
Single or in combination?
Blocking activity in the roadblock and sink models is an intrinsic characteristic of the boundary element and is entirely independent of the presence or absence of other nearby boundaries. In contrast, in the topological loop model, boundary function depends upon which boundary element combination is used to delimit the regulatory domain. For this reason, the activity of a boundary in the loop model will be context dependent and determined by the location and nature of other nearby boundaries. It should be possible to further distinguish between these models by testing whether boundaries function as single units or in combination. For this purpose, we generated a modified version of the domain definition transgene (see Figure 7, A–C1,2), which has the Fab-7 boundary in between the ftz enhancers and the hsp70:LacZ reporter and an scs′ element downstream of the reporter that is flanked by SceI sites and can be excised in situ. In the roadblock and sink models, removing the downstream scs′ boundary will expose the hsp70:LacZ reporter to enhancers/silencers located in the chromosomal DNA segment adjacent to the insertion site; however, this should have no effect on Fab-7 blocking of the ftz enhancer (see Figure 7, A2 and B2). In the loop model, the blocking activity of Fab-7 following the removal of scs′ will depend upon how well it functions in combination with endogenous boundary elements upstream (E1) or downstream (E2) of the transgene. As illustrated in Figure 7, C2a, blocking activity could be conserved if Fab-7 is a good match for one of these boundaries; however, if the boundaries located to either side of the transgene combine preferentially with each other, and not with Fab-7, blocking activity will be reduced or lost (Figure 7, C2b).
We compared Fab-7 blocking before and after in vivo excision of scs′, with I-SceI nuclease, in two transgenic lines. As illustrated in Figure 7, D and E, Fab-7 blocking of the UPS stripe enhancer is partially compromised in both lines when scs′ is removed. While blocking of the NE enhancer is retained in both lines after excision of scs′, β-galactosidase is expressed in a nonspecific pattern in older embryos (not shown). Presumably this expression pattern is generated by regulatory elements in the chromosomal DNA segment downstream of each transgene insertion site.
Boundary resurrection:
In the domain definition assay we found that Fab-7 blocking of the ftz enhancers could be compromised by introducing a Fab-8 or su(Hw) boundary upstream of the ftz enhancers. This finding is not readily explained by either the roadblock or sink/decoy model and as noted above would require some novel explanation or mechanism. One mechanism, which would fit best with the roadblock model, is that the loss of Fab-7 blocking activity could be a consequence of confining excessive ftz enhancer activity between two closely spaced boundaries. In the absence of the upstream Fab-8 or su(Hw) boundary, any excess enhancer activity not used to activate white would be dissipated by the tracking of the activator complex through the white gene into the adjacent chromosomal DNA. However, when the ftz enhancers are confined by two closely flanking boundaries, excess activity might build up until it overwhelmed the weaker boundary, (which in these experiments would be Fab-7) and activated the nearby reporter. (In a recast version of the sink/decoy model, boundaries would repel rather than capture the looping enhancer. In this case, when the ftz enhancers are flanked by two boundaries, the “weaker” boundary, Fab-7, would be overcome by the combination of boundary repulsion and enhancer activity.) Alternatively, it is possible that certain boundary elements are able to neutralize other nearby boundaries. This would be a “novel” activity as the properties of boundaries postulated in either the roadblock or sink/decoy model do not require or include such a deactivation function. Moreover, since the normal position dependence for boundary action (the boundary must be placed between the regulatory element and the target gene) is violated when the Fab-7 boundary is neutralized, this novel activity would have to involve mechanisms distinct from those deployed when an interposed boundary blocks enhancer action.
To test these novel mechanisms, we asked whether Fab-7 blocking activity in a domain definition transgene, which contains an upstream Fab-8 element, can be restored by removing the scs′ boundary downstream of the hsp70:LacZ reporter. If Fab-7 blocking activity is lost because of an accumulation of excess enhancer activity, removing scs′ would not be expected to restore Fab-7 blocking activity. The Fab-8 and Fab-7 boundaries would still confine the ftz enhancers and excessive enhancer activity should still accumulate. Similarly, if Fab-7 is inactivated by the upstream Fab-8 boundary, it should still be inactivated when scs′ is removed. In fact, the loss of blocking activity might be even more severe as the data presented in Figure 7 argue that Fab-7 blocking activity depends upon the downstream scs′ boundary. In contrast, in the loop model, the bypass of Fab-7 induced by the upstream Fab-8 or su(Hw) boundary occurs because these boundaries preferentially combine with scs′ (and/or endogenous boundaries near the site of insertion) to generate the looped domain (see Figure 8, C1). As indicated in Figure 8, C2a and C2b, removal of scs′ changes the neighborhood and this should lead to new combinatorial interactions. These combinations could restore Fab-7 blocking activity (Figure 8, C2a). Alternatively, Fab-7 could still be bypassed (Figure 8, C2b).
