Abstract
The restricted expression of epidermal growth factor (EGF) family ligands is important for proper development and for preventing cancerous growth in mammals. In Caenorhabditis elegans, the class A and B synthetic multivulva (synMuv) genes redundantly repress expression of lin-3 EGF to negatively regulate Ras-mediated vulval development. The class B synMuv genes encode proteins homologous to components of the NuRD and Myb-MuvB/dREAM transcriptional repressor complexes, indicating that they likely silence lin-3 EGF through chromatin remodeling. The two class A synMuv genes cloned thus far, lin-8 and lin-15A, both encode novel proteins. The LIN-8 protein is nuclear. We have characterized the class A synMuv gene lin-56 and found it to encode a novel protein that shares a THAP-like C2CH motif with LIN-15A. Both the LIN-56 and LIN-15A proteins localize to nuclei. Wild-type levels of LIN-56 require LIN-15A, and wild-type levels and/or localization of LIN-15A requires LIN-56. Furthermore, LIN-56 and LIN-15A interact in the yeast two-hybrid system. We propose that LIN-56 and LIN-15A associate in a nuclear complex that inhibits vulval specification by repressing lin-3 EGF expression.
TUMORIGENESIS requires misregulation of pathways controlling cell proliferation, differentiation, and apoptosis and likely involves multiple mutations that result in the activation of proto-oncogenes and the inactivation of tumor-suppressor genes. A particularly frequent target of misregulation in human cancers is the epidermal growth factor (EGF) and Ras-signaling pathway that controls cell proliferation. The EGF/Ras pathway can be overactivated by misexpression of EGF-like ligands, mutation or overexpression of EGF receptors, or constitutive mutational activation of Ras (reviewed by Normanno et al. 2001, 2006; Downward 2003).
In Caenorhabditis elegans, an EGF/Ras pathway plays a central role in vulval development (reviewed by Kornfeld 1997; Moghal and Sternberg 2003). Six multipotent cells, P(3–8).p, of the ventral ectoderm each can express either the 1° or 2° vulval fates or the 3° nonvulval fate. The LIN-3 EGF ligand is expressed in the anchor cell of the somatic gonad and activates the LET-23 EGF receptor (EGFR) in the closest P(3–8).p cells. LET-23 EGFR subsequently signals through a Ras/MAP kinase pathway to specify vulval fates (Aroian et al. 1990; Han and Sternberg 1990; Hill and Sternberg 1992; Lackner et al. 1994; Kornfeld et al. 1995; Wu et al. 1995). P6.p assumes the 1° vulval fate, dividing three times to produce eight descendants, and P5.p and P7.p assume the 2° vulval fate, generating seven descendants each. The 22 progeny of P(5–7).p undergo morphogenesis to generate the adult vulva. P3.p, P4.p, and P8.p assume the nonvulval 3° fate and divide once and fuse with a multinucleate hypodermal cell, hyp7. Loss-of-function mutations in components of the EGF/Ras pathway cause P(5–7).p to adopt the nonvulval 3° fate and result in a vulvaless (Vul) phenotype. Gain-of-function mutations in let-23 EGFR or let-60 Ras or overexpression of lin-3 EGF cause P3.p, P4.p, and P8.p to adopt vulval 1° or 2° fates and result in a multivulva (Muv) phenotype (Beitel et al. 1990; Han and Sternberg 1990; Hill and Sternberg 1992; Katz et al. 1996). Muv animals produce extra vulval tissue that forms ectopic ventral protrusions.
The EGF/Ras pathway, which is essential for C. elegans vulval induction, is antagonized by the functionally redundant class A and B synthetic multivulva (synMuv) genes (Ferguson and Horvitz 1989). Hermaphrodites carrying only a single synMuv mutation generally appear as wild type for vulval induction, while hermaphrodites carrying mutations in both a class A and a class B synMuv gene exhibit a Muv phenotype. Four class A synMuv genes and at least 25 class B genes have been identified (Horvitz and Sulston 1980; Ferguson and Horvitz 1985, 1989; Lu and Horvitz 1998; Hsieh et al. 1999; Solari and Ahringer 2000; von Zelewsky et al. 2000; Ceol and Horvitz 2001, 2004; Couteau et al. 2002; Unhavaithaya et al. 2002; Thomas et al. 2003; Davison et al. 2005; Poulin et al. 2005; Harrison et al. 2006, 2007; Andersen and Horvitz 2007). The synMuv genes likely act upstream of EGF/Ras signaling, as loss-of-function of components of the EGF/Ras pathway can suppress the synMuv phenotype (Ferguson et al. 1987; Huang et al. 1994; Lu and Horvitz 1998; Ceol and Horvitz 2001, 2004; Cui et al. 2006). In class A and class B synMuv double mutants, lin-3 EGF is overexpressed, likely ectopically, indicating that the synMuv genes negatively regulate EGF/Ras signaling by repressing expression of lin-3 EGF (Cui et al. 2006). All synMuv genes tested, including all four class A synMuv genes, repress lin-3 EGF expression (Cui et al. 2006; Andersen et al. 2008).
Given their molecular identities, the class B synMuv genes likely repress lin-3 via chromatin remodeling. lin-35, efl-1, dpl-1, lin-53, hda-1, let-418, met-2, and hpl-2 encode C. elegans counterparts of mammalian Rb, E2F, DP, the Rb-associated protein RbAp48, histone deacetylase (HDAC), the Mi-2 chromatin-remodeling enzyme, a histone H3 lysine-9 methyltransferase, and the histone H3 methyl-lysine-9-binding protein HP1, respectively (Lu and Horvitz 1998; Solari and Ahringer 2000; von Zelewsky et al. 2000; Ceol and Horvitz 2001; Couteau et al. 2002; Andersen and Horvitz 2007). Studies of these mammalian proteins strongly suggest that an Rb/E2F/DP complex represses transcription of target genes by recruiting HDAC, RbAp48, histone H3 lysine-9 methyltransferase, and HP1 (reviewed by Nielsen et al. 2001; Vandel et al. 2001; Zhang and Dean 2001). RbAp48, HDAC, and Mi-2 are components of the histone deacetylase and chromatin-remodeling NuRD complex (reviewed by Knoepfler and Eisenman 1999) and might be involved in transcriptional repression (reviewed by Richards and Elgin 2002). Furthermore, LIN-35 Rb, LIN-53 RbAp48, DPL-1 DP, and additional class B synMuv proteins form a complex in vivo that resembles the Drosophila Myb-MuvB and dREAM and human LINC/DREAM complexes, which regulate the transcription of many E2F-target genes (Korenjak et al. 2004; Lewis et al. 2004; Harrison et al. 2006; Litovchick et al. 2007; Pilkinton et al. 2007; Schmit et al. 2007).
Although the class A synMuv genes function redundantly with the class B synMuv genes, the mechanism by which the class A synMuv genes repress lin-3 EGF expression to inhibit Ras-mediated vulval development is not known. Of the four class A synMuv genes, the lin-8 and lin-15A loci were cloned previously; both encode novel proteins (Clark et al. 1994; Huang et al. 1994; Davison et al. 2005). Nonetheless, it is likely that the class A synMuv genes also act to regulate gene expression. First, although novel in sequence, LIN-8 is a nuclear protein that can interact physically with LIN-35 Rb in vitro, suggesting that it might be present in vivo at the sites of transcriptional repression complexes (Davison et al. 2005). Second, the class A synMuv genes either directly or indirectly repress the transcription of lin-3 EGF (Cui et al. 2006). We report here the cloning and characterization of the class A synMuv gene lin-56 and propose that LIN-56 and LIN-15A normally associate in a nuclear complex that affects cell-fate determination by negatively regulating the EGF/Ras pathway.
MATERIALS AND METHODS
Strains:
C. elegans strains were cultivated as described (Brenner 1974) and were grown at 20° unless otherwise indicated. Bristol N2 was the wild-type strain. The mutant alleles used in this study are listed below, and a description is presented by Riddle et al. (1997) unless noted otherwise: LGII: dpy-10(e128), lin-8(n2731) (Thomas et al. 2003), lin-38(n751), and lin-56(n2728) (Thomas et al. 2003); LGIII: lin-36(n766); LGIV: let-60(n1876) (Beitel et al. 1990); and LGX: lin-15A(n433, n767, n749), lin-15B(n744), and lin-15AB(e1763). Also used was the chromosomal rearrangement mnC1 [dpy-10(e128) unc-52(e444)] and the following deficiencies: eDf21, mnDf16, mnDf29, mnDf57, mnDf61, mnDf62, mnDf71, and mnDf90 (Sigurdson et al. 1984). lin-56(n3355) was isolated in a noncomplementation screen using lin-56(n2728).
