Abstract
Splicing generates mature transcripts from genes in pieces in eukaryotic cells. Overwhelming evidence has accumulated that alternative routes in splicing are possible for most human and mammalian genes, thereby allowing formation of different transcripts from one gene. No function has been assigned to the majority of identified alternative splice forms, and it has been assumed that they compose inert or tolerated waste from aberrant or noisy splicing. Here we demonstrate that five human transcription units (WT1, NOD2, GNAS, RABL2A, RABL2B) have constant splice-isoform ratios in genetically diverse lymphoblastoid cell lines independent of the type of alternative splicing (exon skipping, alternative donor/acceptor, tandem splice sites) and gene expression level. Even splice events that create premature stop codons and potentially trigger nonsense-mediated mRNA decay are found at constant fractions. The analyzed alternative splicing events were qualitatively but not quantitatively conserved in corresponding chimpanzee cell lines. Additionally, subtle splicing at tandem acceptor splice sites (GNAS, RABL2A/B) was highly constrained and strongly depends on the upstream donor sequence content. These results also demonstrate that unusual and unproductive splice variants are produced in a regulated manner.
GENES are expressed by several multiprotein complexes. This process is guided by a multitude of signals and is multifacetedly controlled (Sutherland and Bickmore 2009; Vaquerizas et al. 2009; Barash et al. 2010). DNA-encoded genetic information is kept separately in the chromosomes of a eukaryotic nucleus and is decoded via RNA intermediates, which are processed and transmitted to their destinations, for example, to the sites of cytoplasmic protein synthesis. Genes in pieces (composed of exons spaced apart by introns) additionally depend on a splice apparatus (spliceosome) that uses splice signals in a primary transcript to recognize exon–intron boundaries (splice sites) and to accurately cut out introns and join exons. Often, splicing generates different mature transcripts from the same gene, a process called alternative splicing (AS). This is achieved by alternative usage of splice sites in precursor RNA transcripts. In this way, the complexity of transcriptomes and proteomes is increased in eukaryotic organisms. Obviously, AS events need control to ensure formation of proper splice forms and ratios. Sequence motifs matching the splice-site consensus are very common in primary transcripts, but only a minute fraction of them are used by the spliceosome. This specificity is achieved by further sequence and structural information within the premature transcript recognized by different proteins. A complex network of highly combinatorial molecular interactions ensures the tissue-, developmental-, and elicitor-specific formation of spliced transcripts and is regulated at multiple points (Hertel 2008; Smith et al. 2008).
Millions of short, single-pass cDNA sequence reads have accumulated in databases as expressed sequence tags supplemented from next-generation transcriptome sequencing data, both of which indicate that AS affects almost any multi-exon gene of any mammalian genome (Mortazavi et al. 2008; Wang et al. 2008).
However, there is evidence from two of the best-characterized transcriptomes (human and mouse) that the majority of AS events in humans are not conserved in mice. Even in a comparative analysis of AS patterns in humans and chimpanzees, hominid species separated by a relatively short period of evolutionary time and having very similar genome sequences, it turned out that 4–6% of genes show differences in splicing of orthologous exons (Calarco et al. 2007). This indicates that AS may contribute to phenotypic differences between humans and chimpanzees (and more generally between all species) by diverged regulation of the splicing process of primary transcripts from genes that are very similar in their sequences (Skandalis et al. 2010). Moreover, aberrations in splicing are also found as a cause of the development or progression of various diseases (Barbaux et al. 1997; Valentonyte et al. 2005), and splice mutations are suspected of contributing significantly to a variety of diseased states (Krawczak et al. 1992).
With respect to biological function, the splicing apparatus must assure not only that the correct splice sites are used, but also that the splice isoforms are produced in a precise stoichiometric abundance. In a detailed study of splice-isoform formation in mice, Chisa and Burke (2007) showed that interindividual variation in isoform ratios was highly constrained for six splice events in five genes. Importantly, these results were obtained from a genetically heterogeneous mice population (four-way cross among different inbred strains) that shows considerable variation for other phenotypes. In view of these data, the authors propose the concept of a “splico-stat” (Chisa and Burke 2007) where a hypothetical regulator is set to distinct levels.
Does the concept of a splico-stat that keeps splice-isoform ratios constant and ensures tissue- and species-specific differences also apply to putative aberrant transcripts resulting from splicing noise? Although nonsense-mediated mRNA decay (NMD) is able to diminish a fraction of aberrant splice forms (Magen and Ast 2005), noisy skipping of symmetrical exons would escape such a control. But why should splice forms—especially if they derive from highly expressed genes and consequently might constitute a considerable part of a cell's transcriptome—not be controlled while others are? In their study, Chisa and Burke (2007) selected only splice events that most probably give rise to alternative transcripts coding for proteins of different functions: exon 13 inclusion in Ezh2, usage of alternative donor of exon 9 of Hnf4a, exon 6 and exon 4A inclusion of Vegfa and Kras, respectively, and two independent splice events for Wt1 (exon 5 inclusion and alternative donor of exon 9). Furthermore, in their examples, AS resulted in substantial fractions of 20–40% for the respective minor forms; therefore, these were quite abundant and all six selected cases were conserved between humans and mice. It has been shown that conservation of alternatively used splice sites may differ, being highest for more evenly used alternatives and also higher when the distance between the alternative sites is divisible by three (Nurtdinov et al. 2007). Further analyses of splicing robustness would therefore need to also consider alternative transcripts of low abundance and splice events that create corrupted open reading frames as potential targets for degradation by NMD.