We examined β-galactosidase expression in Fab-8:Fab-7:scs′ embryos before and after excision of the downstream scs′ boundary and the results for two transgenic lines are shown in Figure 8, D–E and F–G, respectively. In the first line, Fab-7 blocking of the ftz UPS and NE enhancers is compromised by the upstream Fab-8 boundary, and β-galactosidase is expressed in a stripe pattern during early embryogenesis and in the CNS during midembryogenesis (Figure 8D). As can be seen in Figure 8E, excision of the scs′ boundary downstream of the hsp70:LacZ reporter restores Fab-7 blocking of both the UPS and NE enhancers. In the second line, the upstream Fab-8 boundary interferes with Fab-7 blocking of the UPS enhancer, but does not seem to affect blocking of the NE enhancer (though there is background mesoderm/endoderm β-galactosidase expression in this line; Figure 8F). In this case, Fab-7 blocking of the UPS enhancer is restored when scs′ is excised. (Note also that the background β-galactosidase expression in the mesoderm/endoderm seen at later stages also seems to be somewhat enhanced when scs′ is deleted.) Similar results were obtained for two other transgenic lines (not shown). These findings would be inconsistent with the expectations of the “revamped” versions of the roadblock and sink models discussed above, while they would fit the predictions of the loop model.
DISCUSSION
Differentiating between models for boundary element function:
Several findings have linked boundary activity to the higher order organization of the chromatin fiber. The first were studies on the Notch locus, which identified a small deletion, facet-strawberry (faswb), which both altered the structural organization of the N locus in polytene chromosomes, fusing the N chromomere or band 3C7, with the adjacent band, 3C6, and induced a chromosomal position effect that downregulated N activity (Keppy and Welshons 1977; Rykowski et al. 1988). Like the boundaries described here, the faswb sequence functions as a boundary element in transgenic reporter assays (Vazquez and Schedl 2000). The 3C experiments have also shown that the differential regulation of the imprinted H19/Igf2 locus in mouse correlates with the formation of CTCF-dependent parent-of-origin specific loops (Murrell et al. 2004; Engel et al. 2008; Nativio et al. 2009). Similarly, the boundaries flanking the mouse β-globin locus pair with each other to generate a loop domain containing the globin genes and the locus control region (Splinter et al. 2006). In addition, insulator-mediated chromosomal loops have been directly visualized in nuclear halos produced from Drosophila nuclei (Byrd and Corces 2003). It was shown that the protein components of the su(Hw) insulator, Su(Hw) and Mod(mdg4), cofractionate with the nuclear matrix and reside at the bases of loops of DNA. Intriguingly, adding an insulator in the middle of the loop, reorganized the looped domain into two smaller loops, anchored to the nuclear matrix (Byrd and Corces 2003).
Though these and other observations support the idea that boundaries subdivide the chromosome into a series of topologically independent loops, it has generally been assumed that their insulating mechanism will not depend upon this topological isolation, but rather upon how enhancers/silencers act at distance. Current models for enhancer action invoke either a tracking or a looping mechanism (Blackwood and Kadonaga 1998), and in each model boundaries are expected to have rather different properties. In the tracking model, boundaries function passively as roadblocks, preventing regulatory complexes from moving processively along the chromatin fiber toward the promoter. By contrast, in the looping model, boundaries act as sinks or decoys and must actively capture the looping enhancer/silencer before it can make contact with the target gene. While the roadblock and sink/decoy models differ substantially in mechanistic detail, they are similar in that they both presuppose that insulating activity is an intrinsic property of the boundary element and is entirely independent of the nature and location of other nearby boundaries. This is not the case with the topological loop model. Since two boundaries are needed to define the endpoints of the looped domain, this model requires that boundaries function in combination. Because some combinations will necessarily be better than others, the configuration of looped domains, and consequently the insulating activity of a particular boundary, will depend upon the identity and relative location of boundaries in the region of interest. The properties of boundary elements postulated by these three models are distinct and each model makes quite different predictions about how these elements will behave in different experimental contexts. In the studies reported here, we have designed several assays that test whether the attributes of boundary elements conform to those predicted by the three models.
The first, the triple boundary assay (Figure 2), tested for boundary competition. In this assay, a boundary interposed between a white enhancer and a white reporter was challenged by introducing another boundary upstream of the enhancer. We tested five different boundaries placed in different combinations at sites downstream of the reporter, in between the enhancer and reporter, and upstream of the enhancer. Depending upon the combination and configuration of the three boundaries in the transgene, we found that blocking activity was unaffected, weakened, or lost altogether. These findings indicate that boundaries can compete with each other and would only be consistent with the predictions of the topological loop model.
