Abstract
In plastids, the conversion of energy in the form of light to ATP requires key electron shuttles, the c-type cytochromes, which are defined by the covalent attachment of heme to a CXXCH motif. Plastid c-type cytochrome biogenesis occurs in the thylakoid lumen and requires a system for transmembrane transfer of reductants. Previously, CCDA and CCS5/HCF164, found in all plastid-containing organisms, have been proposed as two components of the disulfide-reducing pathway. In this work, we identify a small novel protein, CCS4, as a third component in this pathway. CCS4 was genetically identified in the green alga Chlamydomonas reinhardtii on the basis of the rescue of the ccs4 mutant, which is blocked in the synthesis of holoforms of plastid c-type cytochromes, namely cytochromes f and c6. Although CCS4 does not display sequence motifs suggestive of redox or heme-binding function, biochemical and genetic complementation experiments suggest a role in the disulfide-reducing pathway required for heme attachment to apoforms of cytochromes c. Exogenous thiols partially rescue the growth phenotype of the ccs4 mutant concomitant with recovery of holocytochrome f accumulation, as does expression of an ectopic copy of the CCDA gene, encoding a trans-thylakoid transporter of reducing equivalents. We suggest that CCS4 might function to stabilize CCDA or regulate its activity.
Cytochromes c are ubiquitous molecules functioning as electron carriers. They carry a heme cofactor covalently attached via two thioether linkages between the vinyl groups of heme B (iron protoporphyrin IX) and the cysteine sulfhydryls in the apocytochrome c (Thöny-Meyer 1997). The cysteine sulfhydryls are found in a CXXCH motif, also referred to as the heme-binding site, where histidine acts as one of the axial ligands of heme. CXXCK, CXXXCH, or CXXXXCH motifs are variations to the canonical heme-binding site and are found in some bacterial cytochromes c (Jungst et al. 1991; Rios-Velazquez et al. 2001; Hartshorne et al. 2006). Another variation is found in trypanosomatid where heme is attached via a single thioether bond at a F/AXXCH motif on mitochondrial c-type cytochromes (Allen et al. 2004).
Bacterial cytochromes c are assembled in the periplasm via two different pathways, system I and system II (Ferguson et al. 2008; Hamel et al. 2009; Kranz et al. 2009; Bonnard et al. 2010; Sanders et al. 2010). A thiol-disulfide membrane transporter of the DsbD/CcdA family and a membrane-anchored, periplasm-facing thioredoxin-like protein (CcmG in system I or ResA/CcsX in system II) are the defining components of the disulfide-reducing pathway. They are postulated to act sequentially to reduce the disulfide-bonded CXXCH in apocytochrome c prior to the heme ligation (Ritz and Beckwith 2001; Allen et al. 2003; Kadokura et al. 2003; Mapller and Hederstedt 2006). The need for disulfide reduction in cytochrome c assembly is thought to be necessary because the periplasm is also the compartmentwhere disulfide bond formation takes place (Mapller and Hederstedt 2006; Messens and Collet 2006; Kadokura and Beckwith 2010). Inactivation of the disulfide-reducing pathway in bacteria results in a cytochrome c-deficient phenotype, and it is believed that the apocytochrome c CXXCH then becomes the target of the disulfide bond machinery (Deshmukh et al. 2000; Erlendsson and Hederstedt 2002; Turkarslan et al. 2008).
In photosynthetic eukaryotes, c-type cytochromes are housed in the thylakoid lumen of plastids. Plastid cytochromes c are assembled through a multi-component pathway uncovered in the green alga Chlamydomonas reinhardtii through genetic analysis of the ccs mutants (ccs for cytochrome c synthesis) (Howe and Merchant 1992; Howe et al. 1995; Xie et al. 1998). These mutants are deficient for membrane-bound cytochrome f and soluble cytochrome c6, the two c-type cytochromes required for photosynthesis (Howe and Merchant 1992). In Chlamydomonas, cytochrome f and cytochrome c6 are synthesized in the plastid and cytosol, respectively. The heme attachment takes place in the thylakoid lumen, a compartment topologically analogous to the bacterial periplasm. Pulse-chase analyses in the ccs mutants revealed that apoforms of cytochrome f and c6 are synthesized and further processed in the thylakoid lumen, but not converted to their respective holoforms. This indicates that the CCS loci control the heme attachment reaction (Howe and Merchant 1992; Howe et al. 1995; Xie et al. 1998). The CCS loci do not control the covalent attachment of heme Ci to cytochrome b6 of the cytochrome b6f complex (Stroebel et al. 2003). While heme attachment via the CCS pathway occurs in the lumen, covalent linkage of heme Ci to a cysteine on cytochrome b6 is dependent upon the Cofactor binding, Cytochrome b6f complex, and subunit petB (CCB) factors and takes place on the stromal side of the thylakoid membrane (Kuras et al. 1997, 2007; Lyska et al. 2007; Lezhneva et al. 2008; Saint-Marcoux et al. 2009).
The operation of a disulfide-reducing pathway in the context of plastid cytochrome c assembly was first suspected because of the occurrence of orthologs of the bacterial thiol transporter CCDA that localize to the plastid (Nakamoto et al. 2000; Page et al. 2004). In Arabidopsis thaliana, loss of CCDA impacts photosynthesis and results in a cytochrome b6f assembly defect (Page et al. 2004). However, evidence that heme attachment to apocytochrome f is impaired by ccda mutations is still lacking, and the placement of CCDA in plastid cytochrome c maturation needs to be confirmed (Page et al. 2004). The finding that the Chlamydomonas ccs4 and ccs5 mutants could be rescued by application of exogenous thiols led to the proposal that the corresponding gene products are components of the disulfide-reducing pathway (Page et al. 2004). CCS5, a new locus controlling plastid cytochrome c assembly, was recently identified and shown to encode the algal ortholog of Arabidopsis HCF164. HCF164 is a membrane-anchored, lumen-facing thioredoxin-like protein required for cytochrome b6f assembly (Lennartz et al. 2001; Gabilly et al. 2010). The recombinant form of CCS5/HCF164 can reduce a disulfide at the CXXCH motif of apocytochrome f (Lennartz et al. 2001; Motohashi and Hisabori 2006; Gabilly et al. 2010).
In this article, we report the molecular identification of the CCS4 gene by functional complementation of the ccs4 mutant. CCS4 does not carry any motif indicative of redox chemistry despite the fact that thiol-dependent, partial rescue of ccs4 suggests its involvement in the reducing pathway. Expression of an ectopic copy of the CCDA gene, encoding the plastid thiol-disulfide transporter, partially suppresses the ccs4 mutant. This indicates that CCS4 and CCDA interact in the same redox pathway. We discuss the possible roles of CCS4 in the disulfide-reducing pathway required for cytochrome c maturation.
MATERIALS AND METHODS
Strains and culture conditions:
The ccs4-F2D8 mutant strain (mt−) (Xie et al. 1998) was crossed to a wild-type strain (mt+ arg7-8) to generate the ccs4-F2D8 arg7-8 (mt+) used in the complementation experiments. For the thiol rescue experiments, the ccs4-F2D8 arg7-8 strain was crossed to CC-2677 (cw15 nit1 mt−) and a cw15 ccs4 strain was identified. Wild-type stains were CC124 and CC2677. Strains were grown at 22–25° in tris acetate phosphate (TAP) liquid or solid medium (Harris 1989) with or without copper supplementation under dim light (25 μmol/m2/sec) for ccs4 and ccs5 strains or under standard illumination for wild-type strains (300 μmol/m2/sec) as described in Howe and Merchant (1992). Copper-deficient media are used to induce the expression of cytochrome c6 (Quinn and Merchant 1998).