Transgenic animals:
Germline transformation by microinjection was performed as previously described (Mello et al. 1991). The transformation markers pRF4 and myo-3∷gfp were co-injected with experimental constructs at 80 and 50 ng/μl, respectively. Experimental constructs were injected at 10–100 ng/μl. Stable transformants were analyzed.
Molecular biology:
Standard molecular biology procedures were followed (Sambrook et al. 1989). To disrupt ZK673.3 in pEMD4, an oligonucleotide linker containing an opal stop codon was ligated into an SphI site within the second exon. The resulting construct, pEMD5, contains an insertion of the sequence TGAGACTAGTGCATGC and is predicted to produce a truncated protein containing only the first 120 amino acids of the wild-type 322 amino acids of ZK673.3. The disrupted ZK673.3 open reading frame was transferred to pEMD1 by PCR amplification of the region surrounding the engineered insertion from pEMD5, followed by substitution of the wild-type sequence in pEMD1 with the PCR product, generating pEMD14. To ensure that loss of rescuing activity was caused by the disruption of the ZK673.3 open reading frame, the latter was restored by digestion of pEMD14 with SphI followed by religation, generating pEMD15.
RNA interference:
RNA interference (RNAi) was performed by feeding animals bacteria expressing double-stranded RNA, essentially as previously described (Kamath et al. 2001; Timmons et al. 2001). The bacterial strain expressing double-stranded lin-3 RNA was from Kamath et al. (2003), and the DNA sequence of the insert was determined to ensure that it was correct.
Antibody preparation and immunocytochemistry:
Rabbit anti-LIN-56 antisera were generated using purified GST-LIN-56 (aa 1–322) as the immunogen, affinity-purified against MBP-LIN-56 (aa 1–322) as described by Koelle and Horvitz (1996), and pre-adsorbed against an acetone precipitate of proteins prepared from lin-56(n2728) mixed-stage worms, essentially as described by Harlow and Lane (1988). Rabbit anti-LIN-15A antibodies were generated using purified 6His-LIN-15A (aa 77–324) as immunogen and were purified as with the anti-LIN-56 antisera, using 6His-LIN-15A (aa 77–324) protein for affinity purification and lin-15AB(e1763) mixed-stage worms for pre-adsorption. Affinity-purified anti-LIN-56 antibodies were used at 1:2000 for immunoblots and, following pre-adsorption, at 1:100 for immunocytochemistry. Affinity-purified and pre-adsorbed anti-LIN-15A antibodies were used at 1:25 for immunocytochemistry. Anti-α-tubulin monoclonal antibody DM1A (Sigma, St. Louis) and MH27 (Francis and Waterston 1991), which recognizes the apical borders of C. elegans epithelial cells, were used as positive controls for immunocytochemistry at 1:100 and 1:1000, respectively. Embryos were fixed in 0.8% paraformaldehyde for 20 min, as described by Guenther and Garriga (1996). Larvae and adults were fixed in 2% paraformaldehyde for 15 min, essentially as described by Finney and Ruvkun (1990). Images were obtained by using a Zeiss LSM510 laser confocal microscope and software and were processed with Adobe Photoshop.
Subcellular fractionation of embryos:
Embryos were collected by bleaching gravid hermaphrodites in a 0.8 n NaOH, 8% hypochlorite solution. Fractionation into nuclear and cytosolic fractions was performed as described by Chen et al. (2000). The protein content of each fraction was determined using Bradford reagent, and 10 μg of each fraction was used for immunoblots. Rabbit polyclonal antiserum 3930 generated against C. elegans lamin (Gruenbaum et al. 2002) was used at 1:5000 on immunoblots as a control for nuclear fractionation quality. Anti-SQV-4 antibodies (Hwang and Horvitz 2002) were used at 1:3000 on immunoblots as a control for cytosolic fractionation quality.
RT-PCR analysis:
Total RNA was isolated from mixed-stage worms using TRIZOL (Invitrogen, Carlsbad, CA). Standard RT-PCR (Titan One Tube RT-PCR System, Roche Applied Science, Indianapolis) was used to qualitatively compare levels of lin-56 and lin-15A RNA in total RNA samples derived from wild-type, lin-56(n2728), lin-15A(n767), and lin-15AB(e1763) animals. RT-PCR of hexokinase (H25P06.1) was performed as a control. Primers used were lin-15A Fwd9 (5′-CGAATGTCAAGCTTGGCGAACG-3′), lin-15A Rev5 (5′-CGGTTTACTGAGAGACCC-3′), lin-56 Fwd2 (5′-AGACTGGGCAGAATGCG-3′), lin-56 Rev2 (5′-GCTCCACTTTTTCAGGAAAAC-3′), hexokinase Fwd1A (5′-GAGCTCGGCATTCAATATCG-3′), and hexokinase Rev1B (5′-GCTTCATATGCAGCTGCAACC-3′).
Quantitative real-time RT-PCR (Heid et al. 1996) was used to measure the amount of lin-56 mRNA in poly(A)+ mRNA samples derived from wild-type, lin-15A(n767), and lin-15AB(e1763) animals. Poly(A)+ mRNA was purified from total RNA using Oligotex resin (Qiagen, Valencia, CA). cDNA was produced from the purified poly(A)+ mRNA using oligo(dT) primer and SuperScript II Rnase H− Reverse Transcriptase (Invitrogen). Using an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA), absolute lin-56 and hexokinase mRNA levels were determined relative to a genomic DNA dilution series, and lin-56 mRNA levels were then normalized to hexokinase mRNA levels. Fifty- and 100-ng mRNA equivalents were analyzed in triplicate for each genotype. lin-56 and hexokinase reactions were performed in separate tubes. No significant amplification was observed when reverse transcriptase was omitted from the initial RT reaction. Primers and probes used were lin-56 Fwd (5′-TTGGTGCAAAGTCTACACGATGA-3′), lin-56 Rev (5′-TTGCGCACATCGAACTTTGT-3′), lin-56 probe (5′-6-FAM-TCGATCTTCCCTGGGCGAGCAGT-TAMRA-3′), hexokinase Fwd (5′-CGTGGAGCCGCACTCATC-3′), hexokinase Rev (5′-CAGATCCTTCAGCCGCTTCT-3′), and hexokinase probe (5′-VIC-TCGCTTGACTCTCGAAACGATTGCG-TAMRA-3′).
Yeast two-hybrid system:
Full-length cDNAs for lin-15A, lin-37, lin-53, and lin-56 were cloned into Gateway entry vector pDONR201 (Invitrogen) and then each transferred into two Gateway two-hybrid destination vectors: pDEST-DB, encoding the Gal4 DNA-binding domain (DB), and pDEST-AD, encoding the Gal4 activation domain (AD) (Walhout et al. 2000a,b). Yeast strain Y190 (MATa gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3 leu2-112 URA3∷GAL1-lacZ LYS2∷GAL1-HIS3 cyhr) was cotransformed with DB- and AD-fusion constructs. Interaction in the two-hybrid system was assayed by colony formation on SC-Trp-Leu-His medium containing 25 mm 3-aminotriazole (3-AT).
RESULTS
lin-56 encodes a putative transcription factor containing a THAP-like domain:
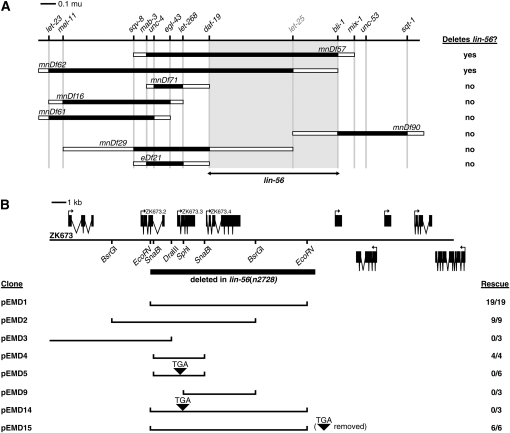
lin-56 was previously mapped close to unc-4 on chromosome II (Thomas et al. 2003). Using deficiencies, we further mapped lin-56 to a region of 554 kb between daf-19 and bli-1 (Figure 1A). Cosmid ZK673 and a 3.5-kb SnaBI–SnaBI subclone of ZK673 (pEMD4) rescued the synMuv phenotype when injected into lin-56(n2728); lin-15B(n744) animals (Figure 1B). The 3.5-kb minimal rescuing fragment contains a single predicted gene, ZK673.3. We found the only existing lin-56 allele, n2728, to contain an 11.2-kb deletion that eliminates not only ZK673.3 but also its downstream neighbor, ZK673.4, and the 3′-end of its upstream neighbor, ZK673.2 (Figure 1B).