In the domain definition assay (Figure 5), we used two reporters, white and hsp70:LacZ, flanking a pair of ftz enhancers to explore how boundary elements define regulatory domains. Consistent with all three models, when Fab-7 is placed between the ftz enhancers and the hsp70:lacZ reporter, the transgene is subdivided into a domain containing the ftz enhancers and white and a domain containing just the lacZ reporter. However, when an su(Hw) or Fab-8 boundary is then placed between white and the ftz enhancers, in most instances two not three regulatory domains are generated, one containing white and the other containing the ftz enhancers, the hsp70:lacZ reporter and a bypassed Fab-7 boundary. These results fit the predictions of the topological loop model, but not the roadblock or sink model. Moreover, consistent with a strong prediction of the loop model, the bypass of Fab-7 requires a functional su(Hw) boundary. When the su(Hw) boundary is inactivated by mutations in the su(Hw) gene, the regulatory domains in the transgene are redefined and Fab-7 blocking activity is restored.
The third assay tested whether boundaries function singly or in combination. In this experiment, we asked whether Fab-7 blocking of the ftz enhancers in the domain definition transgene depends upon the scs′ boundary located downstream of the hsp70:LacZ reporter. We found excising the scs′ boundary can compromise Fab-7 blocking activity. This result is consistent with expectations of the topological loop model and supports the idea that enhancer blocking requires boundaries to function in combination.
Finally we asked whether the disruption of Fab-7 blocking activity when Fab-8 is placed upstream of the ftz enhancers in the domain definition transgene can be rescued by removing the downstream scs′ boundary. In the topological loop model, context is critical for boundary function and for that reason eliminating one of the competing boundaries might be expected to resurrect blocking activity. Indeed this was the case. We found that excising scs′ can reestablish Fab-7 blocking activity.
Of course, it is possible to envision other novel explanations for why the behavior of boundaries in these different assays does not conform to the predictions of the roadblock or sink/decoy models. For example, since Fab-7 boundary activity in the domain definition transgene can be reestablished by removing the downstream scs′ boundary, it could be argued that neutralization activity is only manifested when the target boundary is bracketed by boundary elements. However, since scs can be placed in the middle position in the triple boundary transgene without being inactivated, this rule of three cannot always apply. On a more general level, introducing novel explanations such as a spill over of excess enhancer activity or boundary inactivation to account for results that do not fit the predictions of the roadblock or sink/decoy model is unsatisfying as these ad hoc activities are not intrinsic to the proposed mechanism of insulator action in either model.
Unlike either the roadblock or sink/decoy model, the topological loop model can readily account for all of the properties of boundaries evident in our assays. Other studies also lend support to the idea that insulators function by topological isolation. Using a transient transfection assay, Ameres et al. (2005) generated two topologically independent loops in a plasmid containing a reporter and two sets of TetR binding sites flanking an SV40 enhancer. Loop formation was induced by expressing TetR proteins that can oligomerize and they found that this was sufficient to prevent the SV40 enhancer from activating the reporter. Similarly, in vitro experiments have shown that the formation of a looped domain that topologically isolates the NtrC-dependent enhancer from the Escherichia coli glnAp2 promoter in a closed supercoiled plasmid is sufficient to disrupt transcriptional activation (Bondarenko et al. 2003). Further support for the idea that boundaries function by subdividing the chromosome into topologically independent looped domains comes from studies on how insulators affect FLP-mediated recombination (Krivega et al. 2010). These authors found that placing a single su(Hw) boundary in between two FRT sites substantially suppresses FLP-mediated recombination. On the other hand, when two (appropriately oriented) su(Hw) boundaries are interposed between the FRT sites, FLP-mediated recombination is enhanced. The authors propose that pairing interactions between the two su(Hw) elements (see below) loops out the intervening DNA and brings the two FRT sites into closer proximity with each other.
Implications for boundary function:
Mechanism of loop formation:
A number of different mechanisms for loop formation have been suggested. These include the association of boundaries with the nuclear matrix or some other supporting and ubiquitous nuclear structure, the recruitment of boundaries into specialized boundary-specific structures such as the insulator bodies, and direct boundary:boundary interactions. There is evidence in the literature supporting each of these mechanisms. For example, su(Hw) boundaries are found associated with the nuclear matrix (Nabirochkin et al. 1998) and have also been shown to physically associate with each other in insulator bodies (Gerasimova et al. 2000; Byrd and Corces 2003). This is also true for CTCF insulators (Murrell et al. 2004; Yusufzai et al. 2004; Splinter et al. 2006; Nativio et al. 2009).
