Abstract
B chromosomes are dispensable chromosomes found in >2000 eukaryotic species, usually behaving as genomic parasites. Most B chromosomes seem to be made up of the same kind of DNA sequences present in the A chromosomes. This sequence similarity makes it difficult to obtain specific molecular probes that may permit B-presence diagnosis without cytogenetic analysis. We have developed a sequence-characterized amplified region (SCAR) marker for B chromosomes in the grasshopper Eyprepocnemis plorans, which specifically amplifies a 1510-bp DNA fragment exclusively in B-carrying individuals. Fluorescent in situ hybridization and fiber FISH analyses showed that this marker is a tandemly repeated DNA sequence closely intermingled with 45S rDNA. PCR reactions showed the presence of SCAR-like sequences in the A chromosomes, but in two separate fragments, supporting the intraspecific origin of B chromosomes in this species. SCAR marker DNA sequence showed to be identical in B chromosome variants from several localities from Spain and Morocco, and it was very similar to those found in B chromosome variants from Greece and Armenia. This strongly suggests that this sequence was already present in the ancestral B chromosome of this species. In addition, the scarce sequence variation observed among several B variants from very distant populations suggests either a functional constraint or, more likely, a recent and unique origin for B chromosomes in this species.
B chromosomes (also known as supernumerary or accessory) are dispensable chromosomes found in some individuals of >2000 eukaryotic species (Camacho 2005). These chromosomes are considered genome parasites because they usually diminish host fitness (Östergren 1945; Jones 1985). In spite of their deleterious effects, they manage to maintain themselves in natural populations because they frequently show higher transmission rates (i.e., drive) than A chromosomes (for review, see Camacho 2004, 2005; Jones et al. 2008). The existence of a drive mechanism in the germ line ensures B chromosome invasiveness in natural populations. B chromosome invasion by parasitic B's is thus expected to be fast (Camacho et al. 1997), for which reason B invasions have been reported in only a few cases (e.g., Zurita et al. 1998, Cavallaro et al. 2000; Araujo et al. 2001; 2002; Riera et al. 2004). If B chromosomes inflict a load (i.e., they are deleterious) to the individuals carrying them, modifier genes in the A genome could be selected to counteract B drive and effects, as has been suggested for the B24 chromosome in the grasshopper Eyprepocnemis plorans (Perfectti et al. 2004). Neutralized (nondriving) B chromosomes can persist for long periods of time in natural populations, before eventual extinction, during which time they can be regenerated by mutation (Camacho et al. 1997; Zurita et al. 1998). In addition, B chromosomes typically do not recombine with A chromosomes and they are prone to gene mutational inactivation (McVean 1995). Therefore, the life cycle of a B chromosome can pass through a number of successive parasitic/driving and neutralized/nondriving stages (Camacho 2005), implying that the evolutionary status of a B chromosome may not be useful to estimate the age and origin of a B chromosome.
In spite of being the first selfish genetic elements to be described (Wilson 1907), the origin of B chromosomes is mostly unknown. Two broad hypotheses have been proposed to explain the origin of these chromosomes. Battaglia (1964) first proposed that B chromosomes could arise during interspecies mating (e.g., McAllister and Werren 1997; Perfectti and Werren 2001). But B chromosomes could also arise as a by-product of A chromosome evolution (see Camacho 2005). This intraspecific origin implies that B chromosomes should be made up of the same kind of DNA sequences present in the A chromosomes, making it unlikely to find DNA sequences exclusive to B chromosomes. Although B chromosomes are usually enriched in satellite DNA, these sequences are also abundant in the A genome, particularly in heterochromatin. These repetitive elements contribute to the heterochromatic nature of B chromosomes (Camacho 2005). Since heterochromatinization is a known mechanism of gene silencing and inactivation (Weiler and Wakimoto 1995), the highly heterochromatic nature of these chromosomes could explain why only a few B chromosomes are known to affect phenotypes (Camacho 2005). All these factors can explain the scarcity of genetic and molecular markers associated with B chromosomes. This deficit of markers is hindering the molecular detection of these chromosomes, requiring time-consuming cytogenetic techniques and hampering the elucidation of their origin.
Sequence characterized amplified region (SCAR) markers have been widely used to develop DNA markers for specific DNA regions. A SCAR marker is a genomic DNA fragment located at a single defined locus that is identified by PCR amplification using a pair of specific primers (Paran and Michelmore 1993). These specific primers are usually derived by extending the sequence of random amplified polymorphic DNA (RAPD) or inter-simple sequence repeat (ISSR) primers. Longer primers allow higher annealing temperatures, avoiding multiple priming and site competition among primers, and thus increasing specificity and reproducibility (Hernández et al. 1999). SCAR markers have been developed from RAPD fragments for multiple species and several purposes. For instance, SCARs have allowed genetic mapping (Huo et al. 2009), determination of sex (Oyama et al. 2009), identification of races (Pasquali et al. 2007), populations (Basha and Sujatha 2007), and species (Yau et al. 2002), or detection of association with pest resistance (Whitaker et al. 2010). They have also been used to detect B chromosomes. Tosta et al. (2004) found a RAPD marker associated with presence of the B chromosome of the wasp Partamona helleri, and subsequently they developed a SCAR marker that permits the analysis of B chromosome presence (Tosta et al. 2007). Here, we present the development of a SCAR marker for the B chromosomes of the grasshopper E. plorans.
E. plorans harbors a relatively well-characterized B chromosome system (reviewed in Camacho et al. 2003), although only the E. plorans plorans subspecies appears to carry B chromosomes (López-León et al. 2008). This subspecies inhabits the Mediterranean coasts, the Caucasus, Turkey, Turkmenistan, Iran, and the southwest of the Arabian Peninsula (Dirsh 1958). B chromosomes in this grasshopper species show extensive variation, with >50 cytogenetically different types (López-León et al. 1993a), some of them found in the same population. E. plorans B chromosomes are mitotically stable, implying that the same number of B chromosomes is found in all cells from the same individual. The frequency with which B's are found in natural populations varies widely between populations (Camacho et al. 2003), with individual grasshoppers carrying usually zero to three B chromosomes. These B chromosomes are usually of the same type, but some individuals can carry two different B chromosome variants. As a result of the coevolutionary arms race with the host (A) chromosomes (Camacho et al. 1997), most of the B chromosome variants lack drive. It is also possible that a given B-variant shows drive in some populations but not in others (e.g., Bakkali et al. 2002), or that B's in the same population may lose drive in a short time period, e.g., B-drive was apparent for B24 in a 1992 sample from the Torrox (Málaga, Spain) population (Zurita et al. 1998) but not in a sample taken 6 years later (Perfectti et al. 2004). The most widespread B-variants (B1, B2, and B5) lack both drive and deleterious fitness effects (López-León et al. 1992; but see Herrera et al. 1996; Muñoz et al. 1998). B2 has recently been replaced by the parasitic (driving) B24 in the Torrox population. This last B chromosome significantly decreases egg fertility, although it has spread thanks to being transmitted to 70% of progeny through females (Zurita et al. 1998).