Molecular cloning of the CCS4 gene:
The ccs4-F2D8 arg7-8 strain was transformed by electroporation using the indexed cosmid library, and phototrophic transformants were recovered on minimal medium under high light (300 μmol/m2/sec). An 8-kb BamHI fragment and a 1-kb SacII fragment containing the CCS4 gene were isolated from a complementing cosmid and cloned in pBluescript SK vector yielding the pSK-CCS4 BamHI and pSK-CCS4 SacII plasmids, respectively. The coding sequence of CCS4 (from ATG to stop) was cloned at EcoRI and XbaI sites of pSL18 (Pollock et al. 2004) between the PSAD promoter and terminator using Pccs4-ORF2-NdeI (5′-AACCCATATGTCGACTGGCATTGAGG-3′) and L-Pccs4-ORF2-XbaI (5′-AACTCTAGATCACTTGGTTGCCTGC-3′) as primers and pSK-CCS4 SacII as a template. The resulting plasmid is pSL18-CCS4(ORF1). The coding sequence corresponding to the truncated form of CCS4 (from M32 to stop) was cloned at EcoRI and XbaI sites between the PSAD promoter and terminator of pSL18 via in-fusion technology (Clontech) using PORF5-F-EcoRI (5′-CGATAAGCTTGATATCGAATTCATGGCTATTTCAAAAGGCATTGAGG-3′) and PORF5-R-XbaI (5′-GGTCCAGCTGCTGCCATCTAGATCACTGGTTGCCTGCTCCTGG-3′) as primers and pSK-CCS4 SacII as a template. The final construct is pSL18-CCS4 (ORF2).
Construction of CCDA-expressing plasmid:
The CCDA ORF was cloned between the PSAD promoter and terminator of pSL18. The cloned cDNA (Nakamoto et al. 2000) was used as a template in a PCR reaction with ccdA-NdeI (5′-GGGAATTCCATATGCGAACCGCCATGCATTTAG-3′) and ccdA-EcoRI (5′-CGGAATTCTCACGAGGGCACCAGGCGCG-3′) as NdeI- and EcoRI-engineered primers, respectively. The PCR product was cloned at the NdeI and EcoRI sites and yielded pSL18-CCDA.
RNA extraction and real time PCR:
Wild-type CC124, mutant strains ccs4-F2D8, or ccs4-F2-D8 arg7-8 transformed with the empty cosmid pCB412 or cotransformed with pCB412 and pSK-CCS4 SacII (ccs4-Sac) or with pCB412 and pSK-CCS4 BamHI (ccs4-Bam) were grown in TAP medium at 25° with 25 μmol/m2/sec of light. At ∼6 × 106 cells/ml, total RNA from triplicate cultures per strain was prepared as in Quinn and Merchant (1998). Samples were prepared and real time PCR was performed as in Allen et al. (2007). Gene-specific primers used for amplification were 5′-GCTTCCTCCCTGCAGCCGTCCT-3′ and 5′-GCGGGATCAAGCAGCGACAAGT-3′ for CCS1; 5′-TGGTTGCCTGCTCCTTGGAC-3′ and 5′-GCACGGGCTCAGATGAATGG-3′ for CCS4; and 5′-GCGGGGTCGAGAGGTTATGG-3′ and 5′-CCCTCGTCAGCCCTCTGTGT-3′ for CCDA. Primer efficiencies for CCS1, CCS4, and CCDA were 102, 100, and 99%, respectively. All data were analyzed together with LinRegPCR 11.x to obtain the mean PCR efficiency for each gene (Ruijter et al. 2009). Transcript levels for the genes of interest (gi) were normalized to the transcript levels of the CBLP gene encoding the Chlamydomonas β-subunit-like polypeptide. Relative transcript level (RTL) was calculated as follows: RTL = 1000 × [mean PCR efficiency for CBLP]CtCBLP × [mean PCR efficiency for gi]Ctgi.
Protein preparation and analysis:
Supernatant and pellet fractions were obtained by freeze–thaw fractionation and subsequent centrifugation. Fractions were electrophoretically separated and cytochromes c were revealed by immunodetection or by a heme-staining procedure (Howe and Merchant 1992). Polyclonal antisera raised against Chlamydomonas cytochrome c6, cytochrome f GST-fusion protein, CCS5, CF1, and plastocyanin were used for immunodetection by alkaline phosphatase-conjugated secondary antibodies.
RESULTS
CCS4 gene product may participate in disulfide reduction:
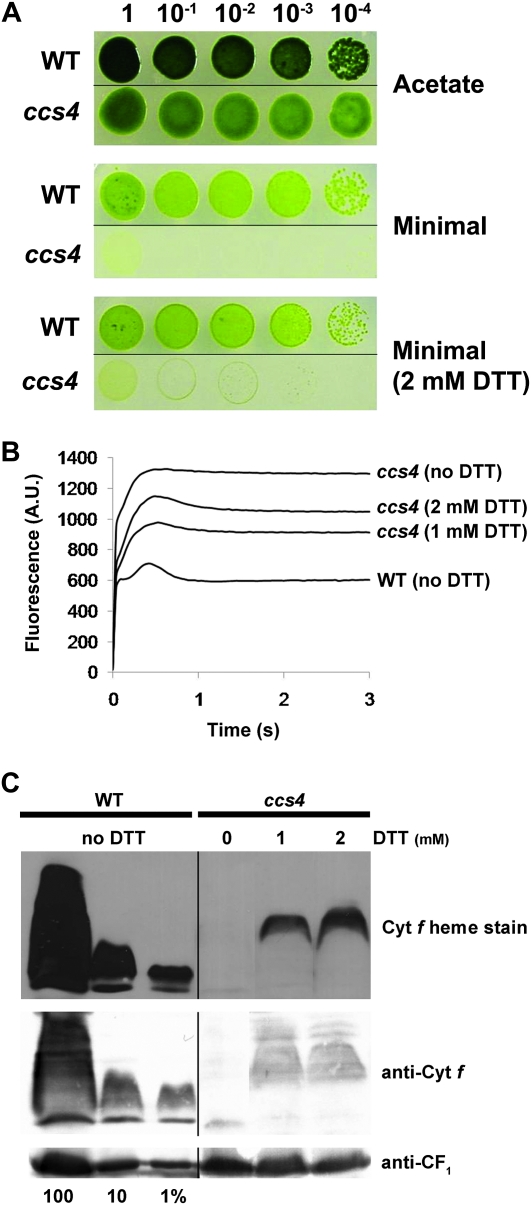
On the basis of our understanding of the biochemistry of cytochrome c maturation, it is expected that some of the CCS loci control disulfide reduction. In an attempt to functionally categorize the gene products corresponding to the genetically defined ccs mutants, we tested for the rescue of the ccs4 mutant by exogenous thiols. Our approach is driven by precedence in bacteria (Beckett et al. 2000; Deshmukh et al. 2000; Bardischewsky and Friedrich 2001; Erlendsson and Hederstedt 2002; Feissner et al. 2005). Moreover, we have shown that ccs5 could be rescued by DTT (Page et al. 2004; Gabilly et al. 2010). As shown in Figure 1A, addition of DTT to minimal medium can rescue the photosynthetic deficiency of ccs4. We noted that 2-mercaptoethane sulfonate, a reduced thiol, is also able to rescue ccs4 to the same extent as DTT (data not shown). This partial rescue is dose dependent and correlates with a restoration of the cytochrome b6f function, as evidenced by fluorescence rise and decay kinetics (Figure 1B). Heme stain and immunoblot analysis confirmed that levels of holocytochrome f are increased in DTT-treated cells (Figure 1C). Consistent with the partial restoration of the photosynthetic growth, accumulation of holocytochrome f is only marginally increased in DTT-treated ccs4 cells.