Our assays were not designed to distinguish between the different mechanisms of loop formation; however, the competition between boundary elements evident in the triple boundary and domain definition assays is most easily explained by direct boundary:boundary interactions or by interactions that are mediated by boundary-specific insulator bodies rather than for binding to sites in the matrix. If boundaries were competing for the same sites in the nuclear matrix or some other ubiquitous nuclear structure, then the competition should follow a consistent hierarchical pattern that reflects the relative affinity of the different boundaries for this structure. This does not seem to be the case. In the triple boundary assay, for example, an upstream scs boundary is less effective in competing with Fab-8 in the blocking position than su(Hw). Accordingly, su(Hw) would be expected to have a higher affinity for the matrix/scaffold than scs and this should be reflected in the relative activity of these elements in other experiments. However, an upstream su(Hw) does not compete with scs in the blocking position and is less effective in disrupting blocking than scs when competing against a Fab-7 boundary in the blocking position.
Further support for the idea that specific interactions rather than an association with a ubiquitous structure are responsible for loop formation comes from studies on the “pairing-dependent” bypass of boundaries. This phenomenon was first observed for su(Hw) (Cai and Shen 2001; Muravyova et al. 2001). While a single su(Hw) element interposed between an enhancer and a promoter blocks activation, blocking is lost when there are two su(Hw) elements. Subsequently, many other boundaries have been found to exhibit pairing-dependent bypass; however, bypass appears to require some type of specific interactions as it is largely restricted to homologous boundary pairs (Kuhn et al. 2003; Gruzdeva et al. 2005; Kyrchanova et al. 2007) or to boundaries like su(Hw) and 1A2 that depend upon the same set of protein factors (Maksimenko et al. 2008). Consistent with this conclusion, Kyrchanova et al. (2008) found that bypass is observed for homologous pairs of multimerized Zw5, CTCF, and su(hw) binding sites, but is not observed for heterologous multimer pairs. This specificity for homologous multimer pairs would not be readily explained by interactions of the different boundary proteins with the same ubiquitous matrix/scaffold. With the possible caveat that the boundary bypass phenomenon could reflect some function of boundary elements (such as mediating pairing between sister chromosomes) not directly related to loop formation, these findings would also argue that the protein complexes associated with boundaries must be interacting directly with each other or interacting with their own unique set of cofactors.
Pairwise or multipartite?
The effects of adding and removing boundaries from the transgenes in our assays are mostly easily understood if loop formation tends to involve pairwise boundary combinations. If simultaneous multipartite combinations between the boundaries in the transgenes were possible, then the configuration of regulatory domains (and the pattern of reporter expression) would likely have resembled that predicted in the roadblock model. For example, in the triple boundary transgene with su(Hw) upstream and Fab-8 in the blocking position, a tripartite combination of all three boundaries in the transgene (either through interactions with each other or via recruitment to insulator bodies) should have generated two regulatory domains, one containing the white enhancer bordered by su(Hw) and Fab-8 and another containing the white reporter bordered by Fab-8 and scs′. The finding that a single interposed boundary element has blocking activity, while two copies are bypassed would also argue that boundary interactions must occur in pairs (Cai and Shen 2001; Muravyova et al. 2001). This view is further supported by the finding that blocking activity is restored when a third boundary is added (Kuhn et al. 2003). On the other hand, an important caveat in all of our experiments is that the distances between the boundaries in the transgenes are rather short, ranging from ∼1 kb to 5 kb. For this reason, it is possible that a less than optimal loop size or steric hindrance makes multipartite interactions between these nearby boundaries unfavorable. Other unanticipated factors could also come into play as the loop size goes from small to large. Clearly, further studies will be required to determine whether our transgene design fortuitously emphasizes boundary competition rather than collaboration.
A boundary combination code?
The studies on pairing-dependent boundary bypass taken together with our findings that the blocking activity of a boundary element depends upon the identity and location of its neighbors would support the view that specific interactions determine how each boundary combines with other boundaries to define looped domains. If boundaries form loops through direct boundary:boundary interactions, then the proteins associated with each boundary could play a key role in determining its partner preference. A possible example of direct protein:protein interactions would be between the scs associated Zw5 and the scs′ associated BEAF (Blanton et al. 2003). If on the other hand, boundaries function by association with specific insulator bodies, partner choice might depend upon interactions of the DNA binding proteins with bridging intermediates. For example, CTCF and Su(Hw) are reported to interact with CP190 (Pai et al. 2004; Mohan et al. 2007). In both of these cases, boundary activity in a given context will depend upon the availability of nearby compatible partners. While many of the boundaries identified in Drosophila would appear to be rather promiscuous in their ability to participate in functional combinations, it is likely that boundary activity of some elements has either been “underestimated” or missed altogether because less than optimal partners were used. For example, a full-length 1.2-kb Fab-7 boundary blocks the white enhancer in only about half the lines when matched with scs′ downstream of the white gene; however, blocking enhancer activity in the eye can be substantially augmented by multimerizing a small 200-bp fragment from the distal half of the Fab-7 boundary (Schweinsberg and Schedl 2004). A plausible explanation for the improved blocking activity is that the factors that interact with the multimer are a better match for scs′ than the factors bound elsewhere in Fab-7 and in the multimer these specific factors are present in multiple copies instead of only once. Similarly, we have not detected any boundary activity for the wari insulator at the 3′ end of the white gene in any of our experiments (e.g., Kellum and Schedl 1991; Gohl et al. 2008); however, insulation activity is observed when wari is assayed with appropriate partners or transgene configurations (Chetverina et al. 2008).