All these types of B chromosomes are enriched in two kinds of repetitive DNA sequences (Cabrero et al. 1999), both of which are shared by the A genome: a 180-bp satellite DNA (satDNA) and ribosomal DNA (rDNA) (López-León et al. 1994). The presence of these two kinds of sequences in the telocentric X chromosome, arranged in the same order (centromere–satDNA–rDNA) as in the B2 chromosome, led López-León et al. (1994) to postulate that B2 derived from the X chromosome. This chromosome arrangement (proximal satDNA and distal rDNA) is also conserved in the main Spanish and Moroccan B chromosomes (Bakkali et al. 1999; Cabrero et al. 1999) suggesting that these B chromosomes share a common origin. However, Eastern Mediterranean populations, including populations from Greece, Turkey, and Armenia, showed B types that differed from the Western Mediterranean B chromosomes by having lower amounts of satDNA and larger amounts of rDNA than Spanish and Moroccan B's (Abdelaziz et al. 2007; López-León et al. 2008). In addition, Eastern B chromosomes seem to contain 5S rDNA, a DNA sequence not detected in Spanish populations (Cabrero et al. 2003b). The different FISH pattern of Western and Eastern Mediterranean B chromosomes led Cabrero et al. (2003b) to postulate a multiregional origin of B chromosomes in this species, and that the two kinds of B chromosomes might have originated from a different A chromosome. However, since the B chromosome DNA sequences hitherto known are repetitive sequences also found in the A genome, we have not yet tested this hypothesis by comparing B chromosome-specific sequences.
Here we report the development of a SCAR marker linked to E. plorans B chromosomes, which allows the molecular detection of B chromosome presence by PCR analysis. The B-SCAR marker appears to be the result of the combination of two DNA sequences also present in A chromosomes, pointing to the intraspecific origin of these B chromosomes. Finally, the presence of the SCAR marker in all B chromosome variants from both Eastern and Western populations of E. plorans suggests a common origin for all these variants, and thus a single origin for B chromosomes in this species. In addition, the low variation observed in the B-SCAR sequence suggests a recent origin for this B chromosome system.
MATERIALS AND METHODS
Development of a SCAR marker for B chromosome presence:
In 2003, we collected male E. plorans grasshoppers from Torrox (Málaga, Spain). For each individual, we dissected out and fixed testes in freshly prepared 3:1 ethanol-acetic acid. Testes were conserved at 4° until cytogenetic analysis was performed to determine the presence, type, and number of B chromosomes carried by each specimen, following the cytogenetic procedure described in Camacho et al. (1991). DNA was extracted from each individual body, using the GenElute Mammalian Genomic DNA Miniprep extraction kit (Sigma-Aldrich) following vendor instructions, and stored at −20°. DNA pools were prepared from eight males lacking B chromosomes (0B pooled sample) and eight males carrying one B24 (+B pooled sample). By also using a combination of several individual samples, we diminished errors due to the amplification of low-frequency alleles present in only a few individuals and not linked to the B chromosome. A similar strategy was used by Griffiths (2000) to detect sex-specific markers in birds.
These mixtures were used to perform the random amplification of polymorphic DNA (RAPD) technique, using 20 nonspecific primers (OPA-1–OPA-20, Table 1) (Operon) in all possible combinations. Each PCR reaction was performed in a total reaction volume of 25 μl with the following composition: 30 ng of DNA from pooled samples, 0.5 μm of each primer, 0.2 mm dNTP (Sigma-Aldrich), 2.5 μl 10X PCR-buffer (NEB) and 2 units Taq (NEB). Each reaction mix was prepared into a vertical laminar flow cabinet to avoid cross-contamination. Reactions were carried out in an MJ Mini Personal Thermal Cycler (Bio-Rad) with the following PCR program: an initial denaturing step at 94° for 2 min, 37 cycles of 94° 1 min; 36° 1 min; 72° 3 min, and a final extension step of 72° 5 min. PCR products were separated in 1.5% agarose gels. When we detected an exclusive DNA band in the amplification of the +B pooled sample, those PCR reactions were repeated three times to assure a positive signal.
TABLE 1.
Primers used in this work
| Primer name | Sequence 5′−3′ |
|---|---|
| OPA-01 | CAGGCCCTTC |
| OPA-02 | TGCCGAGCTG |
| OPA-03 | AGTCAGCCAC |
| OPA-04 | AATCGGGCTG |
| OPA-05 | AGGGGTCTTG |
| OPA-06 | GGTCCCTGAC |
| OPA-07 | GAAACGGGTG |
| OPA-08 | GTGACGTAGG |
| OPA-09 | GGGTAACGCC |
| OPA-10 | GTGATCGCAG |
| OPA-11 | CAATCGCCGT |
| OPA-12 | TCGGCGATAG |
| OPA-13 | CAGCACCCAC |
| OPA-14 | TCTGTGCTGG |
| OPA-15 | TTCCGAACCC |
| OPA-16 | AGCCAGCGAA |
| OPA-17 | GACCGCTTGT |
| OPA-18 | AGGTGACCGT |
| OPA-19 | CAAACGTCGG |
| OPA-20 | GTTGCGATCC |
| BSCAR-1F | TCGGCGATAGGTCTTCGAGG |
| BSCAR-17F | GAGGAGGCGGATATTTCCTCTTGCA |
| BSCAR-540F | CCGGAATGAGATGTCTGCGTTGC |
| BSCAR-622F | TCGTACATGTGTTATGTGAGC |
| BSCAR-967R | TGCTGCCGGCGACAGGTTCACGTA |
| BSCAR-1049F | CGATGCAAAGACCCTGTCACATGC |
| BSCAR-1077R | ACGTTGCATGTGACAGGGTCTT |
| BSCAR-1206R | GCAGAAGCTACTTTTCATGCGGGT |
| BSCAR-1342F | TGCACGGAATCACATTGACC |
| BSCAR-1365R | CACTGGTCAATGTGATTCCGTGCA |
| BSCAR-1492R | TAGCGACTTGTCGTGTGCAGT |
| BSCAR-1510R | TCGGCGATAGTGACGATATA |
| M13F-20 | GTAAAACGACGGCCAGT |
| M13 R | CAGGAAACAGCTATGAC |
| EcoRI-1 | CTCGTAGACTGCGTACC |
| EcoRI-2 | AATTGGTACGCAGTCTAC |
| EcoRI-presel | GACTGCGTACCAATTC |
OPA, oligonucleotides used to amplify RADP markers; BSCAR, primers designed to amplify specific regions of the B-SCAR marker.