Figure 1.—
Cloning of lin-56. (A) Genetic map of the lin-56 region. Deficiency mapping placed lin-56 between the right endpoint of mnDf29 and the left endpoint of mnDf90, as defined by the markers daf-19 and bli-1, respectively (shaded region). Cloned genes are in regular type; genes not cloned are in shaded type. Shaded rectangles indicate regions known to be deleted by each deficiency, and open rectangles indicate regions that contain a deficiency endpoint and might be deleted by each deficiency. (B) Transformation rescue of lin-56. Predicted genes contained within the rescuing cosmid ZK673 (top) and subclones derived from this cosmid (bottom) are shown. The region deleted in lin-56(n2728) mutants is indicated by the solid bar. The sequences flanking the lin-56(n2728) deletion are ACCAAAGGAGGAGGTCAGCC [11,190-bp deletion] CCTTGTGGGGGAACAATGCG. Insertion of a TGA stop codon is indicated by a solid arrowhead. (C) Sequence of the LIN-56 protein. The glutamine mutated to an ochre stop codon in the n3355 allele is shaded. The region aligned in D is underlined. (D) Alignment of the novel C2CH motif found in LIN-56, LIN-15A, LIN-15B, HIM-17, and three additional uncharacterized C. elegans proteins. Y32B12B.4 contains five repeats, denoted r1–r5, of the motif. Conserved cysteines and histidine are in red. Solid and shaded boxes indicate identities and similarities, respectively, with LIN-56. Positions of missense mutations found in lin-15A(n433) and lin-15A(n749) mutants are marked with arrowheads.
Since ZK673.3 and ZK673.4 share a small region of similarity (see below), we were concerned that the class A synMuv phenotype of the n2728 mutant might be caused not by loss of ZK673.3, as the rescue experiments suggest, but either by loss of ZK673.4 or by loss of both ZK673.3 and ZK673.4. Disruption of ZK673.3 in a 10.7-kb rescuing subclone of ZK673 (pEMD1) that contains both ZK673.3 and ZK673.4 resulted in loss of rescuing activity (pEMD14) (Figure 1B), and expression of a ZK673.3 cDNA under the control of heat-shock promoters efficiently rescued the synMuv phenotype of lin-56(n2728); lin-15B(n744) animals (Table 1). Furthermore, RNAi (Fire et al. 1998) of ZK673.3 but not of ZK673.4 resulted in a Muv phenotype in lin-15B(n744) animals (data not shown). Finally, we isolated a second lin-56 allele, n3355, in a noncomplementation screen and found n3355 to correspond to a glutamine-to-ochre nonsense mutation at amino acid 61 of ZK673.3 (Figure 1C). The strengths of the class A synMuv phenotypes of lin-56(n2728) and lin-56(n3355) mutants were indistinguishable, as both caused a fully penetrant Muv phenotype in the weak class B synMuv mutant background lin-15B(n2245) at 15°. Therefore, we conclude that ZK673.3 is lin-56.
TABLE 1.
lin-56 overexpression rescues the lin-56(lf); lin-15B(lf), but not the lin-15AB(lf), synMuv phenotype
| Genotype | Transgene | Line | Heat shock | % Muva ± SD (n)b |
|---|---|---|---|---|
| lin-56(n2728); lin-15B(n744) | Phs-lin-56 | 1 | − | 97.9 ± 0.9 (490) |
| + | 16.5 ± 6.8 (340) | |||
| 2 | − | 99.7 ± 0.3 (608) | ||
| + | 17.5 ± 15.3 (244) | |||
| lin-56(n2728); lin-15B(n744) | Phs-gfp | 1 | − | 100 ± 0 (319) |
| + | 100 ± 0 (365) | |||
| lin-15AB(e1763) | Phs-lin-56 | 1 | − | 100 ± 0 (280) |
| + | 100 ± 0 (211) | |||
| 2 | − | 100 ± 0 (117) | ||
| + | 100 ± 0 (112) | |||
| 3 | − | 100 ± 0 (172) | ||
| + | 100 ± 0 (319) | |||
| 4 | − | 100 ± 0 (140) | ||
| + | 100 ± 0 (226) | |||
| 5 | − | 100 ± 0 (153) | ||
| + | 100 ± 0 (107) | |||
| lin-15AB(e1763) | Phs-gfp | 1 | − | 100 ± 0 (301) |
| + | 100 ± 0 (158) |
lin-56(n2728); lin-15B(n744) or lin-15AB(e1763) hermaphrodites carrying an extrachromosomal array of either a lin-56 cDNA (Phs-lin-56) or the gfp-coding sequence (Phs-gfp) fused to the C. elegans heat-shock promoters and a dominant Rol marker were synchronized by bleaching and starvation-induced L1 arrest. Expression from the heat-shock promoters was induced at the early L2 stage (24–25 hr post starvation) by incubation at 33° for 1 hr. Animals were assayed in at least three individual batches, with at least 100 Rol worms analyzed in total for each line.
% Muv, percentage of Rol animals that were Muv.
n, number of animals examined.
Blumenthal et al. (2002) have suggested that ZK673.2 and ZK673.3 may be expressed as an operon, because these two genes are separated by <500 bp and because they detected the C. elegans SL2 trans-spliced leader sequence at the 5′-end of ZK673.3 mRNAs. However, no sequence upstream of the ZK673.2 open reading frame was required for rescue by ZK673.3 of the lin-56(n2728) class A synMuv phenotype (Figure 1B), consistent with the hypothesis that ZK673.2 and ZK673.3 are at least not always expressed as an operon.
LIN-56, a novel acidic protein of 322 amino acids, contains a domain similar to the THAP domain (Figure 1C), a zinc-finger-like motif that can bind to DNA (Clouaire et al. 2005). Specifically, LIN-56 shares a C-X-[ILV]-C-X(33,38)-A-X(11,13)-C-X(2)-H motif with several C. elegans proteins (Figure 1D), including the class A synMuv protein LIN-15A, the class B synMuv protein LIN-15B, the protein encoded by its downstream genomic neighbor ZK673.4, and HIM-17, a protein required for meiotic recombination and histone H3 lysine-9 methylation in the germline (Reddy and Villeneuve 2004). The conservation of cysteine and histidine residues suggests that this motif may bind a zinc ion and mediate binding to DNA, RNA, protein, or a small molecule (reviewed by Laity et al. 2001). Although the interval between the two internal cysteines is greater than that found in typical zinc fingers (Krishna et al. 2003), the spacing of the cysteine and histidine residues within the C2CH motif described here matches the spacing characteristic of the THAP domain, which can mediate sequence-specific DNA binding (Roussigne et al. 2003; Clouaire et al. 2005; Cayrol et al. 2007; Sabogal et al. 2010). Additional features of the THAP domain, including an invariant tryptophan between the second and third cysteine residues and a C-terminal AVPTIF motif, are not conserved. This THAP-like motif is likely functionally significant, as two lin-15A alleles contain missense mutations therein, and one of these mutations affects a conserved alanine residue (Figure 1D).
The Muv phenotypes of many synMuv double mutants are suppressed by RNAi of lin-3, suggesting that the synMuv genes act upstream of lin-3 in vulval development. To determine if lin-56 also acts upstream of lin-3, we inactivated lin-3 by feeding lin-56(n2728); lin-15B(n744) animals bacteria expressing double-stranded lin-3 RNA. lin-56(n2728); lin-15B(n744) animals fed bacteria with an empty vector exhibited a Muv phenotype with 100% penetrance (n = 209). lin-56(n2728); lin-15B(n744) animals fed bacteria expressing lin-3 double-stranded RNA exhibited a Muv phenotype with a lower 85% penetrance (n = 179) (P < 0.00001), and the expressivity of the Muv animals, as determined by the number of ectopic pseudovulvae, was notably lower than animals fed control bacteria. Therefore, lin-3 is required for the class A synMuv phenotype of lin-56 mutants, consistent with the hypothesis that lin-56 acts upstream of lin-3.