Is proximity important?
So far, more than eight proteins have been implicated in boundary function in flies, and it is reasonable to expect that at least several additional proteins will be identified. While there may be a sufficient number of fly boundary proteins to generate a combinatorial code that could be used to help guide preferential boundary combinations, it is not clear whether this is true in vertebrates as fewer proteins have as yet been implicated in boundary function. It is also possible that partner choice is dictated by relative proximity. Supporting this possibility is the finding that scs is more successful in competing with Fab-8 in the triple boundary assay when it is located closer to scs′.
Modulation of regulatory domains:
Intriguingly, stage/tissue-specific effects on Fab-7 blocking activity are evident in the domain definition transgene. In the absence of a challenging boundary, Fab-7 blocks both the ftz UPS stripe enhancer and the NE CNS enhancer. When challenged by either Fab-8 or su(Hw) boundary, Fab-7 blocking of the UPS enhancer is almost always disrupted. However, this is not true for the blocking of the NE enhancer. In most of the Fab-8 lines, Fab-7 blocking of the NE enhancer is maintained, while it is lost in only half of the su(Hw) lines. Fab-7 is not the only example of a boundary that exhibits a chromosomal context-dependent developmental regulation of boundary function. When the Fab-7 boundary at its endogenous location in BX-C is replaced by su(Hw), the boundary activity of su(Hw) becomes stage and tissue dependent. It is active in the ectoderm of early embryos and in adults, but is not active in the CNS of older embryos (Hogga et al. 2001). Since a CNS-specific inactivation of su(Hw) blocking activity is not evident when this boundary is tested in other chromosomal contexts with different enhancers/reporters (e.g., Hagstrom et al. 1996), this effect seems to be specific to insertion into BX-C.
In the case of Fab-7, the stage/tissue-specific alterations in boundary activity likely arise because different cis-acting sequences and trans-acting factors are important for boundary activity at different points in development (Schweinsberg and Schedl 2004). Blocking of the UPS enhancer is mediated by a multiprotein complex, Elba, which is present in 0- to 6-hr embryos (Aoki et al. 2008; T. Aoki, unpublished data). Elba is not, however, found in older embryos and factors that are expressed at later stages and bind elsewhere in Fab-7 are responsible for blocking the NE enhancer in the CNS. Our results would argue that Elba activity is much more sensitive to competition by Fab-8 or su(Hw) than the factors that confer Fab-7 blocking activity later in development. A similar explanation is not likely to apply to the su(Hw) element inserted into BX-C as the proteins known to be important for su(Hw) boundary activity are expressed ubiquitously. The loss of su(Hw) insulator activity in the CNS could be due to tissue-specific changes in the factors associated with the nearby BX-C boundaries, which make them less compatible with su(Hw) insulator. It could also reflect a tissue and chromosomal locus-specific modulation in Su(Hw) DNA binding activity. This possibility is suggested by the differences in the chromosomal distribution of DNA binding boundary proteins like Su(Hw) that are evident in different tissue culture cell lines (Bushey et al. 2009). Additionally, it is known that boundary proteins are subject to a variety of post-translational modifications that could influence their activity (Yu et al. 2004; Capelson and Corces 2005, 2006). However, whatever the mechanism, the redefinition of regulatory domains seen in our experiments [and with su(Hw) inserted into BX-C] raises the possibility that changing the domain organization of the chromosome is used as a mechanism for regulating gene activity during development.
Acknowledgments
We thank Victor Corces, Jumin Zhou, and the Bloomington Drosophila Stock Center for flies and reagents. We also thank Girish Deshpande, Martha Klovstad, Eric Wieschaus, Jim Broach, and the members of the Schedl lab for advice and helpful discussions. This work was funded by GM-043432 from the National Institutes of Health (P.S.), and a predoctoral fellowship from the New Jersey Commission on Cancer Research (D.G.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.123752/DC1.