Three primer pairs showed different patterns in the +B pooled sample compared with the 0B pooled sample, suggesting a possible B chromosome-specific marker. We chose the OPA-12/OPA-12 primer combination (Table 1) for further analysis, since it yielded the largest amplified fragment (∼1500 bp). This distinctive gel band, only present in the +B pooled sample, was isolated from gel and purified using Gen Elute Gel Extraction kit (Sigma-Aldrich), following the instructions provided by the manufacturer. Isolated DNA was ligated to a vector and cloned in Escherichia coli using the TOPO TA Cloning kit (Invitrogen), following vendor instructions. Plasmid DNA was purified (using Perfectprep Plasmid Mini; Eppendorf) and sequenced using BigDye-Terminator-v3.1 (Applied Biosystems) with universal M13 primers (Table 1) and an ABI PRISM 3100-avant. Chromatograms were analyzed with FinchTV (http://www.geospiza.com/Products/finchtv.shtml) and Bioedit (Hall 1999) and inspected by eye for evidences of heterozygous positions.
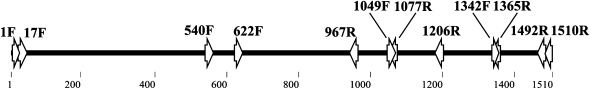
We looked for open reading frames (ORFs) in this DNA sequence using ORF Finder at the National Center for Biotechnology (http://www.ncbi.nlm.nih.gov/gorf/), and searched for homologous sequences in GenBank using Blastn (http://www.ncbi.nlm.nih.gov/blast/). On the basis of the DNA sequence of the amplified fragment, we designed a new set of primers (BSCAR-17F and BSCAR-1492R, Table 1) using Fast PCR (Kalendar et al. 2009). These primers selectively amplify a B chromosome SCAR marker. PCR reactions with this new set of primers were performed with 0.05 μm dNTP, 0.1 μm each primer, and 0.01 units/μl Taq. The selective PCR program was 94° 2 min, 30 cycles at 94° 1 min; 55° 1 min; 72° 1 min, and a final extension step of 5 min at 72°. Using these conditions, we first tested the accuracy of PCR reaction with B-SCAR primers by detecting previously identified B chromosome carriers. For this first screening we used 58 individuals from the same population, 28 of them carrying the B24 variant chromosome, whereas the other 30 individuals lacked B chromosomes (Table 2). We also tested the presence of the B-SCAR in DNA obtained by microdissection of the B24 chromosome and amplified using a ϕ-29 enzyme (GenomiPhi v2 DNA amplification kit; GE). Once the B-SCAR marker sequence was known, we designed 10 additional primers to compare 0B and +B amplification patterns (Table 1, Figure 1).
TABLE 2.
Origin of the DNA samples used in this study, with indication of the type of B chromosome
| Population | B variant | Capture years | No. of individuals analyzed |
No. of individuals with B-SCAR sequenced | B-SCAR haplotype and accession no. | |
|---|---|---|---|---|---|---|
| 0B | +B | |||||
| Algarrobo (Málaga, Spain) | B24 | 2006 | 0 | 1 | 1 | Spain–Morocco (FR681612) |
| B2 | 2006 | 0 | 2 | 2 | ||
| Calasparra (Murcia, Spain) | B1 | 2006 | 0 | 1 | 1 | |
| B1iso | 2006 | 0 | 1 | 1 | ||
| Gallego (Albacete, Spain) | B2 | 2008 | 0 | 1 | 1 | |
| Maro (Málaga, Spain) | B2e | 2004 | 0 | 1 | 1 | |
| B2 | 2006 | 0 | 2 | 2 | ||
| Nerja (Málaga, Spain) | B24 | 2006 | 0 | 2 | 2 | |
| B2 | 2006 | 0 | 4 | 4 | ||
| Salobreña (Granada, Spain) | B2 | 2006 | 5 | 6 | 3 | |
| San Lúcar (Cádiz, Spain) | B1 | 2008 | 0 | 1 | 1 | |
| Tetuán (Málaga, Spain) | B2 | 2006 | 0 | 3 | 3 | |
| Torrox (Málaga, Spain) | B24 | 2003–2006 | 30 | 28 | 3 | |
| Vicorto (Albacete, Spain) | — | 2006 | 1 | 0 | 0 | — |
| Variko (Greece) | BL | 2003 | 0 | 1 | 1 | Variko I (FR681610) |
| BM | 2003 | 0 | 1 | 1 | ||
| BS | 2003 | 0 | 1 | 1 | Variko II (FR681611) | |
| Smir (Morocco) | B1M | 1998 | 0 | 1 | 1 | Spain–Morocco (FR681613) |
| Arax (Armenia) | Ba1 | 2006 | 0 | 1 | 1 | Arax (FR681607) |
| Razdan (Armenia) | Ba2 | 2006 | 0 | 1 | 1 | Razdan (FR681608) |
| Megri (Armenia) | Ba3 | 2006 | 0 | 1 | 1 | Megri (FR681609) |
| Total | 31 | 60 | 32 | |||
Figure 1.—
A depiction of the B-SCAR sequence showing location of the primers used in the present study.
To explore the flanking regions surrounding the B-SCAR marker, we performed Southern hybridization using the central region of the B-SCAR as probe. We amplified the central B-SCAR sequence using BSCAR-570F and BSCAR-1077R primers (Table 1). PCR products were purified, using GenElute PCR Clean-UP kit (Sigma-Aldrich), and labeled, using DIG DNA Labeling and Detection kit (Roche). Since the B-SCAR sequence does not contain any EcoRI targets, we used this restriction enzyme to perform exhaustive restrictions of B-carrier individual DNA. We tested several restriction conditions: 100 units EcoRI for 18 hr, 60 units EcoRI for 135 min, and 60 units EcoRI for 75 min. In all cases, we used 1 μg of +B DNA in a final reaction volume of 50 μL maintained at 37°. Restriction products were visualized in 0.7% agarose gels, then transferred to a nylon membrane, and hybridized with the B-SCAR probe.