In this study, we addressed these questions by analyzing 15 human lymphoblastoid cell lines (LCLs). This allows for analysis under controlled cell culture conditions. In detail, we looked at the exclusion/inclusion of exonic sequences through (i) the frame-preserving, alternative donor usage in WT1 (Wilms' tumor suppressor gene) already analyzed by Chisa and Burke (2007), (ii) the frame-corrupting skipping of exon 3 of NOD2 (nucleotide-binding oligomerization domain containing protein 2), (iii) a combination of frame-preserving exon skipping and alternative tandem acceptor usage at GNAS (GNAS complex locus) whereby the tandem acceptor includes a noncanonical splice site, and (iv) multiple splice events of the closely related paralogs RABL2A and RABL2B (RAB, member of RAS oncogene family-like 2A and 2B) leading to both frame preserving and corrupting splice isoforms (Figure 1). Additionally, for WT1 and GNAS, we compared splice-isoform ratios with those from five corresponding chimpanzee cell lines to evaluate interspecies differences. Altogether, we found further evidence for a splico-stat that ensures rather constant splice-isoform formation independent of the type and amount of splice isoforms.
Figure 1.—
Genes and measured mRNA transcripts. mRNA transcripts of four genes (WT1, NOD2, GNAS, paralogs RABL2A/B) have been evaluated for splice-isoform frequencies in human and chimpanzee LCLs. Oligonucleotides (solid arrows) for PCR amplification are located in the flanks of the alternative splice events. PCR amplifies at least two products generated by exon skipping/inclusion and/or splicing at alternative donors/acceptors (shaded). The tandem acceptor splice sites are discriminated by E (part of tandem exonic) and I (tandem intronic) according to the nomenclature of Hiller et al. (2004).
MATERIALS AND METHODS
Cell lines and cell culture:
All used cell lines were Epstein-Barr virus (EBV)-transformed lymphoblastoid cells cultured in RPMI 1640 medium supplemented with 15% FCS and l-glutamine (2 mm, Gibco, Eggenstein, Germany) at 37°, 5% CO2, and 95% humidity. The human cell lines GM10847, GM12760, GM12864, GM12870, GM12871, GM15215, GM15324, GM15386, GM18502, GM18552, GM18858, GM18972, GM19140, and GM19204 were obtained from the Coriell Cell Repository (Camden, NJ) and C0766 from the European Collection of Cell Cultures (Salisbury, UK). All five chimpanzee cell lines (L2008, L2369, L2433, L2649, L2736) were kindly provided by W. Schempp (Medical Faculty, University of Freiburg, Germany).
RNA isolation and reverse transcription:
For each cell line, RNA was isolated on three consecutive days using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. cDNA synthesis was performed with Sprint RT Complete-Random Hexamer first-strand cDNA synthesis kit (Clontech-Takara Bio Europe, Saint-Germain-en-Laye, France) according to the manufacturer's protocol. Five micrograms of total RNA was used for reverse transcription.
PCR amplification:
PCR was performed with BioMix white (Bioline, Randolph, MA) according to the manufacturer's protocol in a total volume of 25 μl, including 3 μl DNA template and 10 pmol primer. In general, PCR reaction started with an initial denaturation at 93° for 1 min, followed by five cycles of 30 sec denaturation (95°), 30 sec with annealing temperature 1, and 1 min elongation at 72°. An additional 25 cycles were performed with annealing temperature 2 followed by a final elongation of 20 min at 72°. Primers and corresponding annealing temperatures (1/2) were as follows: NOD2ex24.f (5′-Fam-ATTGTCAGGAGGCTCCACAG-3′) and NOD2ex24.r (5′-TGTCCGCATCGTCATTGAG-3′) at 57°/59°, GNASex24.f (5′-Fam-GGTAAAAGCACCATTGTGAAGC-3′) and GNASex24.r (5′-GTCAAAGTCAGGCACGTTCAT-3′) at 56°/58°, WT1ex810.f (5′-AGACCAGCTCAAAAGACACCA-3′) and WT1ex810.r (5′-Fam-CATGTTGTGATGGCGGACTA-3′) at 57°/59°, and RABL2ex47.f (5′-CCCTGACCCTGTACAAGCA-3′) and RABL2ex47.r (5′-Fam-AACATTGGTACCATCAGCAGC-3′) at 57°/59°. Amplification of GNAS mRNA transcripts containing alternative first exons (numbering of the first exons is according to genomic positions:1A, NM_016592; 1B, NM_001077490; 1C, NR_003259; 1D, NM_000516) was performed as described above with a 57°/59° annealing temperature. Human cDNA from brain, pancreas, and leukocyte tissue (lot 7080210, human MTC panel I, Clontech) was used as template for the following primers: GNASex1A5.f (5′-AAGAGTCGAAGGAGCCCAA-3′), GNASex1B5.f (5′-ACGCAGTAAGCTCATCGACA-3′), GNASex1C5.f (5′-TTAGAAGCTCTGCTCCCCG-3′), GNASex1D5.f (5′-CGTGAGGCCAACAAAAAGAT-3′), and GNASex15.r (5′-Fam-GTCAAAGTCAGGCACGTTCAT-3′).
Capillary electrophoresis with laser-induced fluorescence analysis:
PCR amplification was carried out with 5′-6-carboxyfluorescein (FAM)-labeled forward or reverse primers (Metabion, Martinsried, Germany). The FAM-labeled PCR products were appropriately diluted (up to 1/40), and 1 μl was supplemented with 10 μl of formamide (Roth, Karlsruhe, Germany) and 0.5 μl of GeneScan ROX 500 (Applied Biosystems, Foster City, CA). The mixture was denatured at 94° for 3 min and subsequently cooled on ice. The denatured products were then separated on an ABI 3730 capillary sequencer and analyzed with the Gene Mapper 4.0 software (Applied Biosystems). The amount of each isoform within a PCR reaction was calculated by the area under the curve. Subsequent comparisons of isoform proportions were made by standardized calculation of percentage. All values are given as the mean of triplicate PCRs including standard deviation.
Statistical analysis:
To allow comparison among measurements of isoform percent fractions that differ in mean values (standardization of the variance), we calculated the coefficient of variation (CV), which is the standard deviation divided by the mean. Correlation and regression analysis of CV and splice isoform percent fractions and pairwise comparison of splice-isoform groups were done with Sigma Plot 11.0 (Systat Software, Chicago).