Figure 1.—
The ccs4 mutant is partially rescued by exogenous thiols. (A) DTT-dependent photosynthetic rescue of ccs4. Ten-fold dilution series of wild type (cw15 nit1-305) (WT) and ccs4 (ccs4-F2D8 cw15 arg7-8) (ccs4) were plated on acetate and minimal medium with or without 2 mm DTT. Cells grown heterotrophically were incubated at 25° for 7 days with 20 μmol/m2/sec of light. Cells grown phototrophically with or without DTT were incubated at 25° for 14 days with 250 μmol/m2/sec of light. Cells grown phototrophically showed the best rescue with 2 mm DTT. (B) Fluorescence kinetics indicate a partial restoration of cytochrome b6f in DTT-treated ccs4 cells. The fluorescence induction and decay kinetics observed in a dark-to-light transition of ccs4 grown in the absence or presence of 1 and 2 mm DTT are shown compared to those of WT. Fluorescence transients were measured using Handy Fluorcam from Photon System Instruments. The fluorescence is in arbitrary units (A.U.) and recorded over a 3-sec illumination period. The rise and plateau curve for ccs4 is a signature of a specific block in electron transfer at the level of the cytochrome b6f complex because of its impaired assembly in the absence of membrane-bound holocytochrome f. When the energy absorbed by the chlorophyll cannot be utilized because of a block in photosynthetic transfer through cytochrome b6f, an increase in the chlorophyll fluorescence is observed. In wild type, the decay phase corresponds to the re-oxidation of the quinone pool, the primary electron acceptor of the photosystem II, by the cytochrome b6f complex. (C) DTT-dependent partial restoration of holocytochrome f assembly in ccs4. Cytochrome f heme staining and anti-cytochrome f immunoblot analyses were performed on total protein extracts from ccs4 (ccs4-F2D8 cw15 arg7-8) and wild-type (cw15) strains. Cells were grown heterotrophically (on acetate and in low light) in the absence or presence of 1 or 2 mm DTT. Samples of WT and ccs4 strains corresponding to 18 μg of chlorophyll were separated in SDS-containing acrylamide (12%) gel. The gel was then transferred to a PVDF membrane to perform heme staining and immunodecoration with antisera against cytochrome f and CF1 of the ATPase that serves as a loading control. Dilutions of the wild-type sample serve to estimate the cytochrome f abundance.
These results indicate that the CCS4 gene product may participate in disulfide reduction. We have ruled out the possibility that the CCS4 gene encodes for CCDA because the CCDA locus was intact in the ccs4 mutant (Page et al. 2004). Because CCS4 is genetically distinct from the CCS5 locus (Page et al. 2004; Gabilly et al. 2010), we concluded that CCS4 must encode a novel redox component involved in cytochrome c maturation.
Cloning of the CCS4 gene by functional complementation of the ccs4-F2D8 mutant:
We sought to clone the CCS4 gene by complementation of the photosynthetic deficiency of a ccs4-F2D8 arg7-8 strain using an indexed ARG7-based cosmid library (Purton and Rochaix 1994). Three cosmids with overlapping inserts were identified as restoring the photosynthetic competence when introduced into the ccs4-F2D8 arg7 strain (not shown). The complementing activity could be isolated to a 1-kb SacII fragment, suggesting that the CCS4 gene is very small (Figure 2A). This 1-kb fragment restored photosynthetic growth (Figure 2A), fluorescence rise, and decay kinetics, indicating that the cytochrome b6f complex is functional (Figure 2B) and the accumulation of holoforms of cytochrome f and c6 to wild-type levels (Figure 2C).
Figure 2.—
Complementation of the ccs4 mutant. (A, B, and D) The ccs4-F2D8 arg7-8 strain was transformed with pSL18 (ccs4), pSL18 carrying a 1-kb genomic fragment with the CCS4 gene (CCS4), and pSL18 expressing the full-length CCS4 coding sequence (CCS4, ORF1) or a truncated form of the CCS4 protein (CCS4, ORF2). (C) The ccs4-F2D8 arg7-8 strain was transformed with pCB412 (ccs4) or cotransformed with pCB412 and pSK-CCS4-SacII carrying a 1-kb genomic fragment with the CCS4 gene [ccs4 (CCS4)]. Only one representative transformant is shown in A–C. In D, two representative transformants (CCS4, ORF2) are shown. In B–D, CC124 is the wild-type strain (WT). (A) Restoration of the photosynthetic growth of ccs4 by full-length and truncated CCS4. Ten-fold dilution series of each transformant were plated on acetate (under heterotrophic conditions, 20 μmol/m2/sec of light) and minimal medium (phototrophic conditions, 250 μmol/m2/sec of light) and incubated at 25° for 1 and 3 weeks, respectively. (B) Fluorescence kinetics indicate restoration of cytochrome b6f in ccs4 complemented with the full-length and truncated CCS4 gene. Fluorescence transients were measured on colonies grown for 1 day on solid acetate medium after a short dark adaptation using Handy Fluorcam (Photon System Instruments). The fluorescence is in arbitrary units (A.U.) and recorded over a 3-sec illumination period. (C) Plastid c-type cytochromes accumulation is restored in ccs4 complemented with the CCS4 gene.Strains were analyzed for cytochrome f and cytochrome c6 accumulation by heme stain and immunoblot. Samples corresponding to 18 μg of chlorophyll were separated in 12% SDS acrylamide gel to detect cytochrome f and CF1 that serves as loading control. Samples corresponding to 16 μg of chlorophyll were separated in 15% native acrylamide gel to detect cytochrome c6. For an estimation of the protein abundance in the ccs4-complemented strain, dilutions of the wild-type sample were loaded on the gel. Gels were transferred to PVDF membranes prior to heme staining and immunodetection with antisera against cytochrome f, cytochrome c6, and CF1. (D) Cytochrome f accumulation is partially restored in ccs4 complemented by a truncated form of the CCS4 gene. Strains were analyzed for cytochrome f accumulation via heme stain and immunoblot. Experimental conditions are the same as described in C.