To characterize the flanking regions outside the B-SCAR sequence, we used a modified ligation-anchored PCR using EcoRI digestion (Troutt et al. 1992). First, +B DNA was cut with EcoRI (37°, 60 units EcoRI for 135 min), followed by adaptor ligation of EcoRI-adapt1 and EcoRI-adapt2 (see sequences in Table 1). Ligation was performed with 1.7 units T4 ligase (NEB), 2 μl A-Buffer 10X (NEB), 1 μg PCR amplified DNA, and 0.2 μm each EcoRI adaptors in 20 μl total volume. Then we used ligated products as template DNA in a PCR reaction, using EcoRI-presel/BSCA-R622F and EcoRI-presel/BSAC-R967R primer pairs to amplify the B-SCAR sequence flanking regions.
To analyze DNA sequence variation in the B-SCAR marker, we used DNA from individuals captured in different years in 15 natural populations of E. plorans, including 10 Spanish, a Greek, a Moroccan, and three Armenian populations (Table 2). In addition, 0B individuals from some of these populations were used as negative controls to check the absence of amplification in individuals lacking B chromosomes. PCR products were purified by precipitation with sodium acetate and ethanol. Direct sequencing of the PCR products yielded high-quality chromatograms. Sequences were aligned with Geneious Pro (Drummond et al. 2010) using clustalW and adjusted by hand. Polymorphism analysis was performed with DnaSP (Librado and Rozas 2009).
Cytogenetic analysis:
To check that the B-SCAR marker is located in the B chromosome, we performed fluorescent in situ hibridation (FISH), following the procedure described by Cabrero et al. (2003a), on embryos obtained from cultures maintained in our laboratory as described in Camacho et al. (1991). FISH was performed with two different DNA probes: (1) E. plorans rDNA probe generated by PCR amplification of rDNA internal transcribed spacer (ITS) regions, and (2) B-SCAR DNA amplified by PCR. Probe DNA was labeled by nick translation with either tetramethylrhodamine-11-dUTP or fluorogreen-11-dUTP, using standard techniques. Chromosome preparations were observed using an Olympus BX41 fluorescence microscope connected to a refrigerated digital camera. Images were optimized for brightness and contrast with the Gimp freeware.
For fiber-FISH, cerebral ganglia were homogenized in 60% acetic acid and centrifuged for 10 min at 1000 rpm, removing the supernatant. Then, we added 500 μl of 1:3 acetic acid:ethanol and resuspended the cells. To relax the chromatin, we used the technique described in Fidlerová et al. (1994), with modifications suggested by J. J. Pasantes (personal communication). Fixed cells were spread on clean moist slides and, before complete evaporation of the fixative, slides were placed in PBS pH = 7 solution for 1 min. Thereafter, the slides were drained on a paper towel and immediately treated with 0.05 m NaOH in 30% ethanol as follows: 200 μl solution was placed on one end of a long coverslip and then moved along the horizontal slide with the help of the coverslip held at an angle of ∼30°. Then we added a few drops of absolute ethanol (500 μl) and left it dry. Finally, to dehydrate the material, the slides were placed in a 70, 90, and 100% ethanol series and left them to dry. Double FISH was performed as described in Cabrero et al. (2003a).
RESULTS
B-SCAR marker identification:
To find a B chromosome-specific DNA sequence, we amplified 210 RAPD primer combinations to screen for band differences in the two pooled samples (0B and +B samples). Most primer combinations yielded the same pattern in both 0B and +B pooled samples. However, three primer pairs gave different patterns in the two types of samples, suggesting B specificity. We isolated and purified a putative B-specific band, thereafter B-SCAR marker, and sequenced its DNA (see materials and methods). The B-SCAR marker DNA sequence spans 1510 bp, with 53% G + C content and does not show significant similarity with other sequences in the National Center for Biotechnology Information (NCBI) databases (blastn E values >0.40). The longest open reading frame spans 360 nucleotides with no homology with known protein sequences. This sequence includes a microsatellite-like region with a GATTA motif repeated six times between positions 903 and 932. Therefore, this sequence appears to be noncoding and a good candidate for being a specific marker of B chromosome presence. The B-SCAR sequences were uploaded to GenBank with accession nos. FR681607–FR681613 (Table 2).
To verify B specificity of the B-SCAR marker, we did PCR amplifications in 58 individuals from the Torrox population, after cytogenetic determination of the number and type of B chromosomes (see Table 2). For these PCR reactions, we used a new and more specific set of primers (BSCAR-17F and BSCAR-1492R, see Table 1). The B-SCAR marker amplified in all 28 B-carrying individuals but not in the 30 B-lacking individuals, suggesting that this DNA sequence is specific to the B chromosome. Alternatively, this marker might map in the A chromosomes, with sample frequency 28/58 = 0.48. However, the likelihood of finding a sample of 28 B-carrying individuals showing the B-SCAR marker and 30 B-lacking individuals carrying it is negligible [0.4828 × (1−0.48)30 = 3.59 × 10−18]. We also analyzed five 0B and six +B individuals from the Salobreña (Granada, Spain) population. In Salobreña, the most common B chromosome variant is B2, the ancestor of the B24 variant carried by individuals from Torrox (Zurita et al. 1998). The presence of the B-SCAR marker only in the B2-carrying individuals also supports the B chromosome specificity of this DNA fragment.
We also tested for B-SCAR presence in the DNA obtained from a microdissection of the B24 chromosome (Teruel 2009; Teruel et al. 2009). Amplification with BSCAR-17F and BSCAR-1492R yielded a band of the expected size, and direct sequencing confirmed the identity of this fragment.
In addition, we tested the presence of the B-SCAR marker in individuals from other populations carrying different B chromosome variants (Table 2). We found the B-SCAR marker in all individuals carrying a B chromosome of whatever B chromosome type. Sequencing demonstrated that these B chromosome variants (B24, B2, B2e, B1, B1iso, BL, BM, BS, Ba1, Ba2, Ba3, and B1M) all carry the B-SCAR sequence, and thus most likely share common ancestry, despite coming from very distant geographic areas (see Table 2). Direct sequencing of PCR amplifications did not show evidence of heterozygous positions. For further details on sequence differences, see below.