The Shapiro–Wilk test was used to assess normality within groups, each consisting of the 15 percentage ratios for a certain splice-isoform. The smallest P-value was 0.082 for GNAS Δ3 while all other groups were tested with a P-value >0.1. Thus, hypothesized normality could not be rejected for any of the groups on a 5% confidence level. The confidence intervals were calculated using the sample means and sample standard deviations of the groups using the Minitab 15 statistical software. The Levene test rejected the assumption of equal variances of the groups (value of test statistic: 11.74, P < 0.001). Consequently, the Welch test was carried out to check for equality of means between two groups. For the same reason, one-way ANOVA was replaced by Welch–ANOVA (Welch 1951), a generalization of the two-sample Welch test to the case of an arbitrary number of samples. The test was performed using the function oneway.test of the R software package (R Project for Statistical Computing). A method for calculating the power of the Welch–ANOVA was not available in any of the software packages available to the authors. Therefore, the power was estimated with the algorithm for a balanced ANOVA, using the average variance of the five groups, which is ∼2.5. The power was calculated using the Minitab software package.
Pairwise comparison of percent isoform fractions between human and chimpanzee was performed with the Mann–Whitney rank-sum test instead of the Student's t-test because the normality test (Shapiro–Wilk) failed for at least one group within the data set. The Kruskal–Wallis one-way analysis of variance on ranks was used to determine potential differences in percent isoform fractions between GNAS transcripts harboring several first exons.
Gene expression analysis:
SYBR green-based quantitative real-time reverse transcription-PCR for WT1, NOD2, and GNAS was performed on a BioRad iCycler iQ 582BR (Bio-Rad Laboratories, Hercules, CA). GAPDH was used as the reference gene. PCR reaction was carried out with “BioMix white” (Bioline) according to the manufacturer's protocol in a total volume of 25 μl including 10 μl cDNA dilution (1:30, equimolar pooled from RNA isolations on three consecutive days), 5 pmol primer, as well as 0.38 μl 1× ROX and 0.21 μl 1× SYBR Green QPCR (Stratagene, La Jolla, CA) according to the manufacturer's instructions. In general, PCR reaction started with an initial denaturation at 95° for 2 min, followed by 40 cycles of 20 sec of denaturation (95°), 30 sec of annealing at 63°, and 20 sec of elongation at 72°, plus an additional 80° step for 15 sec. Melt curve data were obtained by an initial denaturation at 95° for 30 sec, followed by 56 cycles with an increasing temperature of 0.5° per cycle. Primers were as follows: RT-WT1ex24.f (5′-CCAACCACTCATTCAAGCATG-3′) and RT-WT1ex24.r (5′-GTGGCTCCTAAGTTCATCTGA-3′), RT-NOD2ex910.f (5′-ATCACCAGAGCTTGAGGTGG-3′) and RT-NOD2ex910.r (5′-CTTCAGTCCTTCTGCGAGAGA-3′), RT-GNASex56.f (5′-TGAACGTGCCTGACTTTGAC-3′) and RT-GNASex56.r (5′-CTGGTACTCGTTGGAGCGTT-3′), and RT-GAPDH.f (5′-GGAGGGGAGATTCAGTGTGGT-3′) and RT-GAPDH.r (5′-AACAGCGACACCCACTCCTC-3′). Relative gene expression was calculated for all cell lines and reported as x-fold expression compared to a reference cell line. The cell line with the lowest expression of the respective gene was used as reference. Relative expression of RABL2A and RABL2B was determined as previous described by Kramer et al. (2010).
RESULTS
Quantification of splice isoforms:
In this study, splice-isoform ratios were determined for WT1, NOD2, GNAS, and the paralogs RABL2A and RABL2B. Relevant gene fragments and splice events are shown in Figure 1. Initially, we evaluated the levels of variation in splice-isoform ratios in 15 LCLs derived from blood of human donors with varied ethnicity (supporting information, Table S1). For this purpose, we used fluorescence-based quantification of electrophoretically separated reverse transcription PCR (RT-PCR) products.
In WT1, the alternative usage of a tandem donor of exon 9 results in the insertion/deletion of nine nucleotides (three amino acids, KTS). The mean value of the +KTS isoform fraction among the human LCLs is 53.0 ± 2.0% (Figure 2A), revealing only a very low level of interindividual variation. Surprisingly, we detected a novel splice variant (Δ9), where the complete exon 9 is skipped (verified by cloning and sequencing). Its fraction was 5.3 ± 2.4% (CV = 0.434) among the human LCLs (Figure 2A) with a higher coefficient of variation compared to +KTS splice ratios (CV = 0.021).
Figure 2.—
Splice-isoform and expression levels of WT1. (A) Percent fraction of WT1 splice isoforms are shown for human LCLs. Solid dots represent the +KTS isoform and open dots display skipping of exon 9 (Δ9). (B) WT1 expression of each human LCL is represented by shaded bars relative to the lowest measured expression. Values represent the mean ± standard deviation of triplicate RNA isolations.
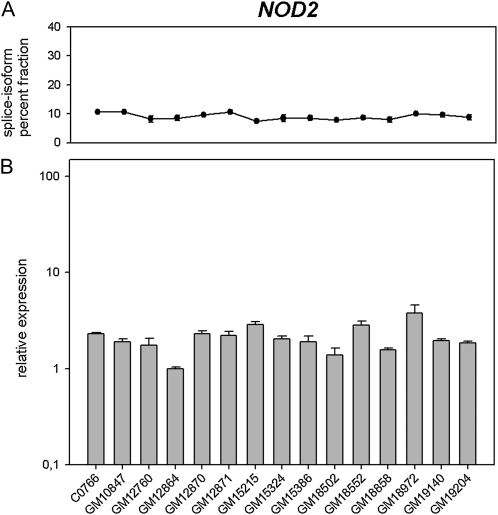
NOD2-S, a short isoform of the NOD2 main transcript (Rosenstiel et al. 2006) caused by skipping of exon 3 (Δ3), was found with a 9.0 ± 1.1% fraction in human LCLs (CV = 0.122, Figure 3A).