References
- Alfert, M., 1954. Composition and structure of giant chromosomes. Int. Rev. Cytol. 3 131–176. [Google Scholar]
- Ameres, S. L., L. Drueppel, K. Pfleiderer, A. Schmidt, W. Hillen et al., 2005. Inducible DNA-loop formation blocks transcriptional activation by an SV40 enhancer. EMBO J. 24 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, T., S. Schweinsberg, J. Manasson and P. Schedl, 2008. A stage-specific factor confers Fab-7 boundary activity during early embryogenesis in Drosophila. Mol. Cell. Biol. 28 1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barges, S., J. Mihaly, M. Galloni, K. Hagstrom, M. Muller et al., 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127 779–790. [DOI] [PubMed] [Google Scholar]
- Bellard, M., M. T. Kuo, G. Dretzen and P. Chambon, 1980. Differential nuclease sensitivity of the ovalbumin and beta-globin chromatin regions in erythrocytes and oviduct cells of laying hen. Nucleic Acids Res. 8 2737–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen, H. J., C. J. O'Kane, C. Wilson, U. Grossniklaus, R. K. Pearson et al., 1989. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 3 1288–1300. [DOI] [PubMed] [Google Scholar]
- Berger, S. L., 2007. The complex language of chromatin regulation during transcription. Nature 447 407–412. [DOI] [PubMed] [Google Scholar]
- Bi, X., and J. R. Broach, 1999. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 13 1089–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood, E. M., and J. T. Kadonaga, 1998. Going the distance: a current view of enhancer action. Science 281 60–63. [DOI] [PubMed] [Google Scholar]
- Blanton, J., M. Gaszner and P. Schedl, 2003. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 17 664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko, V. A., Y. I. Jiang and V. M. Studitsky, 2003. Rationally designed insulator-like elements can block enhancer action in vitro. EMBO J. 22 4728–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey, A. M., E. Ramos and V. G. Corces, 2009. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 23 1338–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd, K., and V. G. Corces, 2003. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 162 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, H. N., and P. Shen, 2001. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291 493–495. [DOI] [PubMed] [Google Scholar]
- Capelson, M., and V. G. Corces, 2005. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol. Cell 20 105–116. [DOI] [PubMed] [Google Scholar]
- Capelson, M., and V. G. Corces, 2006. SUMO conjugation attenuates the activity of the gypsy chromatin insulator. EMBO J. 25 1906–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverina, D., E. Savitskaya, O. Maksimenko, L. Melnikova, O. Zaytseva et al., 2008. Red flag on the white reporter: a versatile insulator abuts the white gene in Drosophila and is omnipresent in mini-white constructs. Nucleic Acids Res. 36 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, J. H., M. Whiteley and G. Felsenfeld, 1993. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74 505–514. [DOI] [PubMed] [Google Scholar]
- Cockerill, P. N., and W. T. Garrard, 1986. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell 44 273–282. [DOI] [PubMed] [Google Scholar]
- Engel, N., A. K. Raval, J. L. Thorvaldsen and S. M. Bartolomei, 2008. Three-dimensional conformation at the H19/Igf2 locus supports a model of enhancer tracking. Hum. Mol. Genet. 17 3021–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall, J. G., 1956. On the submicroscopic structure of chromosomes. Brookhaven Symp. Biol. 8 17–32. [PubMed] [Google Scholar]
- Gaszner, M., and G. Felsenfeld, 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7 703–713. [DOI] [PubMed] [Google Scholar]
- Gaszner, M., J. Vazquez and P. Schedl, 1999. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 13 2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova, T. I., K. Byrd and V. G. Corces, 2000. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6 1025–1035. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T. I., E. P. Lei, A. M. Bushey and V. G. Corces, 2007. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol. Cell 28 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer, P. K., 1997. The role of insulator elements in defining domains of gene expression. Curr. Opin. Genet. Dev. 7 242–248. [DOI] [PubMed] [Google Scholar]
- Geyer, P. K., and V. G. Corces, 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6 1865–1873. [DOI] [PubMed] [Google Scholar]
- Gohl, D., M. Muller, V. Pirrotta, M. Affolter and P. Schedl, 2008. Enhancer blocking and transvection at the Drosophila apterous locus. Genetics 178 127–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruzdeva, N., O. Kyrchanova, A. Parshikov, A. Kullyev and P. Georgiev, 2005. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol. Cell. Biol. 25 3682–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyurkovics, H., J. Gausz, J. Kummer and F. Karch, 1990. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 9 2579–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom, K., M. Muller and P. Schedl, 1996. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 10 3202–3215. [DOI] [PubMed] [Google Scholar]