CCS4 gene encodes a unique protein with no known motif:
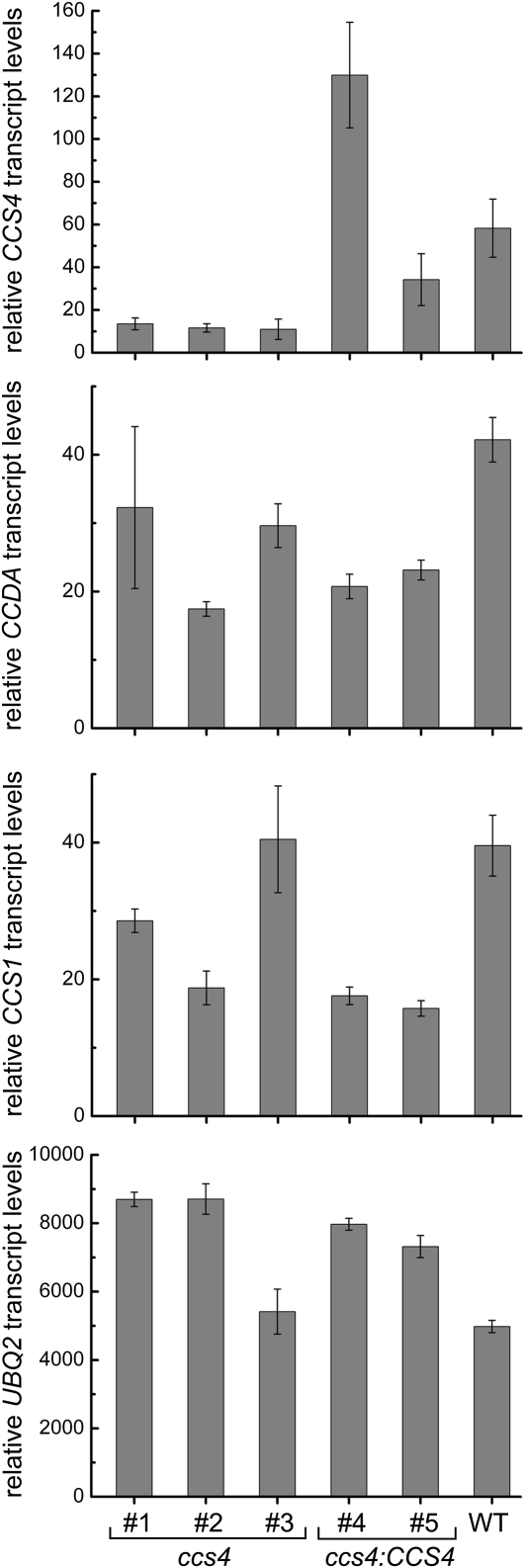
RT-PCR experiments showed that the genomic region corresponding to the 1-kb SacII complementing fragment is transcriptionally active (not shown). However, the size of the full-length transcript could not be determined, as RNA hybridization failed to detect the mRNA, presumably because of its low abundance (not shown). A 285-bp cDNA sequence was assembled from sequencing of RT-PCR products. Interestingly, the CCS4 pre-mRNA contains two small introns of 88 and 104 bp, a rare occurrence as most Chlamydomonas genes contain an average intron size of 373 bp (Merchant et al. 2007). Quantitative RT-PCR experiments, using primers that map to the transcript, evidenced a sixfold reduction in the accumulation of the mRNA in the ccs4 mutant compared to the wild-type strain but increased levels in the ccs4 strain complemented with genomic fragments containing the CCS4 gene (Figure 3). One ORF was identified from sequencing of the RT-PCR products (Figure 4; accession no. ADL27744). This ORF encodes a 93-amino-acid protein with no motifs or residues indicative of redox (e.g., cysteine) or any other biochemical activity. The predicted protein contains an N-terminal hydrophobic stretch that could serve as a membrane anchor and a C-terminal domain rich in charged residues (12 negatively charged and 9 positively charged). On the basis of the positive-inside rule that governs the topology of bacterial and thylakoid membrane proteins (Von Heijne 1989; Gavel et al. 1991), the C-terminal domain of CCS4 is predicted to be exposed to the stromal side of the thylakoid membrane. Standard protein targeting algorithms failed to predict plastid localization, an intriguing finding considering that we expect the protein to act in the plastid. Moreover, the only structural homologs in the database corresponded to predicted proteins in Volvox carteri and Dunaliella salina, two algae closely related to Chlamydomonas (Herron et al. 2009; Prochnik et al. 2010). The predicted Volvox protein is 60% identical to its Chlamydomonas counterpart (Figure 4), which reflects considerable divergence compared to sequences of cytochrome assembly factors CCS1 (Inoue et al. 1997), CCDA (Nakamoto et al. 2000), and CCB1 (Kuras et al. 2007), which are 79, 83, and 85% identical, respectively. The fact that Volvox CCS4 has diverged from its Chlamydomonas counterpart indicates that CCS4-like proteins might not be easily recognizable on the basis of sequence similarity in other photosynthetic eukaryotes. Sequencing of the 1-kb SacII genomic fragment in the ccs4 mutant strain identified one molecular lesion (C to T) in the coding region of the CCS4 gene. This change results in a non-sense mutation at residue Q50 in the predicted sequence (CAG to TAG) and presumably produces a nonfunctional truncated protein (Figure 4). To ascertain that we identified the correct ORF for the CCS4 gene, we cloned the genomic sequence from ATG (M1) to stop (ORF1) in an expression vector (pSL18) containing a paromomycin-resistance (PmR) cassette as a selectable marker. The resulting construct (pSL18/CCS4-ORF1) was introduced in the ccs4 mutant. Of 22 PmR transformants, 12 were able to grow photosynthetically and displayed wild-type fluorescence rise and decay kinetics (Figure 2, A and B). As expected from the restoration of the photosynthetic growth, cytochrome f assembly is also restored to wild-type levels (Figure 2D). The level of complementation is identical to that of the transformants carrying the 1-kb SacII genomic fragment, suggesting that ORF1 encodes the CCS4 gene product (Figure 2, compare A, B and C, D). To determine which of the two methionines (M1 and M32) serve as an intiation codon (Figure 4), we performed site-directed mutagenesis and tested the ability of the mutant forms to complement the ccs4 mutation. Mutagenesis of M1 abolished complementation while alteration of M32 did not (not shown). This confirms that M1 is the initiation codon of the CCS4 gene. We took advantage of the presence of the second methionine (M32) to generate a modified version of the CCS4 gene expressing a truncated form of the CCS4 protein, missing the first 31 amino acids, including the predicted transmembrane domain (Figure 4). We cloned the truncated sequence from ATG to stop (ORF2) in the same expression vector used for our complementation experiments. Of 103 PmR transformants, 43 exhibited partial complementation of the photosynthetic growth defect and pseudo-wild-type fluorescence rise and decay kinetics (Figure 2, A and B). Enhanced levels of cytochrome f accumulated in the partially rescued transformants compared to the ccs4 mutant strain, suggesting that the truncated form of the CCS4 protein retained some activity (Figure 2D). Note that the level of holocytochrome f restoration upon expression of the truncated CCS4 gene is similar to that observed in the DTT-rescued ccs4 cells (Figure 1C). As a control, we showed that transformation of the ccs4 mutant with the empty plasmid yielded no photosynthetic clones among 98 PmR transformants tested. This ruled out the possibility that the partial rescue depended upon the genomic site of integration or was caused by reversion of the photosynthetic deficiency. Unfortunately, despite several attempts, we could not generate a functional tagged version of the CCS4 gene to assess the localization of the gene product within the cell.
Figure 3.—
Relative CCS4, CCDA, and CCS1 mRNA abundance in ccs4 and ccs4 (CCS4)-complemented strains. RNA was isolated and analyzed by real time PCR. Strains are wild-type CC124 (WT), ccs4-F2D8 arg7-8 (#1), ccs4-F2D8 arg7-8 strain transformed by cosmid pCB412 (#2), ccs4-F2D8 mutant (#3), ccs4-F2D8 arg7-8 strain cotransformed by pCB412 and pSK-CCS4 BamHI (#4), or cotransformed by pCB412 and pSK-CCS4 SacII (#5). Relative transcript levels (RTLs) represent the mean levels of three independent experiments, each analyzed in technical triplicates. RTL values are relative to the CBLP levels and were calculated as described in materials and methods. The abundance of UBQ2 is shown as a control.
Figure 4.—
Alignment of Chlamydomonas, Volvox, and Dunaliella CCS4 proteins. Sequences of C. reinhardtii (Crein, accession no. ADL27744), V. carteri (Vcart, accession no. FD920844.1), D. salina (Dsali1, accession no. BM447122.1; Dsali2, accession no. BM448413.1) CCS4 were aligned using the CLUSTALW algorithm (Blosum62 scoring matrix) in Bioedit. The alignment was edited using the GeneDoc multiple alignment editor. Strictly conserved or similar amino acids are on a solid background. The putative membrane anchor is boxed. The solid downward arrowhead indicates the position of the methionine in the truncated CCS4 form and the shaded downward arrowhead indicates the Q residue that is mutated to a stop codon in the ccs4-F2D8 strain.