LIN-56 is a ubiquitous nuclear protein:
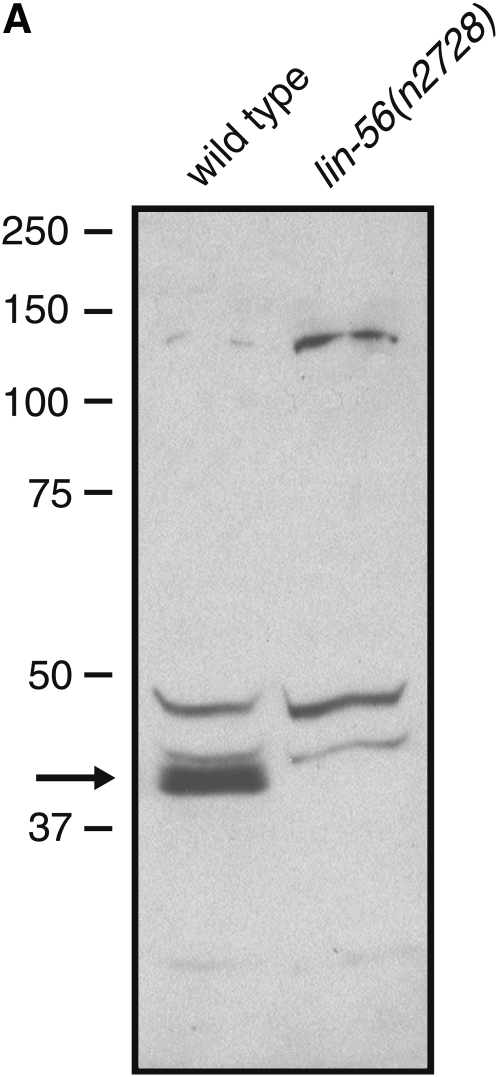
To determine the expression pattern of the LIN-56 protein, we generated anti-LIN-56 antibodies, which recognized a protein of ∼40 kDa in wild-type but not in lin-56(n2728) protein extracts (Figure 2A); the predicted size of LIN-56 is 37 kDa. The anti-LIN-56 antibodies sometimes appear to recognize a doublet (data not shown), which could represent alternative splice variants of lin-56 or products resulting from distinct transcriptional initiation sites. Although the three lin-56 cDNA clones (courtesy of Y. Kohara) analyzed are identical in sequence and lack any evidence of trans-splicing, SL2 trans-splicing of the ZK673.3 mRNA has been observed by others (Blumenthal et al. 2002; Hwang et al. 2004). Alternatively, LIN-56 may be post-translationally modified. The anti-LIN-56 antibodies also recognized several other proteins (Figure 2A), but those proteins were a different size than predicted for LIN-56 and were present in both wild-type and lin-56(n2728) protein extracts and are therefore nonspecific.
Figure 2.—
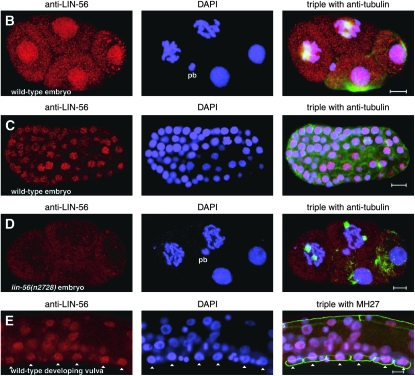
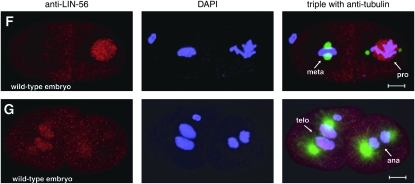
LIN-56 is broadly expressed and localized to nuclei. (A) Immunoblot of wild-type and lin-56(n2728) protein extracts probed with anti-LIN-56 antibodies. The arrow indicates the band that corresponds to LIN-56. Molecular weights of marker proteins are indicated at left in kilodaltons. (B–E) Whole-mount staining with anti-LIN-56 antibodies (red), DAPI (blue), and either anti-tubulin antibody (green) in embryos or MH27 antibody (green) in larvae as a fixation control. Scale bars, 5 μm. Anterior is to the left in all images. pb, polar body. (B and C) LIN-56 staining is observed in the nuclei of most if not all cells throughout embryonic development. Shown are (B) four-cell and (C) late-stage wild-type embryos. (D) Nuclear staining is never observed with anti-LIN-56 antisera in lin-56(n2728) animals. Shown is a four-cell lin-56(n2728) embryo. (E) LIN-56 is present in the P(3–8).p vulval equivalence group and in their descendants during wild-type vulval development. Shown is the midbody of a wild-type L2 larva. Arrowheads point to nuclei of P(3–8).p. MH27 stains the apical borders between the Pn.p cells. (F and G) LIN-56 does not colocalize with chromatin during metaphase or anaphase. F and G show two-cell wild-type embryos. ana, anaphase; meta, metaphase; pro, prophase; telo, telophase.
The anti-LIN-56 antibodies were used for whole-mount staining of worms. Starting at the late one-cell or early two-cell stage, LIN-56 staining was observed in the nuclei of most if not all somatic cells throughout embryonic and larval development as well as in adulthood (Figure 2, B, C, and E; data not shown). Nuclear staining was never observed with the anti-LIN-56 antibodies in the somatic cells of lin-56(n2728) animals of any stage (Figure 2D; data not shown), demonstrating specificity. LIN-56 is present in the P(3–8).p vulval equivalence group and their descendants throughout vulval development (Figure 2E). LIN-56 did not colocalize with chromatin during metaphase or anaphase (Figure 2, F and G). Germline staining was observed in both wild-type and lin-56(n2728) animals and thus was not specific.
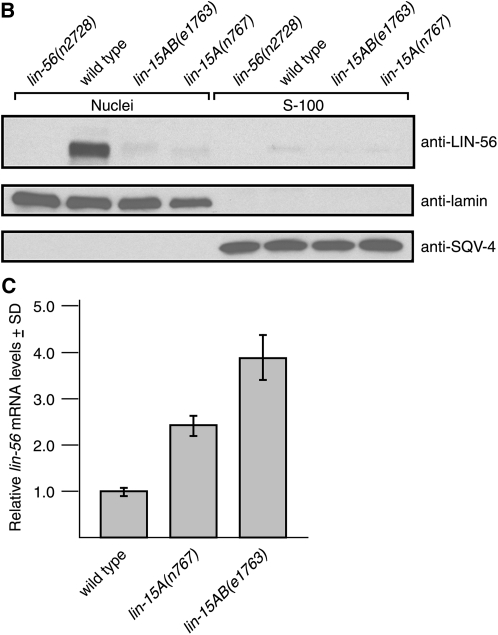
To confirm that LIN-56 is localized to nuclei, we performed subcellular fractionation of embryo lysates. We used lamin as a marker for nuclear fractions (Liu et al. 2000; Gruenbaum et al. 2002) and SQV-4 as a marker for cytosolic material (Hwang and Horvitz 2002). The majority of the LIN-56 protein cofractionated with nuclear lamin; LIN-56 protein was barely detectable in the SQV-4-containing cytosolic fraction (see Figure 3B). LIN-56 is thus stably associated with nuclei.
Figure 3.—
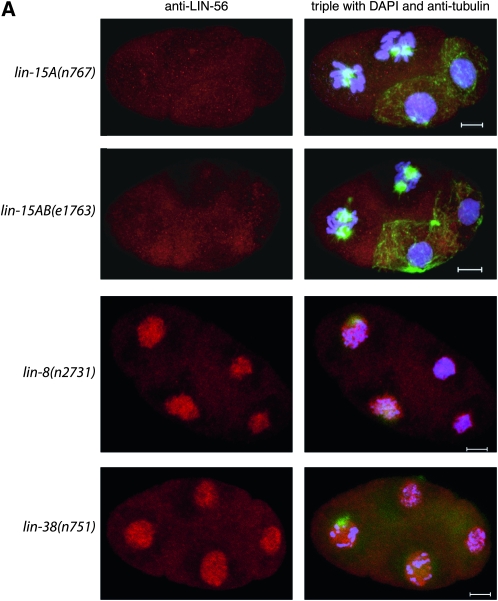
LIN-56 protein but not lin-56 mRNA levels are greatly reduced in lin-15A(lf) mutants. (A) LIN-56 nuclear staining is greatly reduced in lin-15A(n767) and lin-15AB(e1763) mutants, but not in lin-8(n2731) or lin-38(n751) mutants. The posterior two cells in the lin-15A(n767) and lin-15AB(e1763) panels are dividing and therefore would not be expected to display LIN-56 nuclear expression, while the anterior two cells are not dividing. Shown are four-cell embryos stained with anti-LIN-56 antibody (red), DAPI (blue), and anti-tubulin antibody (green). Scale bars, 5 μm. Anterior is to the left in all images. (B) Immunoblot of nuclear and S-100 cytosolic fractions derived from wild-type, lin-56(n2728), lin-15AB(e1763), and lin-15A(n767) embryos probed with anti-LIN-56 antibody. Shown for reference are the same immunoblot probed with anti-lamin and anti-SQV-4 antibodies as markers for nuclear and cytosolic proteins, respectively. (C) lin-56 mRNA levels are not reduced in lin-15A(lf) mutants as compared to the wild type. Quantitative real-time RT-PCR (Heid et al. 1996) was used to measure lin-56 mRNA levels relative to hexokinase mRNA levels. Results for each genotype are normalized to those for the wild type. Error bars, SD.