Chromosome mapping of the B-SCAR marker:
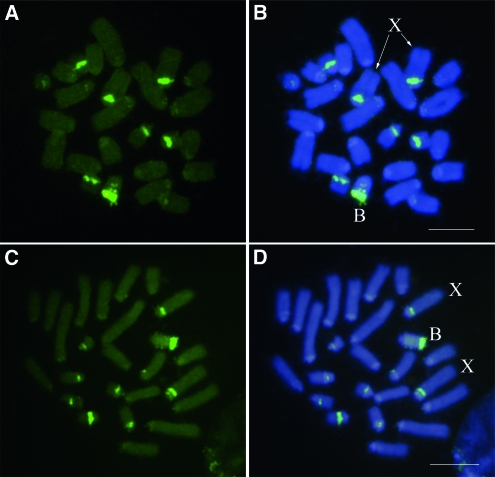
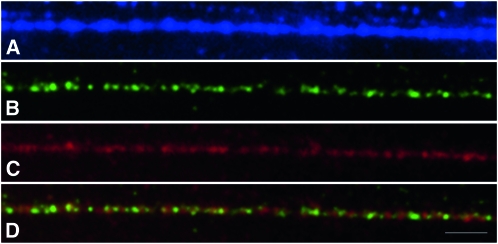
To further test the B-SCAR marker location on the B chromosome, we performed FISH analysis using the B-SCAR sequence as a DNA probe. This probe hybridized with several regions of the B24 chromosome, showing the highest intensity signal in the distal region of B24, where the rDNA is located in this B variant (Cabrero et al. 2003b), although it is also present in two interstitial regions, where we have no evidence for the presence of rDNA (Cabrero et al. 2003b, and see Figure 2). This DNA probe also hybridized intensely with paracentromeric regions in the A chromosomes containing the major rDNA clusters (X, 9, 10, and 11). Faint hybridization was also observed in paracentromeric regions of some other autosomes. Since the SCAR marker colocalized with rDNA, at least at the resolution provided by mitotic metaphase chromosomes, we tried to align the B-SCAR sequence with that of the rDNA internal transcribed spacers (ITS1 and ITS2) of E. plorans (Teruel 2009). However, we found that the B-SCAR has no homology with E. plorans ITS regions. We also performed Fiber-FISH to analyze the order of these sequences (B-SCAR and ITS) in the rDNA region. ITS and B-SCAR probes alternate along the DNA fiber (Figure 3) implying that these sequences are repetitive and arranged in tandem, and that the B-SCAR probably localizes to the ribosomal intergenic regions.
Figure 2.—
Fluorescence in situ hybridization, performed on embryo mitotic cells, with the B-SCAR (A and B) and 45S rDNA (C and D). Note the presence of large B-SCAR and rDNA clusters in B and X chromosomes, as well as the six smaller autosomes. In addition, there are small clusters in paracentromeric regions of most other chromosomes. Bar, 5 μm.
Figure 3.—
Fiber double-FISH for the B-SCAR sequence (green) and rDNA (red) showing the alternated arrangement of these sequences. (A) DAPI, (B) B-SCAR, (C) rDNA, and (D) merged images of B-SCAR and rDNA. Bar, 1 μm.
Finding similar DNA sequences in the 0B genome:
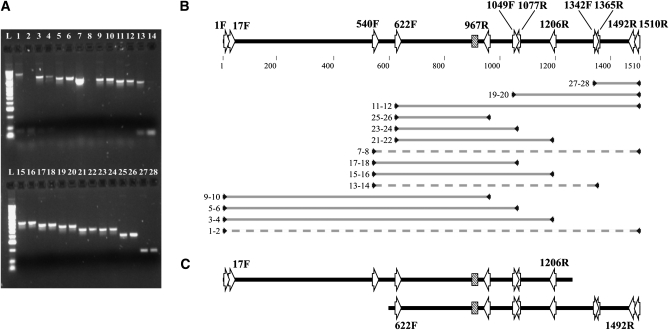
Since BSCAR-17F and BSCAR-1492R primers did not amplify in 0B individuals, we used a series of internal nested primers (Table 1 and Figure 1) to amplify B-SCAR like sequences present in genomic DNA from 0B individuals from the Torrox population. The BSCAR-622F and BSCAR-967R primers amplified a 345-bp central sequence of the B-SCAR (Figure 4, A and B). When we tried this primer combination in 0B individuals, we obtained a band of the same size and identical sequence. Thus, at least part of the B-SCAR is shared between the A and B chromosomes.
Figure 4.—
Results and interpretation of PCR amplifications using primers anchored in the B-SCAR sequence. (A) 1.5% agarose gel showing PCR products of +B (odd lanes) and 0B (even lanes) individuals. All but three combinations (1-2, 7-8, and 13-14) amplify a similar size band in both kinds of individuals. (B) Schematic view of primer positions (arrows) into B-SCAR marker. The microsatellite region is represented as a box. Shaded lines represent PCR products of each line shown in A. Dashed lines represent primer combinations with no amplification in 0B individuals. (C) Interpretation of B-SCAR–like region in A chromosomes to justify the pattern observed in A and B parts of the figure.
Is the B-SCAR marker derived from DNA sequences present in the A chromosomes but in a different arrangement? To test this possibility, we designed several other internal primers (BSCAR-540F, BSCAR-1342F, BSCAR-1049F, BSCAR-1365R, BSCAR-1206R, and BSCAR-1077R, see Figure 1) and tried a series of primer combinations, including the external BSCAR-17R and BSCAR-1492R primers, in 0B individuals. Some primer combinations did amplify in 0B samples (BSCAR-540F/BSCAR-1077R, BSCAR-1342F/BSCAR-1492R, BSCAR-1049F/BSCAR-1492R; see Figure 4), but other combinations did not (BSCAR-540F/BSCAR-1365R, BSCAR-540F/BSCAR-1365R; see Figure 4). This pattern of amplification (Figure 4, A and B) suggests that the A chromosome complement includes two separate regions that jointly span the complete B-SCAR sequence. The longest DNA sequence extends 1207 bp from the 5′ start of the B-SCAR to the BSCAR-1206R primer anchoring site. An additional shorter region spans from the 622 nucleotide to the B-SCAR 3′ end (Figure 4C). These DNA sequences explain the positive FISH hybridization of the B-SCAR probe with A chromosomes and suggest the possibility that the B-SCAR marker could have originated by recombination of two different regions in the A chromosomes sharing a DNA sequence in about the BSCAR-1049F/BSCAR-1077R region (Figure 4C).