- Hebbes, T. R., A. W. Thorne, A. L. Clayton and C. Crane-Robinson, 1992. Histone acetylation and globin gene switching. Nucleic Acids Res. 20 1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogga, I., J. Mihaly, S. Barges and F. Karch, 2001. Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol. Cell 8 1145–1151. [DOI] [PubMed] [Google Scholar]
- Holdridge, C., and D. Dorsett, 1991. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell. Biol. 11 1894–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan, E. E., C. Kwong, B. Adryan, M. Bartkuhn, M. Herold et al., 2007. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 3 e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum, R., and P. Schedl, 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64 941–950. [DOI] [PubMed] [Google Scholar]
- Kellum, R., and P. Schedl, 1992. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 12 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppy, D. O., and W. J. Welshons, 1977. The cytogenetics of a recessive visible mutant associated with a deficiency adjacent to the notch locus in Drosophila melanogaster. Genetics 85 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T. H., Z. K. Abdullaev, A. D. Smith, K. A. Ching, D. I. Loukinov et al., 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivega, M., E. Savitskaya, I. Krivega, M. Karakozova, A. Parshikov et al., 2010. Interaction between a pair of gypsy insulators or between heterologous gypsy and Wari insulators modulates Flp site-specific recombination in Drosophila melanogaster. Chromosoma 119 425–434. [DOI] [PubMed] [Google Scholar]
- Kuhn, E. J., and P. K. Geyer, 2003. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 15 259–265. [DOI] [PubMed] [Google Scholar]
- Kuhn, E. J., M. M. Viering, K. M. Rhodes and P. K. Geyer, 2003. A test of insulator interactions in Drosophila. EMBO J. 22 2463–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurshakova, M., O. Maksimenko, A. Golovnin, M. Pulina, S. Georgieva et al., 2007. Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol. Cell 27 332–338. [DOI] [PubMed] [Google Scholar]
- Kyrchanova, O., S. Toshchakov, A. Parshikov and P. Georgiev, 2007. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on enhancer-promoter communication. Mol. Cell. Biol. 27 3035–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis and G. Felsenfeld, 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293 2453–2455. [DOI] [PubMed] [Google Scholar]
- Loc, P. V., and W. H. Stratling, 1988. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 7 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimenko, O., A. Golovnin and P. Georgiev, 2008. Enhancer-promoter communication is regulated by insulator pairing in a Drosophila model bigenic locus. Mol. Cell. Biol. 28 5469–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovitch, J., M. E. Mirault and U. K. Laemmli, 1984. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell 39 223–232. [DOI] [PubMed] [Google Scholar]
- Mohan, M., M. Bartkuhn, M. Herold, A. Philippen, N. Heinl et al., 2007. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 26 4203–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, H., G. Filippova, D. Loukinov, E. Pugacheva, Q. Chen et al., 2005. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 6 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova, E., A. Golovnin, E. Gracheva, A. Parshikov, T. Belenkaya et al., 2001. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291 495–498. [DOI] [PubMed] [Google Scholar]
- Murrell, A., S. Heeson and W. Reik, 2004. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 36 889–893. [DOI] [PubMed] [Google Scholar]
- Nabirochkin, S., M. Ossokina and T. Heidmann, 1998. A nuclear matrix/scaffold attachment region co-localizes with the gypsy retrotransposon insulator sequence. J. Biol. Chem. 273 2473–2479. [DOI] [PubMed] [Google Scholar]
- Nativio, R., K. S. Wendt, Y. Ito, J. E. Huddleston, S. Uribe-Lewis et al., 2009. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2–H19 locus. PLoS Genet. 5 e1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre, N., C. D. Brown, P. K. Shah, P. Kheradpour, C. A. Morrison et al., 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6 e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma, K., C. D. Allis and S. I. Grewal, 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293 1150–1155. [DOI] [PubMed] [Google Scholar]
- Oki, M., and R. T. Kamakaka, 2002. Blockers and barriers to transcription: Competing activities? Curr. Opin. Cell Biol. 14 299–304. [DOI] [PubMed] [Google Scholar]
- Pai, C. Y., E. P. Lei, D. Ghosh and V. G. Corces, 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 16 737–748. [DOI] [PubMed] [Google Scholar]
- Paulson, J. R., and U. K. Laemmli, 1977. The structure of histone-depleted metaphase chromosomes. Cell 12 817–828. [DOI] [PubMed] [Google Scholar]