LIN-56 protein, but not lin-56 mRNA, is reduced in lin-15A(lf) mutants:
We analyzed the impact of mutations in other class A synMuv genes on the expression of LIN-56. After staining with anti-LIN-56 antibodies we observed that the LIN-56 staining pattern of lin-8(n2731) and lin-38(n751) animals was not different from that of the wild type (Figure 3A; data not shown). lin-8(n2731) causes an early nonsense mutation and is likely a protein null allele (Davison et al. 2005). lin-38(n751) is a non-null allele, as null mutations in lin-38 are inviable (A. M. Saffer and H. R. Horwitz, unpublished results). By contrast, LIN-56 nuclear staining was greatly reduced in lin-15A(n767) and lin-15AB(e1763) individuals at all stages (Figure 3A; data not shown). lin-15A(n767) is a small deletion predicted to result in a truncated LIN-15A protein, and lin-15AB(e1763) is a large deletion that eliminates lin-15A and lin-15B (Clark et al. 1994; Huang et al. 1994). Fractionation experiments and immunoblot analyses confirmed that LIN-56 protein was barely detectable in the nuclear fractions of lin-15A(n767) and lin-15AB(e1763) embryos (Figure 3B). Also, the cytosolic fractions of these lin-15A(lf) mutants contained no more LIN-56 protein than the cytosolic fraction of wild-type embryos (Figure 3B), indicating that the reduced amount of LIN-56 in the nuclei of lin-15A(lf) mutants was not the result of mislocalization of this protein to the cytosol. Instead, the total amount of LIN-56 protein in lin-15A(lf) animals was greatly reduced in comparison to that in the wild type, suggesting that lin-15A is required for the expression or stability of lin-56 mRNA or LIN-56 protein.
To determine if the low level of LIN-56 protein in lin-15A(lf) animals was the result of decreased transcription or stability of lin-56 mRNA, we used quantitative real-time RT-PCR (Heid et al. 1996) to measure lin-56 mRNA levels in wild-type, lin-15A(n767), and lin-15AB(e1763) animals relative to hexokinase mRNA levels. Relative lin-56 mRNA levels appeared to be somewhat higher in lin-15A(n767) and lin-15AB(e1763) animals than in wild-type animals (Figure 3C; see also Figure 4B). The reason for the apparent increase in lin-56 mRNA levels in the two lin-15A(lf) mutants analyzed was not determined. The decreased amount of LIN-56 protein in lin-15A(lf) as compared to wild-type animals thus likely results from a post-transcriptional event, suggesting that LIN-15A is required for either translation or stability of LIN-56 protein.
Figure 4.—
LIN-15A nuclear accumulation but not lin-15A mRNA is reduced in lin-56(lf) mutants. (A) LIN-15A nuclear staining is greatly reduced in lin-56(n2728) and lin-56(n3355) mutants. Multicellular embryos were stained with anti-LIN-15A antibodies (green), anti-tubulin antibody (red), and DAPI (blue). The genotype of each embryo is indicated on the left. Scale bars, 5 μm. Anterior is to the left in all images. (B) lin-15A RNA levels are not reduced in lin-56(n2728) mutants compared to the wild type. RT-PCR was used to determine levels of lin-15A, lin-56, and hexokinase RNA in total RNA derived from wild-type, lin-56(n2728), lin-15A(n767), and lin-15AB(e1763) animals. Marker DNA fragments are indicated at left. The number of PCR cycles is indicated at right.
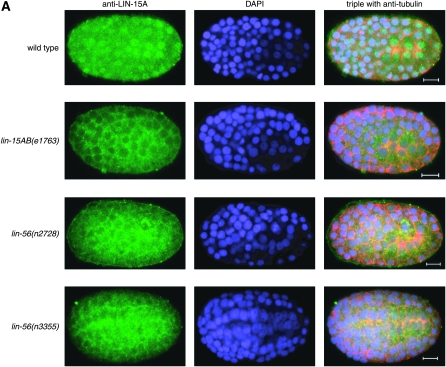
LIN-15A protein nuclear accumulation, but not lin-15A mRNA, is reduced in lin-56(lf) mutants:
Antisera directed against the LIN-15A protein revealed broad nuclear staining (Figure 4A; data not shown). Cytoplasmic staining was observed in both wild-type animals and in mutants completely lacking LIN-15A and therefore represents nonspecific cross-reactivity and not LIN-15A protein. Since LIN-56 protein levels are reduced in lin-15A(lf) mutants, we examined whether LIN-15A protein levels are reduced in lin-56(lf) mutants. LIN-15A nuclear staining was greatly reduced in lin-56(n2728) and lin-56(n3355) embryos (Figure 4A). This loss of nuclear LIN-15A accumulation indicates that lin-56 is required for the expression, stability, or nuclear localization of LIN-15A protein. Loss of lin-56 function reduced LIN-15A nuclear accumulation more in late than in early embryos. LIN-15A nuclear accumulation appeared greater in early lin-56(n3355) embryos than in early lin-56(n2728) embryos (data not shown), suggesting that n3355 may not be as strong an allele of lin-56 as is n2728 or that another gene deleted by n2728 may also have an effect on LIN-15A levels. LIN-15A expression and localization in lin-8(n2731) and lin-38(n751) embryos were indistinguishable from those in wild type (data not shown).
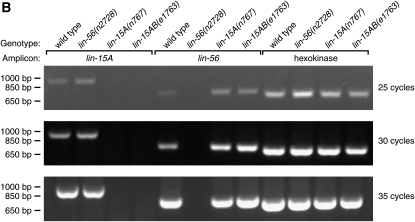
We used RT-PCR to measure the amount of lin-15A RNA in wild-type, lin-56(n2728), lin-15A(n767), and lin-15AB(e1763) animals. lin-15A RNA was amplified from both the wild-type and lin-56(lf) RNA samples but not from the lin-15A(lf) RNA samples (Figure 4B). The decreased amount or mislocalization of LIN-15A protein in lin-56(lf) as compared to wild-type animals thus likely results from a post-transcriptional event, consistent with the hypothesis that LIN-56 is required for the translation, stability, or nuclear localization of LIN-15A protein.
Overexpression of lin-56 does not rescue the lin-15AB(lf) synMuv phenotype, nor does overexpression of lin-15A rescue the lin-56(lf) synMuv phenotype:
If lin-15A acts only to control the translation or stability of LIN-56 protein, then restoring the level of LIN-56 protein should rescue the lin-15A(lf); synMuvB(lf) synMuv phenotype. To test this possibility, we induced a lin-56 cDNA under the control of heat-shock promoters shortly after L1 lethargus in lin-56(n2728); lin-15B(n744) animals and rescued the synMuv phenotype (Table 1). The same treatment failed to rescue the synMuv phenotype of lin-15AB(e1763) animals (Table 1). As assayed by immunoblot, LIN-56 protein was produced under these conditions at levels at least 10-fold greater than those of endogenous LIN-56 in the wild type (supporting information, Figure S1). The mechanism that normally prevents LIN-56 protein accumulation in lin-15AB(e1763) animals is likely overwhelmed by the level of LIN-56 produced by heat-shock overexpression. Also, LIN-56 produced from the heat-shock promoters in lin-15AB(e1763) animals did not obviously differ in electrophoretic mobility from that in lin-56(n2728); lin-15B(n744) animals by immunoblot (Figure S1). Since overexpression of LIN-56 failed to rescue the synMuv phenotype of a lin-15A(lf); synMuvB(lf) mutant, lin-15A cannot function only to positively regulate the translation or stability of LIN-56 protein.