Beyond B-SCAR marker borders:
To analyze the sequences neighboring to the B-SCAR marker, we performed a modified ligation-anchored PCR (Troutt et al. 1992). After cutting +B genomic DNA with EcoRI (since there are no restriction sites for EcoRI within the B-SCAR sequence), a Southern hybridization with the B-SCAR probe was performed. This procedure showed that the B-SCAR marker is inserted within a 4-kb region flanked by EcoRI restriction sites. We linked EcoRI adaptors to the EcoRI restriction fragments and performed a PCR using EcoRI-presel/BSCAR-622F and EcoRI-presel/BSCAR-967R as primer pairs (Table 1). An ∼1300-bp fragment was amplified using the EcoRI-presel/BSCAR-967R primer pair. This sequence encompasses the initial 330 bp of the 5′ flanking region of the B-SCAR. This implies that the 3′ flanking EcoRI restriction site is located 2157 bp downstream of B-SCAR. However, when we tried to amplify the 3′ flanking region (using EcoRI-presel/BSCAR-622F) no amplification was observed, even using long PCR, betaine, or DMSO to inhibit secondary structures in the DNA template. In addition, we tried to amplify the flanking regions using several primer combinations in the 18S and 28S rDNAs and in the B-SCAR sequence. However, these PCR reactions were unsuccessful.
DNA sequence of the SCAR marker in different B chromosome variants:
To analyze the variability of this sequence in the B chromosome system of E. plorans, we amplified the complete B-SCAR sequence in a sample of individuals from different geographic origins, covering the main B chromosome types (see Table 2). All 32 individuals carrying B chromosomes carried the B-SCAR sequence. DNA of +B individuals from 10 Iberian populations with cytogenetically different B chromosome types were used as template to amplify the B-SCAR. Remarkably, all DNA sequences were identical. We also amplified this marker in +B specimens from Armenia, Greece, and Morocco. From 25 individuals (all from Spain) we obtained the complete (1510 bp) B-SCAR sequence, but for the other seven sequences, the regions around the primers were not clearly resolved because of the typical noise at the start of the sequencing reactions (Table 3). The total length of the alignment was 1526 nucleotides, including a minimum sequence of 1200 bp and a maximum of 1510 bp, a mean ungapped length for the 32 sequences of 1456.8 ± 16.1 bp. Sequences show 52.5% of GC content. In these 32 B chromosome sequences, we found six different haplotypes (Table 2), but only three haplotypes after excluding the microsatellite region (Table 3). Only three variable sites appeared in this set of sequences, implying a nucleotide diversity of π = 0.0002 and a low haplotype diversity (Hd = 0.123) (Table 3). The B1M from Morocco showed a B-SCAR identical in DNA sequence to Spanish Bs. However, the B-SCAR sequences from Armenia were all slightly different but shared an insertion of 15 nucleotides, showing the same motif found in the microsatellite region (GAKTA). The same insertion was also present in one of the Greek samples. The other two individuals from Greece also showed an insertion at that site, but of only one repetition of the same motif (GATTA). In addition, a few substitutions were found in these sequences from Armenia and Greece (three singleton substitutions in the Armenian sequences and a common unique nucleotide insertion in the Greek sequences). It is remarkable that even B chromosomes from distant populations showed the same B-SCAR sequence, with only slight differences in a highly variable region. This suggests a recent common origin for all B chromosomes in E. plorans.
TABLE 3.
Polymorphism of the B-SCAR sequences analyzed (excluding the microsatellite region)
| Origin of the sequences | No. of sequences | No. of sites | Total no. of sites (excluding gaps/missing data) | No. of variable sites (S) | No. of haplotypes (h) | Haplotype diversity (Hd) | Nucleotyde diversity (π) | Average no. of nucleotide differences (k) |
|---|---|---|---|---|---|---|---|---|
| Spain | 23 | 1526 | 1510 | 0 | 1 | 0 | 0 | — |
| Spain (including incomplete sequences) | 25 | 1526 | 1365 | 0 | 1 | 0 | 0 | — |
| Morocco | 1 | 1526 | 1392 | 0 | 1 | 0 | 0 | — |
| Armenia | 3 | 1526 | 1196 | 3 | 3 | 1 | 0.0017 | 2 |
| Greece | 3 | 1526 | 1276 | 0 | 1 | 0 | 0 | — |
| West Mediterranean (Spain + Morocco) | 26 | 1526 | 1362 | 0 | 1 | 0 | 0 | — |
| East Mediterranean (Armenia + Greece) | 6 | 1526 | 1172 | 3 | 3 | 0.6 | 0.0009 | 1 |
| All | 32 | 1526 | 1167 | 3 | 3 | 0.123 | 0.0002 | 0.1875 |
DISCUSSION
We have developed a molecular tool for investigating B chromosome presence in E. plorans, which may be very useful in a number of situations. To date, B chromosome presence in E. plorans could only be determined by cytogenetic techniques, usually requiring animal sacrifice, since they are based on gonad analysis. The introduction of testis biopsy analysis (López-León et al. 1993b) permitted determining B chromosome presence in living males, and the development of a technique for determining B chromosome presence in interphase nuclei of hemocytes permitted analyzing B presence even in living females (Cabrero et al. 2006). A PCR test for B-specific SCAR marker is applicable to live, dead, and preserved collection specimens. Although we cannot presently determine B chromosome number with the SCAR marker, a quantitative PCR may be possible.
If B chromosomes are evolutionarily young, it should be possible to trace the origins of these chromosomes by analyzing and comparing their DNA sequences with those in the A chromosomes, although this may be difficult because B chromosome sequences are expected to evolve rapidly through mutation as long as they remain dispensable. In the plant Plantago lagopus, the rise of a supernumerary chromosome was witnessed in the laboratory in the course of a few generations, and the evolutionary pathway toward the B chromosome derivation was inferred to involve aneuploidy, the formation of a ring chromosome, chromosome fragmentation, massive amplification of 5S rDNA, and centromere misdivision (Dhar et al. 2002). But, as time goes on, it becomes more difficult to ascertain what molecular events took place during B chromosome differentiation. In the plant Brachychome dichromosomatica, B chromosomes are made up of a collection of tandem repeat DNA sequences derived from several A chromosome sites (Houben et al. 2001). Jones et al. (2008) suggested that they might have arisen from founder sequences derived from a polymorphic A chromosome region, that were stabilized by the addition of extrachromosomal DNA and telomeric and centromeric sequences. Similarly, in rye, there are two B chromosome-specific repetitive DNA sequences, E3900 and D1100, that seem to have arisen from A chromosome DNA sequences (including some mobile elements) that have undergone a complex series of rearrangements in the B chromosome DNA (Langdon et al. 2000). However, in Nasonia vitripennis, the analysis of the retrotransposon NATE provided strong evidence for the origin of the paternal sex ratio B chromosome by interspecific hybridization (McAllister and Werren 1997). The molecular organization of a 18-kb linear region of this B chromosome suggested that it may be in the late stages of molecular degeneration, due to the accumulation of repetitive elements, deletions, and duplications. This would be compatible with the interspecific transfer of an old B chromosome (McAllister and Werren 1997).