Genetic interaction with CCDA indicates the involvement of CCS4 in the disulfide-reducing pathway:
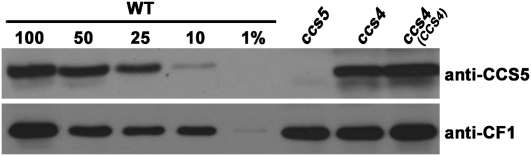
The thiol-based rescue of the ccs4 mutant is an intriguing finding, considering that the CCS4 protein does not display any motif or cysteine residue to indicate reducing activity. We reasoned the thiol-based rescue of the ccs4 mutant must be indirect, operating via redox components interacting with CCS4. One possible scenario is that the ccs4 mutation inactivates the transfer of reducing equivalents to the thylakoid lumen. In plastids, this transfer requires the activity of thiol-disulfide transporter CCDA and thioredoxin-like CCS5/HCF164 (Page et al. 2004; Motohashi and Hisabori 2006; Gabilly et al. 2010; Motohashi and Hisabori 2010). Real time PCR experiments showed no reduction in the abundance of the CCDA and CCS5 transcripts in response to the ccs4 mutation (Figure 3; data not shown). Therefore, we do not envision CCS4 as a regulator of the expression of either CCS5 or CCDA. Nevertheless, an impact on the abundance of the corresponding polypeptides is a possibility. We could not test the abundance of the CCDA protein in ccs4 because of the lack of antibodies, but immunoblot analyses with an anti-CCS5 antibody (Gabilly et al. 2010) showed that the level of CCS5 is unchanged in the ccs4 mutant (Figure 5).
Figure 5.—
Accumulation of the CCS5 protein in ccs4. Total protein (corresponding to 20 μg of chlorophyll) from wild-type CC124 (WT), T78.15b− (ccs5), ccs4-F2D8 arg7-8 mutant (ccs4), and ccs4-F2D8 arg7-8 complemented with pSL18-CCS4 (ORF1) (CCS4) was analyzed by SDS-PAGE (12%) and immunoblotting with antisera against CCS5 or CF1 of the ATPase that serves as a loading control. For an estimation of the protein abundance, dilutions of the wild-type sample were loaded on the gel.
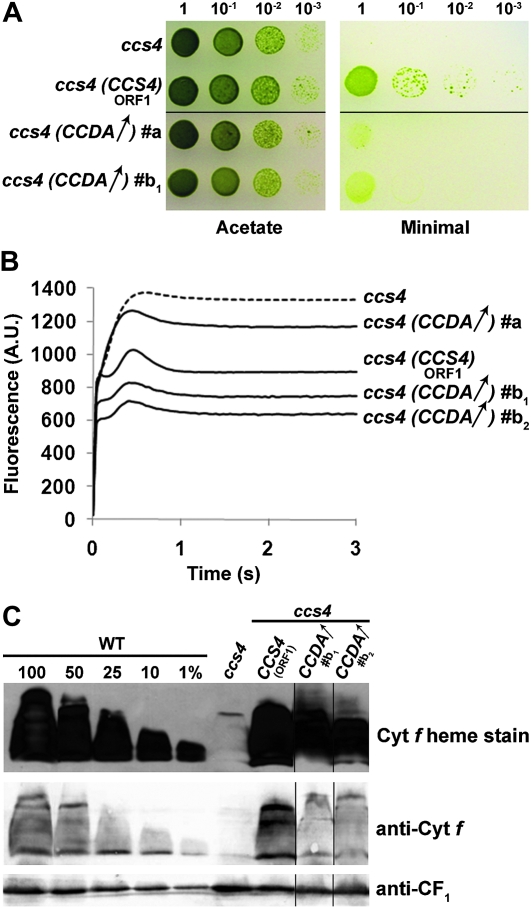
In parallel work, we noted that ccs4 could be rescued by plasmids carrying the promoter-less, full-length CCDA cDNA when we selected for phototrophic colonies following transformation of ccs4 (not shown). The frequency was low, but because recovery of photosynthetic proficiency was linked to the introduced cDNA in the rescued strain, the observation was genuine. We reasoned that such transformants must arise from integration of the CCDA cDNA in the vicinity of a promoter and therefore were few in number. This suggests that photosynthetic rescue of the ccs4 mutation would occur only upon increased expression of the CCDA transcript. To test this, we cloned the CCDA cDNA (from ATG to stop) in front of the PSAD promoter, previously used in Chlamydomonas to drive expression of cDNAs (Fischer and Rochaix 2001). The CCDA-expressing construct (pSL18/CCDA) was introduced in the ccs4 mutant and transformants were selected on the basis of their resistance to paromomycin, a trait conferred by the selectable marker on the construct. Of 44 PmR transformants, 14 were considered as suppressed for the CCS phenotype on the basis of the partial restoration of the photosynthetic growth defect (Figure 6A). While 12 transformants were weakly suppressed, 2 displayed an increased level of phototrophic growth (Figure 6A). To confirm our results, we cotransformed the ccs4 mutant with the PmR cassette containing vector pSL18 and a plasmid containing only the full-length CCDA cDNA (lacking promoter and terminator sequences for expression). Of 45 PmR transformants, 15 were weakly suppressed for the photosynthetic defect while 2 displayed a stronger restoration of the photosynthetic growth. As a control, we used a construct expressing the CCS4 gene from the same plasmid and transformed the ccs4 mutant. Of 22 PmR transformants, 12 displayed photosynthetic growth and fluorescence transients indistinguishable from wild type (Figure 6, A and B). No photosynthetic transformants were obtained among the 45 PmR transformants generated with the empty vector pSL18, ruling out the possibility that the two classes of suppressed transformants that we recovered with pSL18/CCDA resulted from reversion to photosynthetic proficiency. The CCDA-dependent suppression correlated with partial restoration of the cytochrome b6f activity, and therefore with holocytochrome f assembly, in the transformed strains (Figure 6B). However, we could only demonstrate enhanced holocytochrome f accumulation in the strongly suppressed transformants (Figure 6C). It is likely that the level of holocytochrome f is only marginally increased in the weakly suppressed strains and falls below the detection limit of our heme stain technique. RT-PCR experiments showed that the ectopic copy of the CCDA gene is expressed in both weakly and strongly suppressed transformants (supporting information, Figure S1).
Figure 6.—
Expression of an ectopic copy of the CCDA gene partially suppresses ccs4. The ccs4-F2D8 arg7-8 strain was transformed with pSL18 (ccs4), pSL18 expressing the full-length CCS4 coding sequence (CCS4, ORF1), and pSL18 expressing the CCDA ORF (CCDA↑, transformants #a and #b1) or cotransformed with pSL18 and the CCDA cDNA cloned (without promoter and terminator sequences) in pBluescript (CCDA↑, transformant #b2). (A) Expression of an ectopic copy of CCDA partially restores the phototrophic growth of ccs4. Ten-fold dilution series were plated on acetate (heterotrophic conditions, 20 μmol/m2/sec of light) and minimal medium (phototrophic conditions, 300 μmol/m2/sec of light) and incubated at 25° for 1 and 3 weeks, respectively. (B) Fluorescence kinetics indicate partial restoration of cytochrome b6f in ccs4 expressing an ectopic copy of CCDA. Fluorescence induction and decay kinetics were measured as described in Figure 1B. (C) Holocytochrome f accumulation is partially restored in the ccs4 mutant expressing an ectopic copy of CCDA. Strains were analyzed for cytochrome f accumulation by heme stain and immunoblot. Experimental conditions are the same as described in Figure 2C.