Likewise, if lin-56 acts only to control the translation or stability of LIN-15A protein, then restoring the level of LIN-15A protein should rescue the lin-56(lf); synMuvB(lf) synMuv phenotype. To test this possibility, we induced a lin-15A cDNA under the control of heat-shock promoters shortly after L1 lethargus in lin-36(n766); lin-15A(n767) animals and rescued the synMuv phenotype (Table 2). The same treatment failed to rescue the synMuv phenotype of lin-56(n2728); lin-36(n766) animals (Table 2). Since overexpression of lin-15A failed to rescue the synMuv phenotype of a lin-56(lf); synMuvB(lf) mutant, lin-56 is unlikely to act only by positively regulating the translation or stability of LIN-15A protein.
TABLE 2.
lin-15A overexpression rescues the lin-36(lf); lin-15A(lf), but not the lin-56(lf); lin-36(lf), synMuv phenotype
| Genotype | Transgene | Line | Heat shock | % Muva (n)b |
|---|---|---|---|---|
| lin-36(n766); lin-15A(n767) | None | − | 99.6 (251) | |
| lin-36(n766); lin-15A(n767) | Phs-lin-15A | 1 | − | 100 (39) |
| + | 16 (96) | |||
| 2 | − | 99 (86) | ||
| + | 12 (93) | |||
| lin-56(n2728); lin-36(n766) | None | − | 99.6 (234) | |
| lin-56(n2728); lin-36(n766) | Phs-lin-15A | 1 | − | 100 (177) |
| + | 99.8 (605) | |||
| 2 | − | 99.6 (227) | ||
| + | 100 (675) |
lin-36(n766); lin-15A(n766) or lin-56(n2728); lin-15A(n766) hermaphrodites with or without an extrachromosomal array of lin-15A cDNA (Phs-lin-15A) fused to the C. elegans heat-shock promoters and a GFP+ marker were synchronized by bleaching and starvation-induced L1 arrest. Expression from the heat-shock promoters was induced at the early L2 stage (24–25 hr post starvation) by incubation at 33° for 1 hr.
% Muv, percentage of animals that were Muv.
n, number of animals examined.
LIN-56 and LIN-15A physically interact:
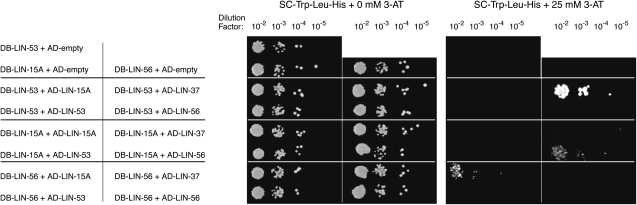
Since LIN-56 protein levels are reduced in a lin-15A mutant background and LIN-15A nuclear accumulation is reduced in a lin-56 mutant background, we hypothesized that LIN-56 and LIN-15A might normally be stabilized by association with each other in a functional complex. To investigate if LIN-56 and LIN-15A can interact, we used the GAL4-based yeast two-hybrid system (Fields and Song 1989). The class B synMuv proteins LIN-37 and LIN-53 were previously shown to interact using this approach (Walhout et al. 2000a; see also Figure 5). The class A synMuv proteins LIN-56 and LIN-15A interacted specifically with each other, as detected by using yeast cotransformed with either constructs DB-LIN-15A and AD-LIN-56 or DB-LIN-56 and AD-LIN-15A (Figure 5). By contrast, neither LIN-56 nor LIN-15A appeared to interact specifically with either LIN-37 or LIN-53 (Figure 5).
Figure 5.—
LIN-15A and LIN-56 interact with each other in the yeast two-hybrid system. Growth of cotransformant Y190 colonies on SC-Trp-Leu-His media in either the absence or the presence of 25 mm 3-AT. Interaction of the DB- and AD-fusion proteins results in increased expression of a GAL1∷HIS3 reporter gene, permitting colony formation in the absence of histidine and presence of 3-AT, a competitive inhibitor of the enzyme encoded by HIS3. Plasmids cotransformed into each strain are indicated at left. Interactions are evident between DB-LIN-15A and AD-LIN-56, DB-LIN-56 and AD-LIN-15A, and DB-LIN-53 and AD-LIN-37, as previously reported (Walhout et al. 2000a). DB, Gal4 DNA-binding domain. AD, GAL4 activation domain.
DISCUSSION
LIN-56 and LIN-15A might function as a complex in vivo:
lin-56 and lin-15A encode putative transcriptional regulators that are present in most or all cells in C. elegans. The LIN-56 and LIN-15A proteins share a THAP-like C2CH motif and are dependent on each other for wild-type levels and/or localization. lin-15A is required post-transcriptionally for LIN-56 protein expression or stability, and lin-56 is required for the expression, stability, or nuclear localization of LIN-15A. As overexpression of LIN-56 did not rescue the lin-15A(lf) synMuv phenotype, it seems unlikely that lin-15A functions only to regulate LIN-56 protein levels. Likewise, expression of lin-15A was unable to rescue the lin-56(lf) synMuv phenotype, suggesting that lin-56 does not function only to regulate LIN-15A levels. Rather, given the ability of LIN-56 and LIN-15A to interact in the yeast two-hybrid system, we favor a model in which LIN-56 and LIN-15A associate in a functional complex required for the inhibition of vulval cell fates, with the absence of one of the complex subunits resulting in the instability or loss of nuclear localization of the other(s). This model is supported by genetic evidence showing that, unlike all other pairs of class A synMuv genes, lin-15A and lin-56 are unable to function independently of each other in vulval development (Andersen et al. 2008).
Class A synMuv genes likely directly regulate gene expression:
The THAP C2CH domain is conserved from C. elegans to humans and was initially proposed to bind DNA on the basis of its similarity to a region of the Drosophila P-element transposase (Roussigne et al. 2003). The THAP domain of the human protein THAP1 has since been shown to possess zinc-dependent sequence-specific DNA-binding activity in vitro (Clouaire et al. 2005). Furthermore, coimmunoprecipitation studies revealed that THAP1 associates in vivo with the promoter of a pRB/E2F cell-cycle target gene (Cayrol et al. 2007).
Several C. elegans proteins contain THAP domains, THAP-like domains, or both (Reddy and Villeneuve 2004; Clouaire et al. 2005). Analysis of a subset of the THAP domain-containing proteins in C. elegans suggests that they might mediate access to chromatin remodeling and chromatin-modifying complexes (Reddy and Villeneuve 2004). Specifically, HIM-17 and the class B synMuv proteins LIN-15B and LIN-36 contain one or more THAP domains. HIM-17 is required both for accumulation of histone H3 methyl-lysine 9 in the germline and for meiotic recombination, suggesting a link between these two processes (Reddy and Villeneuve 2004). The class B synMuv proteins are thought to silence genes required for vulval cell-fate specification through chromatin remodeling, most likely including histone H3 lysine-9 methylation (Lu and Horvitz 1998; Solari and Ahringer 2000; von Zelewsky et al. 2000; Ceol and Horvitz 2001; Couteau et al. 2002; Andersen and Horvitz 2007). Furthermore, the class B synMuv protein LIN-35 Rb acts with HIM-17 in meiotic recombination (Reddy and Villeneuve 2004).
The region of similarity between the class A synMuv proteins LIN-56 and LIN-15A shares the C2CH signature of the THAP domain but lacks additional conserved residues, such as an invariant tryptophan and a C-terminal AVPTIF motif (Clouaire et al. 2005). This variant THAP-like domain is found also in HIM-17 and LIN-15B (Reddy and Villeneuve 2004). Given the nuclear localization of LIN-56 and LIN-15A, we suggest that this variant C2CH motif is also likely to mediate interaction with DNA. Together with the nuclear localization of LIN-8 and its physical interaction with LIN-35 Rb (Davison et al. 2005), these results strongly suggest that class A synMuv proteins inhibit vulval cell-fate specification through the regulation of transcription.
The class A synMuv genes, including both lin-15A and lin-56, repress expression of lin-3 EGF redundantly with the class B synMuv genes (Cui et al. 2006; Andersen et al. 2008). It was previously shown that RNAi of lin-3 can suppress the lin-15A(lf); synMuvB(lf) synMuv phenotype (Cui et al. 2006), and we have shown that RNAi of lin-3 can also suppress the lin-56(lf); synMuvB(lf) synMuv phenotype, consistent with the hypothesis that both lin-15A and lin-56 act upstream of lin-3. Given their molecular identities, the class B synMuv genes likely repress gene expression by chromatin remodeling. On the basis of their potential DNA-binding THAP-like domains and nuclear localization, we propose that the class A synMuv genes also regulate transcription. Both class A and class B synMuv proteins might directly repress expression of lin-3. Alternatively, either one or both classes of synMuv proteins could directly regulate the expression of another protein, which subsequently represses lin-3.