Figure 3.—
Splice-isoform and expression levels of NOD2. (A) Percent fraction of NOD2 exon 3 skipping (Δ3) are represented as solid dots for human LCLs. (B) NOD2 expression of each human LCL is represented as a shaded bar relative to the lowest measured expression. Values represent the mean ± standard deviation of triplicate RNA isolations.
Skipping of GNAS exon 3 (Δ3) is coupled with alternative usage of the intron 3 tandem acceptor. The tandem acceptor motif UGCAG utilizes UG as a noncanonical intron-proximal splice site (Pollard et al. 2002). For such tandem acceptor structures, Hiller et al. (2004) denote the intron-proximal splice site as the “E” acceptor, since part of the tandem will be exonic while the whole tandem is intronic for the intron-distal “I” acceptor (Figure 1). In the case of GNAS, Δ3 accounts for 75.1 ± 1.1% in the human samples (CV = 0.015). The overall E acceptor usage is 54.0 ± 1.0% (CV = 0.019), with 66.8 ± 1.0% (CV = 0.065) and 15.5 ± 1.2% (CV = 0.018) for Δ3 (E) and exon 3 inclusion [+3 (E)], respectively (Figure 4A). Obviously, the usage of the tandem acceptor sites depends on the upstream donor site (P < 0,001, Mann–Whitney rank-sum test).
Figure 4.—
Splice-isoform and expression levels of GNAS. (A) Percent fractions of GNAS splice isoforms are shown for human LCLs. Solid dots represent the skipping of exon 3 (Δ3) and open dots display transcripts with E acceptor (E) usage of exon 4. (B) Percent fractions of GNAS splice isoforms are shown for human LCLs. Shaded dots represent the E acceptor usage for skipping of exon 3 [Δ3 (E)] and open triangles for the inclusion of exon 3 [+3 (E)]. (C) GNAS expression of each LCL is shown as a shaded bar relative to the lowest measured expression. Values represent the mean ± standard deviation of triplicate RNA isolations.
Next, we inspected the complex splicing of the very similar paralogs RABL2A and RABLB, located on human chromosomes 2 and 22, respectively. Our RT-PCR approach detects in parallel isoforms deriving from both genes using exon 4 and 7 primers matching completely both paralogs. We observed skipping of exon 6 (Δ6) and of exons 5 and 6 together (Δ5Δ6) as well as alternative usage of a CAGCAG tandem acceptor of intron 6. Human Δ5Δ6 transcripts account for 14.4 ± 1.1% (CV = 0.076) and Δ6 isoforms for 3.7 ± 0.4% (CV = 0.108). The I acceptor of the tandem was utilized by 12.5 ± 0.8% (CV = 0.064) in the full-length transcripts [+5+6 (I)], whereas values of 8.8 ± 1.0% (CV = 0.144) and 4.8 ± 0.5% (CV = 0.104) in Δ6 (I) and Δ5Δ6 (I), respectively, were found (Figure 5A). The percent fractions of the I acceptor usage are significantly different (P < 0.001, Kruskal–Wallis one-way analysis of variance on ranks) in combination with the three mentioned donor splice sites.
Figure 5.—
Splice-isoform and expression levels of RABL2A/B. (A) Percent fractions of RABL2A and RABL2B splice isoforms for human LCLs. Shaded dots represent the skipping of exon 5 and 6 (Δ5Δ6), open dots the skipping of exon 6 (Δ6), and solid triangles display the inclusion of exons 5 and 6 (+5+6). (B) The percentage of I acceptor usage at exon 7 is shown as solid dots for skipping of exons 5 and 6 [Δ5Δ6 (I)], open triangles for skipping of exon 6 [Δ6 (I)], and as shaded squares for the inclusion of exons 5 and 6 [+5+6 (I)]. Values represent the mean ± standard deviation of triplicate RNA isolations. (C) RABL2A and RABL2B expression of each LCL is represented by lightly shaded (RABL2B) and darkly shaded (RABL2A) bars as Percent fraction of RABL2A/B overall expression. For GM18502 and GM18972, no data (n.d.) are available.
Overall, the determined splice ratios reveal a low interindividual variance (Table S2). The variation of the percent fractions within each isoform is very small compared to the differences between the means. Even for the two isoform combinations with the closest means, NOD2 Δ3 (9.0) and RABL2A/B Δ5Δ6 (14.4), a two-sample t-test (Welch test) yields a statistically significant difference between the means of these groups on a 1% significance level (P-value < 0.01; 99% confidence interval for the difference: −8.81; −1.90).
Variations in human transcript levels:
It is known that transcription and splicing are coordinated processes (Kornblihtt et al. 2004). Therefore, we determined the overall expression levels of the genes under investigation using quantitative real-time PCR. NOD2 and GNAS showed only relatively small differences in expression levels. Relative expression among human LCLs was in the range of up to 3.8- and 2.1-fold, respectively (Figures 3B and 4B). In contrast, up to ∼4000-fold variations in WT1 expression were detected (Figure 2B). In the case of RABL2A/B, only expression percent fractions of the two paralogs were determined (Figure 5C). RABL2B was expressed 1.8- to 3.0-fold higher than RABL2A (Kramer et al. 2010).