The CCDA-dependent suppression was specific for the ccs4 strain. When we tested the ccs5 mutant for rescue by CCDA, none of the 102 transformants screened displayed a restoration of photosynthesis (not shown). Our results suggest that CCDA is a component in the reducing pathway for cytochrome c maturation and can substitute partially for loss of CCS4 function when expressed ectopically.
DISCUSSION
In this article, we have further dissected the plastid disulfide-reducing pathway operating in cytochrome c assembly. We show that (1) the ccs4 mutant is partially rescued by exogenous thiols, (2) the CCS4 gene encodes a novel and unique protein with no motif suggestive of a redox activity, and (3) expression of an ectopic copy of the CCDA gene partially suppresses the ccs4 mutant.
A bacterial-like, trans-thylakoid, disulfide-reducing pathway:
An indication that the CCS4 and CCS5 gene products participate in the disulfide-reducing pathway is inferred from the observation that reduced thiols can rescue the cytochrome c assembly phenotype of the ccs4 (Figure 1) and ccs5 mutants (Gabilly et al. 2010). In bacteria, the disulfide-reducing pathway is defined by a membrane thiol-disulfide transporter (DsbD/CcdA) and a thioredoxin-like protein (CcmG/ResA/CcsX). This pathway is postulated to transfer reducing equivalents across the membrane for reduction of the CXXCH disulfide in apocytochrome c prior to the covalent attachment of heme (Ferguson et al. 2008; Hamel et al. 2009; Kranz et al. 2009; Bonnard et al. 2010; Sanders et al. 2010). The ability of exogenous thiol compounds to bypass mutations inactivating the disulfide-reducing components (Sambongi and Ferguson 1994; Beckett et al. 2000; Deshmukh et al. 2000; Bardischewsky and Friedrich 2001; Erlendsson and Hederstedt 2002; Feissner et al. 2005) and the fact that recombinant ResA and CcsX can participate in thiol-disulfide exchange reactions support this proposal (Monika et al. 1997; Setterdahl et al. 2000). The occurrence of CcdA-like proteins in plastids suggests that a trans-thylakoid, disulfide-reducing pathway, similar to the one found in bacteria, is required for the maturation of cytochromes c in the lumen (Nakamoto et al. 2000; Page et al. 2004). The first component of this pathway was discovered via the identification of the CCS5/HCF164 protein, a membrane-bound, lumen-facing, thioredoxin-like protein shown to act as an apocytochrome f CXXCH disulfide reductase (Lennartz et al. 2001; Motohashi and Hisabori 2006; Gabilly et al. 2010). Our finding that expression of CCDA is able to suppress the ccs4 mutant solidifies the placement of the thiol-disulfide transporter in plastid cytochrome c maturation (Figure 6). Indeed, earlier studies in Arabidopsis support, but do not establish, the requirement of plastid CCDA in the conversion of apo- to holocytochromes c (Page et al. 2004). The working model is that CCS5/HCF164 is maintained in a reduced state via the activity of CCDA, but this awaits experimental confirmation (Page et al. 2004; Motohashi and Hisabori 2006; Gabilly et al. 2010; Motohashi and Hisabori 2010). Thioredoxin-m was postulated as a possible reductant of CCDA on the stromal side based on the observation that both CCDA and CCS5/HCF164 can be reduced in intact Arabidopsis thylakoids by recombinant spinach thioredoxin-m (Motohashi and Hisabori 2006, 2010).
What is the function of the CCS4 protein?:
It is unlikely that CCS4 has a reducing activity in the assembly process because there are no motifs and residues in the protein sequence implying such an activity (Figure 4). One possibility is that the ccs4 mutation results in a loss of CCDA function. This is compatible with the fact that (1) ccs4 can be partially rescued by DTT (Figure 1), as seen in bacterial ccdA/dsbD mutants that are restored for cytochrome c assembly in the presence of exogenous thiols (Sambongi and Ferguson 1994; Beckett et al. 2000; Deshmukh et al. 2000), and (2) expression of CCDA can partially bypass the ccs4 mutation (Figure 6). In one scenario, CCS4 could operate by stabilizing CCDA in the thylakoid membrane. The presence of a putative transmembrane domain in the CCS4 protein is compatible with such a hypothesis. However, this transmembrane domain is not absolutely required for function, as a truncated form of CCS4, lacking the hydrophobic stretch, still retains some activity in the assembly of plastid cytochromes c (Figure 2). Another possibility is that CCS4 controls the activity of CCDA by facilitating the delivery of reducing equivalents from the stroma to the thylakoid lumen. It is conceivable that CCS4 acts as a “holdase” for presentation of the apocytochrome c CXXCH to the CCS5/HCF164 reductase in the thylakoid lumen. However, this model is unlikely because the positive-inside rule predicts a stromal localization for the C-terminal domain of CCS4. Moreover, a direct interaction of the CCS4 C-terminal domain with plastid apoforms of cytochromes c could not be detected via yeast two-hybrid using apocytochrome f as prey (not shown).
We could not determine a subcellular localization for CCS4; therefore, we cannot exclude that CCS4 could act in the cytosol. One possibility is that CCS4 acts as a chaperone or import factor in the cytosol. However, we find this hypothesis unlikely because the ccs4 mutant is specifically deficient in plastid cytochromes c and does not display a pleiotropic phenotype. Hence, we favor a model where CCS4 is localized at the thylakoid membrane and interacts with CCDA by stabilizing the protein and/or controlling its activity, possibly via its C-terminal domain. It is conceivable that loss of CCS4 results in decreased activity and/or destabilization of CCDA, and this is consistent with expression of CCDA partially rescuing the phenotype. There are several examples of polytopic membrane proteins whose stability and activity are influenced by the presence of single transmembrane proteins (Schulz et al. 1999; Yu et al. 1999; Peters et al. 2008). Interestingly, in system I bacteria, CcmD, a small transmembrane protein containing a cytoplasm-facing C-terminal domain with charged residues, controls the activity of cytochrome c assembly factors involved in the heme relay pathway (Goldman et al. 1997; Schulz et al. 2000; Ahuja and Thöny-Meyer 2005; Richard-Fogal et al. 2008). CcmD was shown to physically interact with the heme relay Ccm components and also influence their stability in the membrane (Schulz et al. 2000; Ahuja and Thöny-Meyer 2005; Richard-Fogal et al. 2008). Loss of CcmD can be partially rescued by overexpression of the CcmCE proteins, two key components of the heme delivery complex (Schulz et al. 1999).
Unique features of the CCS4 protein:
If CCS4 acts in the plastid, its import mechanism remains to be understood as the protein does not display a typical N-terminal targeting sequence (Figure 4). Intriguingly, a truncated form of CCS4 lacking the putative transmembrane domain still retains some activity. This indicates that the putative targeting information does not lie in the N-terminal part of the protein. It is conceivable that CCS4 reaches the plastid via internal targeting signals. Recent proteomics data revealed that 20% of plastid resident proteins are devoid of N-terminal targeting sequences and are not processed upon import in the plastid (Kleffmann et al. 2004).
The CCS4 protein does not appear to be evolutionarily conserved at the primary sequence level (Figure 4). We could not find any CCS4 orthologs in other genomes, including genomes of green algae such as Ostreococcus and Chlorella, in addition to V. carteri and D. salina. One possibility is that the function of CCS4 is dependent upon the overall charge of the protein rather than upon a specific primary sequence. The primary sequence of CcmD in bacterial system I cytochrome c maturation does not appear to be conserved, yet CcmD-like proteins can be recognized on the basis of charge conservation in operons containing cytochrome c biogenesis genes (Ahuja and Thöny-Meyer 2005; Richard-Fogal et al. 2008). Another possibility is that CCS4 is restricted to Volvocales, an order of green algae including Chlamydomonas (Merchant et al. 2007), Dunaliella (Oren 2005), and Volvox (Prochnik et al. 2010). Indeed, genomics and proteomics studies have revealed that Volvocales harbor unique proteins in their organelles (Atteia et al. 2009; Prochnik et al. 2010).