In maize, several B-specific sequences have been isolated. For instance, the ZmBs repeat is a long tandem array of ∼1400 bp located in and around the B centromere and near the tip of its long arm (Alfenito and Birchler 1993; Lamb et al. 2005). The CL-1 repeat is ∼1500 bp long (Cheng and Lin 2003, 2004). StarkB is a 22-kb element not arranged in arrays and was characterized by Lamb et al. (2007) after previous detection of a RAPD fragment specifically amplifying only in B-carrying individuals (Stark et al. 1996). This element is composed of repetitive sequences also present in the A chromosomes as well as novel sequences unique to the B chromosome (Lamb et al. 2007). Insertions and deletions are frequent in the StarkB elements, and they also include LTR retrotransposons whose sequence analysis provided an estimate of a minimum of 2 million years of age for these elements and hence for the B chromosome. As pointed out by Lamb et al. (2007), there is a high resemblance between the rye E3900/D1100 and maize StarkB elements since they are composed of a complex mixture of B-specific sequences and ones shared with As and show clear evidence of degeneration of A chromosome sequences, which is the first step in B chromosome formation (Jones and Houben 2003).
In E. plorans, the 1510-bp B-specific SCAR marker probably arose by rearrangement of two DNA sequences that are separate in the A chromosomes. Within the 1510 bp of the B-SCAR sequence, there is a (GAKTA)6-10 microsatellite that could have been involved in the rearrangement producing the B-SCAR fragment from the two A chromosome regions. Microsatellite sequences have also been found in B chromosomes from other species. For instance, in Crepis capillaris, Jamilena et al. (1994) found a (CA)n microsatellite showing some differences between A and B chromosomes. In rye, a short GAAAT microsatellite is also present in the E3900 B-specific sequence (Langdon et al. 2000).
The origin of the B-SCAR sequence must probably have occurred at the B chromosome origin, since all B variants analyzed here, from very distant populations (e.g., Morocco and Armenia) show the same two A chromosome DNA sequences combined into the B-SCAR sequence. The high similarity among B-SCAR sequences suggests that B chromosome variants from distant E. plorans populations have a common origin, despite prior evidence for a possible multiregional origin of B chromosomes in this species (Cabrero et al. 2003b). However, our present results give strong support to a unique B origin, since the B-SCAR was present in B-carrying individuals from all Eastern (Greece and Armenia) and Western (Iberian Peninsula and Morocco) populations analyzed. Although we have not tested for B-SCAR presence in Daghestan (North Caucasus), its presence in Armenia (South Caucasus) suggests that it could also be present in Bs from Daghestan.
The high similarity in DNA sequence among B-SCAR sequences suggests that B chromosomes in E. plorans are rather young. B-SCAR sequences show a low nucleotide diversity (π = 0.0002, see Table 3) probably implying a rather young age for this sequence. We have no nucleotide diversity estimations at population level for other nuclear DNA sequences in E. plorans, but nucleotide diversity for three mtDNA regions [two Cytochrome c oxidase subunit I (COI) region and mitochondrially encoded NADH dehydrogenase subunit 5 (ND5); Manrique-Poyato 2010] is rather higher (π = 0.0244–0.0337). Although direct comparison of these values is difficult due to the disparate evolutionary models of both types of DNA, these data appear to support a young B chromosome hypothesis. Unless the B-SCAR sequence evolution has some functional constraints, a recent B chromosome origin would be more consistent with the scarce nucleotide variation found among the DNA sequences analyzed from Eastern and Western populations. Bearing in mind that B chromosomes in this species follow a near-neutral mode of evolution, by which they begin being parasitic and thus have high invasive ability (Zurita et al. 1998; Riera et al. 2004), the present geographic range of B chromosomes in E. plorans could be the result of a rapid spread. Once B chromosomes are neutralized, their elimination is rather slow (Camacho et al. 1997), which explains their ubiquitous presence in most E. plorans plorans populations (see Camacho et al. 2003).
Acknowledgments
We thank Tatiana López and Mohamed Abdelaziz for technical assistance. Specimens from Morocco, Greece, and Armenia were kindly provided by M. Bakkali, D. Chobanov, and A. Bugrov, respectively. We also thank J. J. Pasantes for his advice on fiber-FISH analysis. We specially thank D. Charlesworth for helpful comments during manuscript edition. This study was supported by grants from the Spanish Ministerio de Ciencia e Inovación (CGL2009-11917) and Plan Andaluz de Investigación (CVI-1664), including European regional development fund (FEDER).