Limited promoter dependence on splice-site selection in GNAS:
GNAS exhibits an intricate genomic organization characterized by several alternative promoters (Figure S1) and offers the possibility to study variation of splice-isoform ratios depending on the promoter used for transcription. We applied upstream primers in the four different first exons (1A–1D) of transcripts deposited in GenBank (1A: NM_016592; 1B: NM_001077490; 1C: NR_003259; 1D: NM_000516) in combination with a common downstream primer in exon 5 of NM_000516. Both LCLs and a commercial cDNA tissue panel were analyzed. However, no exon 1C products were obtained in either sample, and we detected only the three exon 1 A/B/D amplicons simultaneously in the pancreatic sample of the tissue panel. Therefore, we selected the pancreas sample for quantitative analysis. Using exon 1 forward primers, the same splice isoforms (Δ3, +3, and alternative tandem acceptor usage, Figure S1) were detectable as previously observed using the exon 2 forward primer. Among exon 1A/B/D transcript isoforms, 59.4 ± 1.4%, 68.7 ± 0.6%, and 73.5 ± 0.2% represent the Δ3 isoform, respectively. Of the +3 transcripts, 15.8 ± 2.3% (1A), 15.1 ± 1.3% (1B), and 15.2 ± 0.4% (1D) used the E acceptor. Among Δ3 isoforms, this splice site was utilized in 70.9 ± 2.0% (1A), 67.8 ± 0.6% (1B), and 67.9 ± 0.2% (1D) of the cases. Δ3 or E acceptor isoforms ratios were significantly different in transcripts derived from several GNAS promoters (both: P = 0.004, Kruskal–Wallis one way analysis of variance on ranks). However, considering E acceptor usage depending on exon 3 inclusion/skipping, we found no statistically significant differences in the ratios for either +3 (E) (P = 0.481) or Δ3 (E) (P = 0.929, both derived from Kruskal–Wallis one way analysis of variance on ranks) when inspecting transcription from different promoters.
Splicing of GNAS and WT1 in corresponding chimpanzee cell lines:
Finally, five LCLs from chimpanzees were analyzed for AS of the orthologous GNAS and WT1 genes. Both genes are highly conserved. The exons 2, 3, and 4 of GNAS are completely identical in both species while exon 8 in WT1 exhibits a single nucleotide exchange 53 bp upstream of the donor. Most analyzed splice-isoform ratios in chimpanzees were slightly but significantly lowered compared to humans (Figure 6): GNAS Δ3 72.1 ± 1.6% (P < 0.001, Mann–Whitney rank-sum test), WT1 +KTS 53.5 ± 4.6% (P = 0.019), and WT1 Δ9 3.5 ± 1.5% (P = 0.025). Only splice-site usage of the GNAS noncanonical UG (32.0 ± 1.2%) is considerably lowered in chimpanzees (P < 0.001) independently of whether exon 3 is included (41.5 ± 1.0%) or skipped (6.6 ± 1.6%) in the transcripts (both P < 0.001).
Figure 6.—
Alternative splicing in humans vs. chimpanzees. The percent fractions of the measured splice isoforms of either human (n = 15) or chimpanzee (n = 5) cell lines were box plotted. (A) GNAS exon 3 skipping (Δ3) and E acceptor (E) usage as well as E acceptor usage depending on exon 3 skipping [Δ3 (E)] and inclusion [+3 (E)]. (B) WT1s +KTS isoform and skipping of exon 9 (Δ9). Statistical significance (Mann–Whitney rank-sum test): *P < 0.05; **P < 0.001.
DISCUSSION
We are far from having understood the flow of genetic information between the genome and the functional gene products (RNAs, proteins). A high level of coordination is needed to have a complex ribonome (Mansfield and Keene 2009) that maintains the global mRNA landscape in balance but dynamic and sensitive to stimuli. The concept of a splico-stat (Chisa and Burke 2007) that adjusts alternative transcripts from the same gene to pertinent ratios fits into such a system on the level of transcript processing. Along this line, genome-wide studies of AS events have shown an only limited level of splice-form variation among humans. Using exon tiling arrays and a large set of cell lines from European and Yoruba individuals (87 and 89 samples, respectively), it was shown that the proportion of genes showing differential AS between the two populations is quite low (∼8%) compared to the overall percentage of alternatively processed genes (Zhang et al. 2009). In an experimentally similar study, Kwan et al. (2008) found in 57 Utah residents (with Northern and Western European ancestry) from the Centre d'Etude du Polymorphisme Humain collection a significant association between transcript levels and genetic variation. Of 17,897 genes covered by their array, 324 showed SNP-dependent altered expression on the exon and/or transcript level with just 85 of them (26%) due to altered splicing. A recent study (Coulombe-Huntington et al. 2009) showed that differences in splicing between human individuals are mainly due to polymorphisms close to splice sites. Also recent investigations using next-generation sequencing (NGS) to characterize transcriptomes indicate that interindividual differences in splice-isoform expression in the same tissue are less variable than those observed in different tissues of the same individual (Wang et al. 2008). Especially at high coverage, transcript analysis by NGS reveals correlation of any two pairs of exons within a gene (Montgomery et al. 2010) and that SNPs near splice sites and within the spliced exon influence transcript isoform levels (Pickrell et al. 2010). It is therefore doubtless that genetic variation shapes individual transcriptomes, but these data also show the robustness of the splicing process. While such genome-wide studies evaluate whole transcriptomes, to the best of our knowledge, only the study of Chisa and Burke (2007) has focused on the splicing robustness problem in detail. By analyzing mouse tissues, they showed that splice-isoform levels are expressed in constant ratios in genetically diverse animals in a tissue-dependent manner suggestive of a splico-stat with tissue-specific settings.