- Preston, C. R., C. C. Flores and W. R. Engels, 2006. Differential usage of alternative pathways of double-strand break repair in Drosophila. Genetics 172 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin, S. V., V. V. Chernokhvostov, A. V. Roodyn, I. B. Zbarsky and G. P. Georgiev, 1981. Proteins tightly bound to DNA in the regions of DNA attachment to the skeletal structures of interphase nuclei and metaphase chromosomes. Cell 27 65–73. [DOI] [PubMed] [Google Scholar]
- Rippe, K., 2001. Making contacts on a nucleic acid polymer. Trends Biochem. Sci. 26 733–740. [DOI] [PubMed] [Google Scholar]
- Rykowski, M. C., S. J. Parmelee, D. A. Agard and J. W. Sedat, 1988. Precise determination of the molecular limits of a polytene chromosome band: regulatory sequences for the Notch gene are in the interband. Cell 54 461–472. [DOI] [PubMed] [Google Scholar]
- Schedl, P., and J. R. Broach, 2003. Making good neighbors: the right fence for the right job. Nat. Struct. Biol. 10 241–243. [DOI] [PubMed] [Google Scholar]
- Schwartz, Y. B., T. G. Kahn, D. A. Nix, X. Y. Li, R. Bourgon et al., 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38 700–705. [DOI] [PubMed] [Google Scholar]
- Schweinsberg, S., K. Hagstrom, D. Gohl, P. Schedl, R. P. Kumar et al., 2004. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168 1371–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg, S. E., and P. Schedl, 2004. Developmental modulation of Fab-7 boundary function. Development 131 4743–4749. [DOI] [PubMed] [Google Scholar]
- Splinter, E., H. Heath, J. Kooren, R. J. Palstra, P. Klous et al., 2006. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 20 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder, J., A. Larsen, J. D. Engel, M. Dolan, M. Groudine et al., 1980. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell 20 451–460. [DOI] [PubMed] [Google Scholar]
- Tautz, D., and C. Pfeifle, 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98 81–85. [DOI] [PubMed] [Google Scholar]
- Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld and W. de Laat, 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10 1453–1465. [DOI] [PubMed] [Google Scholar]
- Tolhuis, B., E. de Wit, I. Muijrers, H. Teunissen, W. Talhout et al., 2006. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 38 694–699. [DOI] [PubMed] [Google Scholar]
- Udvardy, A., 1999. Dividing the empire: boundary chromatin elements delimit the territory of enhancers. EMBO J. 18 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardy, A., and P. Schedl, 1984. Chromatin organization of the 87A7 heat shock locus of Drosophila melanogaster. J. Mol. Biol. 172 385–403. [DOI] [PubMed] [Google Scholar]
- Udvardy, A., E. Maine and P. Schedl, 1985. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol. 185 341–358. [DOI] [PubMed] [Google Scholar]
- Valenzuela, L., and R. T. Kamakaka, 2006. Chromatin insulators. Annu. Rev. Genet. 40 107–138. [DOI] [PubMed] [Google Scholar]
- Vazquez, J., and P. Schedl, 1994. Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J. 13 5984–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, J., and P. Schedl, 2000. Deletion of an insulator element by the mutation facet-strawberry in Drosophila melanogaster. Genetics 155 1297–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault, A., and J. O. Thomas, 1993. Chromatin structure of the beta-globin chromosomal domain in adult chicken erythrocytes. Cold Spring Harb. Symp. Quant. Biol. 58 15–24. [DOI] [PubMed] [Google Scholar]
- West, A. G., and P. Fraser, 2005. Remote control of gene transcription. Hum. Mol. Genet. 14(Spec No 1): R101–R111. [DOI] [PubMed] [Google Scholar]
- West, A. G., M. Gaszner and G. Felsenfeld, 2002. Insulators: many functions, many mechanisms. Genes Dev. 16 271–288. [DOI] [PubMed] [Google Scholar]
- West, A. G., S. Huang, M. Gaszner, M. D. Litt and G. Felsenfeld, 2004. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell 16 453–463. [DOI] [PubMed] [Google Scholar]
- Worcel, A., and C. Benyajati, 1977. Higher order coiling of DNA in chromatin. Cell 12 83–100. [DOI] [PubMed] [Google Scholar]
- Xie, X., T. S. Mikkelsen, A. Gnirke, K. Lindblad-Toh, M. Kellis et al., 2007. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. USA 104 7145–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, M., H. Yoshida, F. Hirose, Y. H. Inoue, Y. Hayashi et al., 2001. Ectopic expression of BEAF32A in the Drosophila eye imaginal disc inhibits differentiation of photoreceptor cells and induces apoptosis. Chromosoma 110 313–321. [DOI] [PubMed] [Google Scholar]
- Yu, W., V. Ginjala, V. Pant, I. Chernukhin, J. Whitehead et al., 2004. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat. Genet. 36 1105–1110. [DOI] [PubMed] [Google Scholar]
- Yusufzai, T. M., H. Tagami, Y. Nakatani and G. Felsenfeld, 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13 291–298. [DOI] [PubMed] [Google Scholar]
- Zhao, K., C. M. Hart and U. K. Laemmli, 1995. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81 879–889. [DOI] [PubMed] [Google Scholar]