Acknowledgments
We thank R. Kranz and F. Daldal for stimulating scientific discussion on cytochrome c assembly and R. Lamb and S. Cline for critical reading of the manuscript. This work is supported by a Muscular Dystrophy Association grant (4727), a National Science Foundation grant (MCB-0920062) to P.P.H., and National Institutes of Health (GM48350), National Research Initiative of the U. S. Department of Agriculture Cooperative State Research, Education and Extension Service (2004-35318-14953) grants to S.S.M.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.125369/DC1.
Sequence data from this article have been deposited with the GenBank Data Library under accession no. ADL27744.
References
- Ahuja, U., and L. Thöny-Meyer, 2005. CcmD is involved in complex formation between CcmC and the heme chaperone CcmE during cytochrome c maturation. J. Biol. Chem. 280 236–243. [DOI] [PubMed] [Google Scholar]
- Allen, J. W., O. Daltrop, J. M. Stevens and S. J. Ferguson, 2003. C-type cytochromes: diverse structures and biogenesis systems pose evolutionary problems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, J. W., M. L. Ginger and S. J. Ferguson, 2004. Maturation of the unusual single-cysteine (XXXCH) mitochondrial c-type cytochromes found in trypanosomatids must occur through a novel biogenesis pathway. Biochem. J. 383 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, M. D., J. A. del Campo, J. Kropat and S. S. Merchant, 2007. FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron-deficient Chlamydomonas reinhardtii. Eukaryot. Cell 6 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atteia, A., A. Adrait, S. Brugiere, M. Tardif, R. van Lis et al., 2009. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 26 1533–1548. [DOI] [PubMed] [Google Scholar]
- Bardischewsky, F., and C. G. Friedrich, 2001. Identification of ccdA in Paracoccus pantotrophus GB17: disruption of ccdA causes complete deficiency in c-type cytochromes. J. Bacteriol. 183 257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett, C. S., J. A. Loughman, K. A. Karberg, G. M. Donato, W. E. Goldman et al., 2000. Four genes are required for the system II cytochrome c biogenesis pathway in Bordetella pertussis, a unique bacterial model. Mol. Microbiol. 38 465–481. [DOI] [PubMed] [Google Scholar]
- Bonnard, G., V. Corvest, E. H. Meyer and P. P. Hamel, 2010. Redox processes controlling the biogenesis of c-type cytochromes. Antioxid. Redox Signal. 13 1385–1401. [DOI] [PubMed] [Google Scholar]
- Deshmukh, M., G. Brasseur and F. Daldal, 2000. Novel Rhodobacter capsulatus genes required for the biogenesis of various c-type cytochromes. Mol. Microbiol. 35 123–138. [DOI] [PubMed] [Google Scholar]
- Erlendsson, L. S., and L. Hederstedt, 2002. Mutations in the thiol-disulfide oxidoreductases BdbC and BdbD can suppress cytochrome c deficiency of CcdA-defective Bacillus subtilis cells. J. Bacteriol. 184 1423–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feissner, R. E., C. S. Beckett, J. A. Loughman and R. G. Kranz, 2005. Mutations in cytochrome assembly and periplasmic redox pathways in Bordetella pertussis. J. Bacteriol. 187 3941–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, S. J., J. M. Stevens, J. W. Allen and I. B. Robertson, 2008. Cytochrome c assembly: A tale of ever increasing variation and mystery? Biochim. Biophys. Acta 1777 980–984. [DOI] [PubMed] [Google Scholar]
- Fischer, N., and J. D. Rochaix, 2001. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genomics 265 888–894. [DOI] [PubMed] [Google Scholar]
- Gabilly, S. T., B. W. Dreyfuss, M. Karamoko, V. Corvest, J. Kropat et al., 2010. CCS5, a thioredoxin-like protein involved in the assembly of plastid c-type cytochromes. J. Biol. Chem. 285 29738–29749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavel, Y., J. Steppuhn, R. Herrmann and G. Von Heijne, 1991. The ‘positive-inside rule’ applies to thylakoid membrane proteins. FEBS Lett. 282 41–46. [DOI] [PubMed] [Google Scholar]
- Goldman, B. S., D. L. Beckman, A. Bali, E. M. Monika, K. K. Gabbert et al., 1997. Molecular and immunological analysis of an ABC transporter complex required for cytochrome c biogenesis. J. Mol. Biol. 268 724–738. [DOI] [PubMed] [Google Scholar]
- Hamel, P., V. Corvest, P. Giege and G. Bonnard, 2009. Biochemical requirements for the maturation of mitochondrial c-type cytochromes. Biochim. Biophys. Acta 1793 125–138. [DOI] [PubMed] [Google Scholar]
- Harris, E. H., 1989. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego. [DOI] [PubMed]
- Hartshorne, S., D. J. Richardson and J. Simon, 2006. Multiple haemlyase genes indicate substrate specificity in cytochrome c biogenesis. Biochem. Soc. Trans. 34 146–149. [DOI] [PubMed] [Google Scholar]
- Herron, M. D., J. D. Hackett, F. O. Aylward and R. E. Michod, 2009. Triassic origin and early radiation of multicellular volvocine algae. Proc. Natl. Acad. Sci. USA 106 3254–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G., and S. Merchant, 1992. The biosynthesis of membrane and soluble plastidic c-type cytochromes of Chlamydomonas reinhardtii is dependent on multiple common gene products. EMBO J. 11 2789–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G., L. Mets and S. Merchant, 1995. Biosynthesis of cytochrome f in Chlamydomonas reinhardtii: analysis of the pathway in gabaculine-treated cells and in the heme attachment mutant B6. Mol. Gen. Genet. 246 156–165. [DOI] [PubMed] [Google Scholar]
- Inoue, K., B. W. Dreyfuss, K. L. Kindle, D. B. Stern, S. Merchant et al., 1997. CCS1, a nuclear gene required for the post-translational assembly of chloroplast c-type cytochromes. J. Biol. Chem. 272 31747–31754. [DOI] [PubMed] [Google Scholar]
- Jungst, A., S. Wakabayashi, H. Matsubara and W. G. Zumft, 1991. The nirSTBM region coding for cytochrome cd1-dependent nitrite respiration of Pseudomonas stutzeri consists of a cluster of mono-, di-, and tetraheme proteins. FEBS Lett. 279 205–209. [DOI] [PubMed] [Google Scholar]
- Kadokura, H., and J. Beckwith, 2010. Mechanisms of oxidative protein folding in the bacterial cell envelope. Antioxid. Redox Signal. 13 1231–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura, H., F. Katzen and J. Beckwith, 2003. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72 111–135. [DOI] [PubMed] [Google Scholar]
- Kleffmann, T., D. Russenberger, A. von Zychlinski, W. Christopher, K. Sjolander et al., 2004. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14 354–362. [DOI] [PubMed] [Google Scholar]
- Kranz, R. G., C. Richard-Fogal, J.-S. Taylor and E. R. Frawley, 2009. Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol. Mol. Biol. Rev. 73 510–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras, R., C. de Vitry, Y. Choquet, J. Girard-Bascou, D. Culler et al., 1997. Molecular genetic identification of a pathway for heme binding to cytochrome b6. J. Biol. Chem. 272 32427–32435. [DOI] [PubMed] [Google Scholar]