Class A synMuv genes might have functions beyond those in vulval development:
Mutation of the class A synMuv genes, unlike mutation of the class B synMuv genes, has not been reported to result in defects other than those associated with vulval development. Specifically, mutation of the class A synMuv genes does not appear to cause cell-cycle defects (Boxem and van den Heuvel 2002; Fay et al. 2002), reduced gene expression from repetitive transgene arrays (Hsieh et al. 1999), or sterility or lethality (Lu and Horvitz 1998; Beitel et al. 2000; Melendez and Greenwald 2000; von Zelewsky et al. 2000; Ceol and Horvitz 2001; Belfiore et al. 2002; Couteau et al. 2002; Dufourcq et al. 2002; Unhavaithaya et al. 2002). Nonetheless, the broad expression patterns of the LIN-8 (Davison et al. 2005), LIN-15A, and LIN-56 proteins suggest that additional roles might exist for the class A synMuv genes. Perhaps the class A synMuv genes function redundantly with loci other than the class B synMuv genes to regulate biological processes other than the vulval cell-fate decision. This possibility seems particularly likely for lin-8, which is a member of a novel C. elegans gene family with 16 other genes (Davison et al. 2005).
Implications for mammalian tumorigenesis:
The inhibition of vulval development in C. elegans involves multiple redundant pathways: both a class A and a class B synMuv gene must be inactivated for ectopic vulval development to occur (Ferguson and Horvitz 1989). The class B synMuv genes include counterparts of the mammalian tumor-suppressor gene Rb and genes that interact with Rb (Lu and Horvitz 1998; Ceol and Horvitz 2001; Harrison et al. 2006). pRb and the related proteins p107 and p130 play critical roles in mammalian cell-cycle regulation, apoptosis, development, and differentiation (Classon and Harlow 2002), and Rb is often mutated in human cancers (Nevins 2001). Inappropriate activation of EGF/Ras signaling is also a common event in cancers (Normanno et al. 2006). Because the class A synMuv genes function redundantly with an Rb pathway to repress transcription of an EGF ligand and inhibit Ras-mediated vulval cell-fate specification, we propose that analogous THAP domain proteins in mammals might act as tumor-suppressor genes by repressing the expression of EGF-like ligands.
Acknowledgments
We thank Erik Andersen, Craig Ceol, Alison Frand, and Niels Ringstad for editorial comments; Melissa Harrison for help with yeast two-hybrid analysis; Erik Andersen for construction of lin-15A and lin-56 Gateway entry clones; Beth Castor for help with DNA sequence determination; Na An for strain management; Yuji Kohara for EST cDNA clones; Alan Coulson and the C. elegans Sequencing Consortium for cosmid clones and sequences; and Steve Bell for use of the ABI PRISM 7000 Sequence Detection System. The deficiency strains were provided by Theresa Stiernagle of the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health (NIH) National Center for Research Resources. This work was supported by NIH grant GM24663 (to H.R.H.) and a Howard Hughes Medical Institute predoctoral fellowship (to E.M.D.). L.S.H. was supported by a March of Dimes Birth Defects Foundation grant (to P.W.S.). H.R.H. and P.W.S. are Investigators of the Howard Hughes Medical Institute.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.124487/DC1.
References
- Andersen, E. C., and H. R. Horvitz, 2007. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134 2991–2999. [DOI] [PubMed] [Google Scholar]
- Andersen, E. C., A. M. Saffer and H. R. Horvitz, 2008. Multiple levels of redundant processes inhibit Caenorhabditis elegans vulval cell fates. Genetics 179 2001–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroian, R. V., M. Koga, J. E. Mendel, Y. Ohshima and P. W. Sternberg, 1990. The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature 348 693–699. [DOI] [PubMed] [Google Scholar]
- Beitel, G. J., S. G. Clark and H. R. Horvitz, 1990. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature 348 503–509. [DOI] [PubMed] [Google Scholar]
- Beitel, G. J., E. J. Lambie and H. R. Horvitz, 2000. The C. elegans gene lin-9, which acts in an Rb-related pathway, is required for gonadal sheath cell development and encodes a novel protein. Gene 254 253–263. [DOI] [PubMed] [Google Scholar]
- Belfiore, M., L. D. Mathies, P. Pugnale, G. Moulder, R. Barstead et al., 2002. The MEP-1 zinc-finger protein acts with MOG DEAH box proteins to control gene expression via the fem-3 3′ untranslated region in Caenorhabditis elegans. RNA 8 725–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal, T., D. Evans, C. D. Link, A. Guffanti, D. Lawson et al., 2002. A global analysis of Caenorhabditis elegans operons. Nature 417 851–854. [DOI] [PubMed] [Google Scholar]
- Boxem, M., and S. van den Heuvel, 2002. C. elegans class B synthetic multivulva genes act in G(1) regulation. Curr. Biol. 12 906–911. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrol, C., C. Lacroix, C. Mathe, V. Ecochard, M. Ceribelli et al., 2007. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood 109 584–594. [DOI] [PubMed] [Google Scholar]
- Ceol, C. J., and H. R. Horvitz, 2001. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol. Cell 7 461–473. [DOI] [PubMed] [Google Scholar]
- Ceol, C. J., and H. R. Horvitz, 2004. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev. Cell 6 563–576. [DOI] [PubMed] [Google Scholar]
- Chen, F., B. M. Hersh, B. Conradt, Z. Zhou, D. Riemer et al., 2000. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science 287 1485–1489. [DOI] [PubMed] [Google Scholar]
- Clark, S. G., X. Lu and H. R. Horvitz, 1994. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics 137 987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classon, M., and E. Harlow, 2002. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2 910–917. [DOI] [PubMed] [Google Scholar]
- Clouaire, T., M. Roussigne, V. Ecochard, C. Mathe, F. Amalric et al., 2005. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc. Natl. Acad. Sci. USA 102 6907–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau, F., F. Guerry, F. Muller and F. Palladino, 2002. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 3 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, M., J. Chen, T. R. Myers, B. J. Hwang, P. W. Sternberg et al., 2006. SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans. Dev. Cell 10 667–672. [DOI] [PubMed] [Google Scholar]
- Davison, E. M., M. M. Harrison, A. J. Walhout, M. Vidal and H. R. Horvitz, 2005. lin-8, which antagonizes Caenorhabditis elegans Ras-mediated vulval induction, encodes a novel nuclear protein that interacts with the LIN-35 Rb protein. Genetics 171 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward, J., 2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3 11–22. [DOI] [PubMed] [Google Scholar]
- Dufourcq, P., M. Victor, F. Gay, D. Calvo, J. Hodgkin et al., 2002. Functional requirement for histone deacetylase 1 in Caenorhabditis elegans gonadogenesis. Mol. Cell. Biol. 22 3024–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, D. S., S. Keenan and M. Han, 2002. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 16 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, E. L., and H. R. Horvitz, 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, E. L., and H. R. Horvitz, 1989. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, E. L., P. W. Sternberg and H. R. Horvitz, 1987. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature 326 259–267. [DOI] [PubMed] [Google Scholar]
- Fields, S., and O. Song, 1989. A novel genetic system to detect protein-protein interactions. Nature 340 245–246. [DOI] [PubMed] [Google Scholar]
- Finney, M., and G. Ruvkun, 1990. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63 895–905. [DOI] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver et al., 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 806–811. [DOI] [PubMed] [Google Scholar]
- Francis, R., and R. H. Waterston, 1991. Muscle cell attachment in Caenorhabditis elegans. J. Cell Biol. 114 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum, Y., K. K. Lee, J. Liu, M. Cohen and K. L. Wilson, 2002. The expression, lamin-dependent localization and RNAi depletion phenotype for emerin in C. elegans. J. Cell Sci. 115 923–929. [DOI] [PubMed] [Google Scholar]
- Guenther, C., and G. Garriga, 1996. Asymmetric distribution of the C. elegans HAM-1 protein in neuroblasts enables daughter cells to adopt distinct fates. Development 122 3509–3518. [DOI] [PubMed] [Google Scholar]