Chisa and Burke (2007) looked at splicing events in mice that preserved the reading frame (Wt1, Ezh2, Hnf4a, Vegfa) or, in the case of Kras, changed the protein's C terminus but positioned the respective stop codon just seven nucleotides upstream of the last exon–exon junction. Hence, none of the investigated splice forms are putative substrates for NMD. In the this study, we expanded this work by investigating splice-isoform ratios in one cell type, a set of LCLs derived from 15 individuals and five corresponding chimpanzee cell lines. In addition to reevaluating the usage of the biologically important alternative donor sites in WT1, we have chosen complex and unusual splice events currently without any clearly defined functional implication for our investigation (Figure 1). Skipping of the asymmetrical exon 3 (106 nt) of NOD2 and exon 6 (112 nt) of the RABL2A/B paralogs is a frame-disrupting event where the levels of the respective transcripts bearing a premature termination codon might be controlled by NMD (Cheng et al. 1990; Magen and Ast 2005). Furthermore, we included splice isoforms that result from the usage of tandem acceptor and noncanonical acceptor sites. At the very complex imprinted GNAS locus (Plagge et al. 2008) multiple transcripts arise from monoallelic, parental origin-dependent expression from multiple promoters. An exon-skipping event (exon 3 of NM_001077488, 45 nt) occurs in combination with the use of competing acceptor sites spaced apart by 3 nt. This tandem acceptor involves a UG (Pollard et al. 2002) at the E acceptor position. which, although this motif is very common in mammalian genomes, is rarely used in splicing (Szafranski et al. 2007). The most complex situation examined is the splicing of exon 4 to exon 7 of the two highly similar paralogs RABL2A and RABL2B (Wong et al. 1999). Here, six transcripts were formed from each locus. Exon skipping led to transcripts lacking exon 6 or exon 5 together with exon 6. Because intron 6 harbors a 3′ tandem splice site, each donor is spliced to either the proximal (E) or the distal (I) acceptor. Furthermore, we determined expression levels for the genes analyzed.
As expected, for WT1 we found the ratio of the alternative donor-derived ±KTS isoforms in a very narrow range across the different human cell lines, similar to published data for human (Barbaux et al. 1997; Klamt et al. 1998) and mouse tissues (Chisa and Burke 2007). In addition, we found a minor splice form lacking symmetrical exon 9 (Δ9) in both humans and chimpanzees. This isoform is expressed at ∼5% in all human cell lines. A similar situation is found for the skipping of exon 3 of GNAS, which we observed to be rather constant in all cell lines. The minor splice form of NOD2, NOD2-S, results from skipping of an asymmetrical exon within the coding sequence and hence should be an NMD target (Hilleren and Parker 1999; Vilardell et al. 2000). However, also for this form a constant fraction is detected in all human cell lines. For RABL2A and RABL2B, skipping events lead to isoforms that are (Δ6) or are not (Δ5Δ6) potential NMD targets. Although these transcripts derive from two paralogs—i.e., ratios are represented by processed pre-mRNAs from four genomic loci—all show very similar values in the different human cell lines. Our data for human transcripts support the splico-stat model and are statistically confirmed by the calculated 95% confidence intervals for the percent ratios of five selected isoforms (Table S2). The convincingly small variation within each isoform enables a very clear discrimination between the individual isoforms. Not surprisingly, ANOVA rejects the null hypothesis of the equal means of these five isoform fractions with a P-value that is <0.001 and a power that is almost 1. These numbers illustrate how small the within-group variances are when compared to the differences between the group means.
Remarkably, isoform fraction values are negatively correlated with the CV (Spearman's rank correlation coefficient rs = −0.916; P < 0.001). Regression analysis yields an exponential dependency between the two parameters (Figure 7). The CV is proportional to the mean to the power of −0.84 (P < 0.001). Therefore, the CV is approximately a reciprocal function of the mean. This is caused by the fact that the magnitude of the standard deviation apparently does not systematically depend on the mean, but is similar for all values of the mean (Table S2). This can be explained by the assumption that the splico-stat confines the variation of the percent isoform ratio to a low, mean-independent level. Alternatively, the variation within a given isoform might be largely determined by random measurement error rather than by intrinsic differences in ratio between the cell lines, a conclusion that utterly supports the concept of constrained splice-isoform ratios.
Figure 7.—
Correlation of coefficient of variation (CV) and splice-isoform percent fractions. Regression analysis shows a power exponential dependency (curved line) between coefficient of variation and percent isoform fraction with a coefficient of determination (R2) of 0.84. The CV is calculated as the standard deviation divided by the mean. Mean values are given as percent age fraction of the respective isoform measured as triplicates in 15 human LCLs.
For WT1, the observed uniformity of splice-site selection in cell lines from different individuals is independent of the expression level. We found exon 9 inclusion but with particular alternative donor usage (±KTS) very constrained although expression levels varied by several orders of magnitude (Figure 2B). NOD2 and GNAS expression levels as well as relative expression of RABL2A and RABL2B were rather constant. One argument for such a notably strict splicing regulation could be the physiological importance of the ±KTS ratio, because an imbalance is causative for the Frasier syndrome (Klamt et al. 1998).
Given the strong evidence that splice-isoform ratios are set to constant levels across individuals within a species, the question arises as to what would be the reason for evolution favoring this phenomenon? Obviously, if isoforms code for proteins that interfere with each other in their functions, such a regulation is meaningful. An example is the splice-isoform coding for a truncated form of STAT3 that competes with the full-length form for binding to target gene promoters but does not activate transcription (Caldenhoven et al. 1996). However, for most alternative transcripts, and in particular for those of low abundance, it is not obvious. Moreover, their formation often lacks phylogenetic conservation (Skandalis et al. 2010). Isolated cases indicate, however, that low abundance of an alternative transcript and seemingly missing conservation are not sufficient criteria to discard an identified alternative transcript as being nonfunctional. For example, the human galactosidase gene GLB1 is alternatively transcribed into a low-abundance splice isoform missing three exons (Morreau et al. 1989). It turned out that this short isoform codes for an elastin receptor (Privitera et al. 1998) and is involved in inflammatory processes (Baranek et al. 2007).
Modrek and Lee (2003) have also reasoned that splice-isoform levels should be controlled on the basis of comparisons of splice isoforms as “internal paralogs” of duplicated genes. Considerable evidence indicates that a balance of gene dosage is critical for normal phenotype (Veitia and Birchler 2010) and is evolutionarily adjusted, so one would expect that similarly a splico-stat may keep splice dosages in balance.