- Kuras, R., D. Saint-Marcoux, F. A. Wollman and C. de Vitry, 2007. A specific c-type cytochrome maturation system is required for oxygenic photosynthesis. Proc. Natl. Acad. Sci. USA 104 9906–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennartz, K., H. Plücken, A. Seidler, P. Westhoff, N. Bechtold et al., 2001. HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b6f complex in Arabidopsis. Plant Cell 13 2539–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezhneva, L., R. Kuras, G. Ephritikhine and C. de Vitry, 2008. A novel pathway of cytochrome c biogenesis is involved in the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J. Biol. Chem. 283 24608–24616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyska, D., S. Paradies, K. Meierhoff and P. Westhoff, 2007. HCF208, a homolog of Chlamydomonas CCB2, is required for accumulation of native cytochrome b6 in Arabidopsis thaliana. Plant Cell Physiol. 48 1737–1746. [DOI] [PubMed] [Google Scholar]
- Mapller, M., and L. Hederstedt, 2006. Role of membrane-bound thiol-disulfide oxidoreductases in endospore-forming bacteria. Antioxid. Redox Signal. 8 823–833. [DOI] [PubMed] [Google Scholar]
- Merchant, S. S., S. E. Prochnik, O. Vallon, E. H. Harris, S. J. Karpowicz et al., 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messens, J., and J.-F. Collet, 2006. Pathways of disulfide bond formation in Escherichia coli. Int. J. Biochem. Cell Biol. 38 1050–1062. [DOI] [PubMed] [Google Scholar]
- Monika, E. M., B. S. Goldman, D. L. Beckman and R. G. Kranz, 1997. A thioreduction pathway tethered to the membrane for periplasmic cytochromes c biogenesis; in vitro and in vivo studies. J. Mol. Biol. 271 679–692. [DOI] [PubMed] [Google Scholar]
- Motohashi, K., and T. Hisabori, 2006. HCF164 receives reducing equivalents from stromal thioredoxin across the thylakoid membrane and mediates reduction of target proteins in the thylakoid lumen. J. Biol. Chem. 281 35039–35047. [DOI] [PubMed] [Google Scholar]
- Motohashi, K., and T. Hisabori, 2010. CcdA is a thylakoid membrane protein required for the transfer of reducing equivalents from stroma to thylakoid lumen in the higher plant chloroplast. Antioxid. Redox Signal. 13 1169–1176. [DOI] [PubMed] [Google Scholar]
- Nakamoto, S. S., P. Hamel and S. Merchant, 2000. Assembly of chloroplast cytochromes b and c. Biochimie 82 603–614. [DOI] [PubMed] [Google Scholar]
- Oren, A., 2005. A hundred years of Dunaliella research: 1905–2005. Saline Syst. 1 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, M. L. D., P. P. Hamel, S. T. Gabilly, H. Zegzouti, J. V. Perea et al., 2004. A homolog of prokaryotic thiol disulfide transporter CcdA is required for the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J. Biol. Chem. 279 32474–32482. [DOI] [PubMed] [Google Scholar]
- Peters, A., C. Kulajta, G. Pawlik, F. Daldal and H.-G. Koch, 2008. Stability of the cbb3-type cytochrome oxidase requires specific CcoQ-CcoP interactions. J. Bacteriol. 190 5576–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock, S. V., D. L. Prout, A. C. Godfrey, S. D. Lemaire and J. V. Moroney, 2004. The Chlamydomonas reinhardtii proteins Ccp1 and Ccp2 are required for long-term growth, but are not necessary for efficient photosynthesis, in a low-CO2 environment. Plant Mol. Biol. 56 125–132. [DOI] [PubMed] [Google Scholar]
- Prochnik, S. E., J. Umen, A. M. Nedelcu, A. Hallmann, S. M. Miller et al., 2010. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purton, S., and J. D. Rochaix, 1994. Complementation of a Chlamydomonas reinhardtii mutant using a genomic cosmid library. Plant Mol. Biol. 24 533–537. [DOI] [PubMed] [Google Scholar]
- Quinn, J. M., and S. Merchant, 1998. Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 297 263–279. [DOI] [PubMed] [Google Scholar]
- Richard-Fogal, C. L., E. R. Frawley and R. G. Kranz, 2008. Topology and function of CcmD in cytochrome c maturation. J. Bacteriol. 190 3489–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Velazquez, C., R. L. Cox and T. J. Donohue, 2001. Characterization of Rhodobacter sphaeroides cytochrome c2 proteins with altered heme attachment sites. Arch. Biochem. Biophys. 389 234–244. [DOI] [PubMed] [Google Scholar]
- Ritz, D., and J. Beckwith, 2001. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 55 21–48. [DOI] [PubMed] [Google Scholar]
- Ruijter, J. M., C. Ramakers, W. M. Hoogaars, Y. Karlen, O. Bakker et al., 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Marcoux, D., F. A. Wollman and C. de Vitry, 2009. Biogenesis of cytochrome b6 in photosynthetic membranes. J. Cell Biol. 185 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambongi, Y., and S. J. Ferguson, 1994. Specific thiol compounds complement deficiency in c-type cytochrome biogenesis in Escherichia coli carrying a mutation in a membrane-bound disulphide isomerase-like protein. FEBS Lett. 353 235–238. [DOI] [PubMed] [Google Scholar]
- Sanders, C., S. Turkarslan, D. W. Lee and F. Daldal, 2010. Cytochrome c biogenesis: the Ccm system. Trends Microbiol. 18 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, H., R. A. Fabianek, E. C. Pellicioli, H. Hennecke and L. Thöny-Meyer, 1999. Heme transfer to the heme chaperone CcmE during cytochrome c maturation requires the CcmC protein, which may function independently of the ABC-transporter CcmAB. Proc. Natl. Acad. Sci. USA 96 6462–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, H., E. C. Pellicioli and L. Thöny-Meyer, 2000. New insights into the role of CcmC, CcmD and CcmE in the haem delivery pathway during cytochrome c maturation by a complete mutational analysis of the conserved tryptophan-rich motif of CcmC. Mol. Microbiol. 37 1379–1388. [DOI] [PubMed] [Google Scholar]
- Setterdahl, A. T., B. S. Goldman, M. Hirasawa, P. Jacquot, A. J. Smith et al., 2000. Oxidation-reduction properties of disulfide-containing proteins of the Rhodobacter capsulatus cytochrome c biogenesis system. Biochemistry 39 10172–10176. [DOI] [PubMed] [Google Scholar]
- Stroebel, D., Y. Choquet, J. L. Popot and D. Picot, 2003. An atypical haem in the cytochrome b6f complex. Nature 426 413–418. [DOI] [PubMed] [Google Scholar]
- Thöny-Meyer, L., 1997. Biogenesis of respiratory cytochromes in bacteria. Microbiol. Mol. Biol. Rev. 61 337–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkarslan, S., C. Sanders, S. Ekici and F. Daldal, 2008. Compensatory thio-redox interactions between DsbA, CcdA and CcmG unveil the apocytochrome c holdase role of CcmG during cytochrome c maturation. Mol. Microbiol. 70 652–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Heijne, G., 1989. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature 341 456–458. [DOI] [PubMed] [Google Scholar]
- Xie, Z., D. Culler, B. W. Dreyfuss, R. Kuras, F. A. Wollman et al., 1998. Genetic analysis of chloroplast c-type cytochrome assembly in Chlamydomonas reinhardtii: one chloroplast locus and at least four nuclear loci are required for heme attachment. Genetics 148 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L., S.-C. Tso, S. K. Shenoy, B. N. Quinn and D. Xia, 1999. The role of the supernumerary subunit of Rhodobactersphaeroides cytochrome bc1 complex. J. Bioenerg. Biomembr. 31 251–258. [DOI] [PubMed] [Google Scholar]