- Han, M., and P. W. Sternberg, 1990. let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a ras protein. Cell 63 921–931. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane, 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Harrison, M. M., C. J. Ceol, X. Lu and H. R. Horvitz, 2006. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc. Natl. Acad. Sci. USA 103 16782–16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, M. M., X. Lu and H. R. Horvitz, 2007. LIN-61, one of two Caenorhabditis elegans malignant-brain-tumor-repeat-containing proteins, acts with the DRM and NuRD-like protein complexes in vulval development but not in certain other biological processes. Genetics 176 255–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid, C. A., J. Stevens, K. J. Livak and P. M. Williams, 1996. Real time quantitative PCR. Genome Res. 6 986–994. [DOI] [PubMed] [Google Scholar]
- Hill, R. J., and P. W. Sternberg, 1992. The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature 358 470–476. [DOI] [PubMed] [Google Scholar]
- Horvitz, H. R., and J. E. Sulston, 1980. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96 435–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, J., J. Liu, S. A. Kostas, C. Chang, P. W. Sternberg et al., 1999. The RING finger/B-box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans. Genes Dev. 13 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L. S., P. Tzou and P. W. Sternberg, 1994. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol. Biol. Cell 5 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, H. Y., and H. R. Horvitz, 2002. The Caenorhabditis elegans vulval morphogenesis gene sqv-4 encodes a UDP-glucose dehydrogenase that is temporally and spatially regulated. Proc. Natl. Acad. Sci. USA 99 14224–14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, B. J., H. M. Muller and P. W. Sternberg, 2004. Genome annotation by high-throughput 5′ RNA end determination. Proc. Natl. Acad. Sci. USA 101 1650–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R. S., M. Martinez-Campos, P. Zipperlen, A. G. Fraser and J. Ahringer, 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2 RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231–237. [DOI] [PubMed] [Google Scholar]
- Katz, W. S., G. M. Lesa, D. Yannoukakos, T. R. Clandinin, J. Schlessinger et al., 1996. A point mutation in the extracellular domain activates LET-23, the Caenorhabditis elegans epidermal growth factor receptor homolog. Mol. Cell. Biol. 16 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler, P. S., and R. N. Eisenman, 1999. Sin meets NuRD and other tails of repression. Cell 99 447–450. [DOI] [PubMed] [Google Scholar]
- Koelle, M. R., and H. R. Horvitz, 1996. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell 84 115–125. [DOI] [PubMed] [Google Scholar]
- Korenjak, M., B. Taylor-Harding, U. K. Binne, J. S. Satterlee, O. Stevaux et al., 2004. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119 181–193. [DOI] [PubMed] [Google Scholar]
- Kornfeld, K., 1997. Vulval development in Caenorhabditis elegans. Trends Genet. 13 55–61. [DOI] [PubMed] [Google Scholar]
- Kornfeld, K., K. L. Guan and H. R. Horvitz, 1995. The Caenorhabditis elegans gene mek-2 is required for vulval induction and encodes a protein similar to the protein kinase MEK. Genes Dev. 9 756–768. [DOI] [PubMed] [Google Scholar]
- Krishna, S. S., I. Majumdar and N. V. Grishin, 2003. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 31 532–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner, M. R., K. Kornfeld, L. M. Miller, H. R. Horvitz and S. K. Kim, 1994. A MAP kinase homolog, mpk-1, is involved in ras-mediated induction of vulval cell fates in Caenorhabditis elegans. Genes Dev. 8 160–173. [DOI] [PubMed] [Google Scholar]
- Laity, J. H., B. M. Lee and P. E. Wright, 2001. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11 39–46. [DOI] [PubMed] [Google Scholar]
- Lewis, P. W., E. L. Beall, T. C. Fleischer, D. Georlette, A. J. Link et al., 2004. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 18 2929–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick, L., S. Sadasivam, L. Florens, X. Zhu, S. K. Swanson et al., 2007. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell 26 539–551. [DOI] [PubMed] [Google Scholar]
- Liu, J., T. Rolef Ben-Shahar, D. Riemer, M. Treinin, P. Spann et al., 2000. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol. Biol. Cell 11 3937–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X., and H. R. Horvitz, 1998. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 95 981–991. [DOI] [PubMed] [Google Scholar]
- Melendez, A., and I. Greenwald, 2000. Caenorhabditis elegans lin-13, a member of the LIN-35 Rb class of genes involved in vulval development, encodes a protein with zinc fingers and an LXCXE motif. Genetics 155 1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros, 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghal, N., and P. W. Sternberg, 2003. The epidermal growth factor system in Caenorhabditis elegans. Exp. Cell Res. 284 150–159. [DOI] [PubMed] [Google Scholar]
- Nevins, J. R., 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10 699–703. [DOI] [PubMed] [Google Scholar]
- Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison et al., 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412 561–565. [DOI] [PubMed] [Google Scholar]
- Normanno, N., C. Bianco, A. De Luca and D. S. Salomon, 2001. The role of EGF-related peptides in tumor growth. Front. Biosci. 6 D685–D707. [DOI] [PubMed] [Google Scholar]
- Normanno, N., A. De Luca, C. Bianco, L. Strizzi, M. Mancino et al., 2006. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 366 2–16. [DOI] [PubMed] [Google Scholar]
- Pilkinton, M., R. Sandoval and O. R. Colamonici, 2007. Mammalian Mip/LIN-9 interacts with either the p107, p130/E2F4 repressor complex or B-Myb in a cell cycle-phase-dependent context distinct from the Drosophila dREAM complex. Oncogene 26 7535–7543. [DOI] [PubMed] [Google Scholar]
- Poulin, G., Y. Dong, A. G. Fraser, N. A. Hopper and J. Ahringer, 2005. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J. 24 2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, K. C., and A. M. Villeneuve, 2004. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118 439–452. [DOI] [PubMed] [Google Scholar]
- Richards, E. J., and S. C. Elgin, 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108 489–500. [DOI] [PubMed] [Google Scholar]
- Riddle, D. L., 1997. C. Elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Riddle, D. L., T. Blumenthal, B. J. Meyer and J. R. Priess, 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Roussigne, M., S. Kossida, A. C. Lavigne, T. Clouaire, V. Ecochard et al., 2003. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem. Sci. 28 66–69. [DOI] [PubMed] [Google Scholar]
- Sabogal, A., A. Y. Lyubimov, J. E. Corn, J. M. Berger and D. C. Rio, 2010. THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves. Nat. Struct. Mol. Biol. 17 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and J. Sambrook, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schmit, F., M. Korenjak, M. Mannefeld, K. Schmitt, C. Franke et al., 2007. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle 6 1903–1913. [DOI] [PubMed] [Google Scholar]
- Sigurdson, D. C., G. J. Spanier and R. K. Herman, 1984. Caenorhabditis elegans deficiency mapping. Genetics 108 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari, F., and J. Ahringer, 2000. NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr. Biol. 10 223–226. [DOI] [PubMed] [Google Scholar]
- Thomas, J. H., C. J. Ceol, H. T. Schwartz and H. R. Horvitz, 2003. New genes that interact with lin-35 Rb to negatively regulate the let-60 ras pathway in Caenorhabditis elegans. Genetics 164 135–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L., D. L. Court and A. Fire, 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263 103–112. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya, Y., T. H. Shin, N. Miliaras, J. Lee, T. Oyama et al., 2002. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111 991–1002. [DOI] [PubMed] [Google Scholar]
- Vandel, L., E. Nicolas, O. Vaute, R. Ferreira, S. Ait-Si-Ali et al., 2001. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol. Cell. Biol. 21 6484–6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zelewsky, T., F. Palladino, K. Brunschwig, H. Tobler, A. Hajnal et al., 2000. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development 127 5277–5284. [DOI] [PubMed] [Google Scholar]
- Walhout, A. J., R. Sordella, X. Lu, J. L. Hartley, G. F. Temple et al., 2000. a Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287 116–122. [DOI] [PubMed] [Google Scholar]
- Walhout, A. J., G. F. Temple, M. A. Brasch, J. L. Hartley, M. A. Lorson et al., 2000. b GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328 575–592. [DOI] [PubMed] [Google Scholar]
- Wu, Y., M. Han and K. L. Guan, 1995. MEK-2, a Caenorhabditis elegans MAP kinase kinase, functions in Ras-mediated vulval induction and other developmental events. Genes Dev. 9 742–755. [DOI] [PubMed] [Google Scholar]
- Zhang, H. S., and D. C. Dean, 2001. Rb-mediated chromatin structure regulation and transcriptional repression. Oncogene 20 3134–3138. [DOI] [PubMed] [Google Scholar]