Our comparison of human and chimpanzee splicing expands previous knowledge of AS species specificity (Calarco et al. 2007; Skandalis et al. 2010). Although the sequences of the evaluated genes GNAS and WT1 were highly conserved between human and chimpanzee, we found significant interspecies differences in splice-isoform ratios. Importantly, however, splice-isoform ratios between the different individuals were constrained, just as was found in the human cell lines.
The usage of GNAS and RABL2A/B tandem acceptors causing symmetrical changes in exon sizes revealed surprising results. We monitored splicing of tandem acceptors in combination with different donor sites in dependence of skipping either one (GNAS) or one or two (RABL2A/B) upstream exons, respectively. Tandem acceptors are widespread in many genomes and variable in respect to the isoform ratios (Hiller et al. 2006, 2007). It has been proposed that their usage is of a stochastic nature determined by the strength of each acceptor site and surrounding sequence (Chern et al. 2006). Here, however, a strong dependence of the ratio of tandem acceptor-derived isoforms from the donor site used was observed for both GNAS and RABL2A/B tandem acceptors. These observations are in agreement with earlier experimental evidence that a simple scanning mechanism is not sufficient to explain a 3′ splice-site definition of an intron harboring tandem acceptors (Chen et al. 2000). Nevertheless, for GNAS and the RABL2 paralogs, splice ratios resulting from tandem acceptor usage are very constant in combination with an explicit donor in the different human cell lines. Furthermore, the sequence context guiding tandem splice-site selection must be restricted for the upstream exon–intron–tandem region because we found the E and I ratios for exon 3 skipping and inclusion constant and independent of the alternative first exons of GNAS (Figure S1). This is in agreement with a detailed analysis of exon 3 skipping and UG usage in GNAS by applying a minigene approach, which demonstrated that cis-acting regulatory sequence motifs are within the limited sequence of the minigene (Pollard et al. 2002). Most probably, exon 3 skipping and tandem splice-site usage is regulated independently. This view is also supported by the comparative analysis with splicing of GNAS in chimpanzees. While skipping of exon 3 in humans and chimpanzees is in the same range, subtle splicing at the tandem acceptor is rather different. This is very surprising because exons 2–4 and their flanking intron sequences are completely identical in the two species. Further investigation may elucidate the regulatory details and provide more mechanistic insight into the splicing of tandem acceptors.
Altogether, our data support the concept of a splico-stat for unusual splice events. From a future perspective, knowledge of universally valid molecular signatures such as splice-isoform ratios is important, especially because of their role as physiological biomarkers.
Acknowledgments
We thank Sabine Gallert for excellent technical assistance. This work was supported by grants from the Deutsche Forschungsgmeinschaft (DFG Hu498/3 to K.H. and J.H.) and the Bundesministerium für Bildung und Forschung (01GS0809). The funding sources had no influence on study design; collection, analysis, and interpretation of data; writing of the paper; and decision to submit it for publication.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.125096/DC1.
References
- Baranek, T., R. Debret, F. Antonicelli, B. Lamkhioued, A. Belaaouaj et al., 2007. Elastin receptor (spliced galactosidase) occupancy by elastin peptides counteracts proinflammatory cytokine expression in lipopolysaccharide-stimulated human monocytes through NF-kappaB down-regulation. J. Immunol. 179 6184–6192. [DOI] [PubMed] [Google Scholar]
- Barash, Y., J. A. Calarco, W. Gao, Q. Pan, X. Wang et al., 2010. Deciphering the splicing code. Nature 465 53–59. [DOI] [PubMed] [Google Scholar]
- Barbaux, S., P. Niaudet, M. C. Gubler, J. P. Grunfeld, F. Jaubert et al., 1997. Donor splice-site mutations in WT1 are responsible for Frasier syndrome. Nat. Genet. 17 467–470. [DOI] [PubMed] [Google Scholar]
- Calarco, J. A., Y. Xing, M. Caceres, J. P. Calarco, X. Xiao et al., 2007. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 21 2963–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldenhoven, E., T. B. van Dijk, R. Solari, J. Armstrong, J. A. Raaijmakers et al., 1996. STAT3beta, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J. Biol. Chem. 271 13221–13227. [DOI] [PubMed] [Google Scholar]
- Chen, S., K. Anderson and M. J. Moore, 2000. Evidence for a linear search in bimolecular 3′ splice site AG selection. Proc. Natl. Acad. Sci. USA 97 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J., M. Fogel-Petrovic and L. E. Maquat, 1990. Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol. Cell. Biol. 10 5215–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern, T. M., E. van Nimwegen, C. Kai, J. Kawai, P. Carninci et al., 2006. A simple physical model predicts small exon length variations. PLoS Genet. 2 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisa, J. L., and D. T. Burke, 2007. Mammalian mRNA splice-isoform selection is tightly controlled. Genetics 175 1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe-Huntington, J., K. C. Lam, C. Dias and J. Majewski, 2009. Fine-scale variation and genetic determinants of alternative splicing across individuals. PLoS Genet. 5 e1000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel, K. J., 2008. Combinatorial control of exon recognition. J. Biol. Chem. 283 1211–1215. [DOI] [PubMed] [Google Scholar]
- Hiller, M., K. Huse, K. Szafranski, N. Jahn, J. Hampe et al., 2004. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat. Genet. 36 1255–1257. [DOI] [PubMed] [Google Scholar]
- Hiller, M., K. Huse, K. Szafranski, N. Jahn, J. Hampe et al., 2006. Single-nucleotide polymorphisms in NAGNAG acceptors are highly predictive for variations of alternative splicing. Am. J. Hum. Genet. 78 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller, M., S. Nikolajewa, K. Huse, K. Szafranski, P. Rosenstiel et al., 2007. TassDB: a database of alternative tandem splice sites. Nucleic Acids Res. 35 D188–D192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P., and R. Parker, 1999. Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet. 33 229–260. [DOI] [PubMed] [Google Scholar]
- Klamt, B., A. Koziell, F. Poulat, P. Wieacker, P. Scambler et al., 1998. Frasier syndrome is caused by defective alternative splicing of WT1 leading to an altered ratio of WT1 +/−KTS splice isoforms. Hum. Mol. Genet. 7 709–714. [DOI] [PubMed] [Google Scholar]
- Kornblihtt, A. R., M. de la Mata, J. P. Fededa, M. J. Munoz and G. Nogues, 2004. Multiple links between transcription and splicing. RNA 10 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, M., O. Backhaus, P. Rosenstiel, D. Horn, E. Klopocki et al., 2010. Analysis of relative gene dosage and expression differences of the paralogs RABL2A and RABL2B by Pyrosequencing. Gene 455 1–7. [DOI] [PubMed] [Google Scholar]
- Krawczak, M., J. Reiss and D. N. Cooper, 1992. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 90 41–54. [DOI] [PubMed] [Google Scholar]
- Kwan, T., D. Benovoy, C. Dias, S. Gurd, C. Provencher et al., 2008. Genome-wide analysis of transcript isoform variation in humans. Nat. Genet. 40 225–231. [DOI] [PubMed] [Google Scholar]
- Magen, A., and G. Ast, 2005. The importance of being divisible by three in alternative splicing. Nucleic Acids Res. 33 5574–5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, K. D., and J. D. Keene, 2009. The ribonome: a dominant force in co-ordinating gene expression. Biol. Cell 101 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B., and C. J. Lee, 2003. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 34 177–180. [DOI] [PubMed] [Google Scholar]
- Montgomery, S. B., M. Sammeth, M. Gutierrez-Arcelus, R. P. Lach, C. Ingle et al., 2010. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464 773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreau, H., N. J. Galjart, N. Gillemans, R. Willemsen, G. T. van der Horst et al., 1989. Alternative splicing of beta-galactosidase mRNA generates the classic lysosomal enzyme and a beta-galactosidase-related protein. J. Biol. Chem. 264 20655–20663. [PubMed] [Google Scholar]
- Mortazavi, A., B. A. Williams, K. McCue, L. Schaeffer and B. Wold, 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5 621–628. [DOI] [PubMed] [Google Scholar]
- Nurtdinov, R. N., A. D. Neverov, A. V. Favorov, A. A. Mironov and M. S. Gelfand, 2007. Conserved and species-specific alternative splicing in mammalian genomes. BMC Evol. Biol. 7 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell, J. K., J. C. Marioni, A. A. Pai, J. F. Degner, B. E. Engelhardt et al., 2010. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464 768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagge, A., G. Kelsey and E. L. Germain-Lee, 2008. Physiological functions of the imprinted Gnas locus and its protein variants Galpha(s) and XLalpha(s) in human and mouse. J. Endocrinol. 196 193–214. [DOI] [PubMed] [Google Scholar]
- Pollard, A. J., A. R. Krainer, S. C. Robson and G. N. Europe-Finner, 2002. Alternative splicing of the adenylyl cyclase stimulatory G-protein G alpha(s) is regulated by SF2/ASF and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) and involves the use of an unusual TG 3′-splice site. J. Biol. Chem. 277 15241–15251. [DOI] [PubMed] [Google Scholar]
- Privitera, S., C. A. Prody, J. W. Callahan and A. Hinek, 1998. The 67-kDa enzymatically inactive alternatively spliced variant of beta-galactosidase is identical to the elastin/laminin-binding protein. J. Biol. Chem. 273 6319–6326. [DOI] [PubMed] [Google Scholar]
- Rosenstiel, P., K. Huse, A. Till, J. Hampe, S. Hellmig et al., 2006. A short isoform of NOD2/CARD15, NOD2-S, is an endogenous inhibitor of NOD2/receptor-interacting protein kinase 2-induced signaling pathways. Proc. Natl. Acad. Sci. USA 103 3280–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skandalis, A., M. Frampton, J. Seger and M. H. Richards, 2010. The adaptive significance of unproductive alternative splicing in primates. RNA 16 2014–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. J., C. C. Query and M. M. Konarska, 2008. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol. Cell 30 657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, H., and W. A. Bickmore, 2009. Transcription factories: Gene expression in unions? Nat. Rev. Genet. 10 457–466. [DOI] [PubMed] [Google Scholar]
- Szafranski, K., S. Schindler, S. Taudien, M. Hiller, K. Huse et al., 2007. Violating the splicing rules: TG dinucleotides function as alternative 3′ splice sites in U2-dependent introns. Genome Biol. 8 R154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentonyte, R., J. Hampe, K. Huse, P. Rosenstiel, M. Albrecht et al., 2005. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat. Genet. 37 357–364. [DOI] [PubMed] [Google Scholar]
- Vaquerizas, J. M., S. K. Kummerfeld, S. A. Teichmann and N. M. Luscombe, 2009. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 10 252–263. [DOI] [PubMed] [Google Scholar]
- Veitia, R. A., and J. A. Birchler, 2010. Dominance and gene dosage balance in health and disease: Why levels matter! J. Pathol. 220 174–185. [DOI] [PubMed] [Google Scholar]
- Vilardell, J., P. Chartrand, R. H. Singer and J. R. Warner, 2000. The odyssey of a regulated transcript. RNA 6 1773–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, E. T., R. Sandberg, S. Luo, I. Khrebtukova, L. Zhang et al., 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, B. L., 1951. On the comparison of several mean values: an alternative approach. Biometrika 38 330–336. [Google Scholar]
- Wong, A. C., D. Shkolny, A. Dorman, D. Willingham, B. A. Roe et al., 1999. Two novel human RAB genes with near identical sequence each map to a telomere-associated region: the subtelomeric region of 22q13.3 and the ancestral telomere band 2q13. Genomics 59 326–334. [DOI] [PubMed] [Google Scholar]
- Zhang, W., S. Duan, W. K. Bleibel, S. A. Wisel, R. S. Huang et al., 2009. Identification of common genetic variants that account for transcript isoform variation between human populations. Hum. Genet. 125 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]