References
- Abdelaziz, M., M. Teruel, D. Chobanov, J. P. M. Camacho and J. Cabrero, 2007. Physical mapping of rDNA and satDNA in A and B chromosomes of the grasshopper Eyprepocnemis plorans from a Greek population. Cytogenet. Genome Res. 119 143–146. [DOI] [PubMed] [Google Scholar]
- Alfenito, M. R., and J. A. Birchler, 1993. Molecular characterization of a maize B chromosome centric sequence. Genetics 135 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo, S. M., S. G. Pompolo, F. Perfectti and J. P. Camacho, 2001. Integration of a B chromosome into the A genome of a wasp. Proc. Roy. Soc. Ser. B 268 1127–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo, S. M. S. R., S. G. Pompolo, F. Perfectti and J. P. M. Camacho, 2002. Integration of a B chromosome into the A genome of a wasp, revisited. Proc. Roy. Soc. Ser. B 269 1475–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkali, M., J. Cabrero, M. D. Lopez-Leon, F. Perfectti and J. P. M. Camacho, 1999. The B chromosome polymorphism of the grasshopper Eyprepocnemis plorans in north africa. I. B variants and frequency. Heredity 83 428–434. [DOI] [PubMed] [Google Scholar]
- Bakkali, M., F. Perfectti and J. P. M. Camacho, 2002. The B-chromosome polymorphism of the grasshopper Eyprepocnemis plorans in North Africa: II. Parasitic and neutralized B1 chromosomes. Heredity 88 14–18. [DOI] [PubMed] [Google Scholar]
- Basha, S. D., and M. Sujatha, 2007. Inter and intra-population variability of Jatropha curcas (L.) characterized by RAPD and ISSR markers and development of population-specific SCAR markers. Euphytica 156 375–386. [Google Scholar]
- Battaglia, E., 1964. Cytogenetics of B chromosomes. Caryologia 17 245–299. [Google Scholar]
- Cabrero, J., M. D. Lopez-Leon, M. Bakkali and J. P. M. Camacho, 1999. Common origin of B chromosome variants in the grasshopper Eyprepocnemis plorans. Heredity 83 435–439. [DOI] [PubMed] [Google Scholar]
- Cabrero, J., A. Bugrov, E. Warchałowska-Sliwa, M. D. López-León, F. Perfectti et al., 2003. a Comparative FISH analysis in five species of Eyprepocnemidine grasshoppers. Heredity 90 377–381. [DOI] [PubMed] [Google Scholar]
- Cabrero, J., M. Bakkali, A. Bugrov, E. Warchalowska-Sliwa, M. D. López-León et al., 2003. b Multiregional origin of B chromosomes in the grasshopper Eyprepocnemis plorans. Chromosoma 112 207–211. [DOI] [PubMed] [Google Scholar]
- Cabrero, J., M. I. Manrique-Poyato and J. P. M. Camacho, 2006. Detection of B chromosomes in interphase hemolymph nuclei from living specimens of the grasshopper Eyprepocnemis plorans. Cytogenet. Genome Res. 114 66–69. [DOI] [PubMed] [Google Scholar]
- Camacho, J. P. M. (Editor) 2004. B-Chromosomes in the Eukaryote Genome. Ed. 1. Karger AG, Basel, Switzerland.
- Camacho, J. P. M., 2005. B chromosomes, pp. 223–286 in The Evolution of the Genome, Ed. 1, edited by T. R. Gregory. Academic Press, New York.
- Camacho, J. P. M., J. Cabrero, E. Viseras, M. D. Lopez-Leon, J. Navas-Castillo et al., 1991. G banding in two species of grasshopper and its relationship to C, N, and fluorescence banding techniques. Genome 34 638–643. [Google Scholar]
- Camacho, J. P. M., M. W. Shaw, M. D. López-León, M. C. Pardo and J. Cabrero, 1997. Population dynamics of a selfish B chromosome neutralized by the standard genome in the grasshopper Eyprepocnemis plorans. Am. Nat. 149 1030–1050. [DOI] [PubMed] [Google Scholar]
- Camacho, J. P. M., J. Cabrero, M. D. López-León, M. Bakkali and F. Perfectti, 2003. The B chromosomes of the grasshopper Eyprepocnemis plorans and the intragenomic conflict. Genetica 117 77–84. [DOI] [PubMed] [Google Scholar]
- Cavallaro, Z. I., L. A. Bertollo, F. Perfectti and J. P. Camacho, 2000. Frequency increase and mitotic stabilization of a B chromosome in the fish Prochilodus lineatus. Chromosome Res. 8 627–634. [DOI] [PubMed] [Google Scholar]
- Cheng, Y., and B. Lin, 2003. Cloning and characterization of maize B chromosome sequences derived from microdissection. Genetics 164 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y., and B. Lin, 2004. Molecular organization of large fragments in the maize B chromosome: indication of a novel repeat. Genetics 166 1947–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar, M. K., B. Friebe, A. K. Koul and B. S. Gill, 2002. Origin of an apparent B chromosome by mutation, chromosome fragmentation and specific DNA sequence amplification. Chromosoma 111 332–340. [DOI] [PubMed] [Google Scholar]
- Dirsh, V. M., 1958. Revision of the genus Eyprepocnemis Fieber, 1853 (Orthoptera: acridoidea). Proc. Roy. Entomol. Soc. Lond. Ser. B, Taxonomy 27 33–45. [Google Scholar]
- Drummond, A. J, B. Ashton, S. Buxton, M. Cheung, A. Cooper et al., 2010. Geneious v5.1 (http://www.geneious.com).
- Fidlerová, H., G. Senger, M. Kost, P. Sanseau and D. Sheer, 1994. Two simple procedures for releasing chromatin from routinely fixed cells for fluorescence in situ hybridization. Cytogenet. Cell Genet. 65 203–205. [DOI] [PubMed] [Google Scholar]
- Griffiths, R., 2000. Sex identification using DNA markers, pp. 295–321 in Molecular Methods in Ecology, Ed. 1, edited by A. J. Baker. Blackwell Science, Oxford.
- Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41 95–98. [Google Scholar]
- Hernández, P., A. Martín and G. Dorado, 1999. Development of SCARs by direct sequencing of RAPD products: a practical tool for the introgression and marker-assisted selection of wheat. Mol. Breed. 5 245–253. [Google Scholar]
- Herrera, J., M. Lopez-Leon, J. Cabrero, M. Shaw and J. Camacho, 1996. Evidence for B chromosome drive suppression in the grasshopper Eyprepocnemis plorans. Heredity 76 633–639. [Google Scholar]
- Houben, A., D. Verlin, C. R. Leach and J. N. Timmis, 2001. The genomic complexity of micro B chromosomes of Brachycome dichromosomatica. Chromosoma 110 451–459. [DOI] [PubMed] [Google Scholar]
- Huo, H., J. A. Conner and P. Ozias-Akins, 2009. Genetic mapping of the apospory-specific genomic region in Pennisetum squamulatum using retrotransposon-based molecular markers. Theor. Appl. Genet. 119 199–212. [DOI] [PubMed] [Google Scholar]
- Jamilena, M., M. Garrido-Ramos, C. Ruiz Rejon and M. R. Rejon, 1994. Molecular relationship between the A and B chromosomes of Crepis capillaris. Heredity 73 527–531. [Google Scholar]
- Jones, R. N., 1985. Are B chromosomes selfish?, pp. 397–425 in The Evolution of Genome Size, Ed. 1, edited by T. Cavalier-Smith. Wiley, London.
- Jones, R. N., and A. Houben, 2003. B chromosomes in plants: Escapees from the A chromosome genome? Trends Plant Sci. 8 417–423. [DOI] [PubMed] [Google Scholar]
- Jones, R. N., W. Viegas and A. Houben, 2008. A century of B chromosomes in plants: So what? Ann. Bot. 101 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalendar, R., D. Lee and A. H. Schulman, 2009. FastPCR Software for PCR Primer and Probe Design and Repeat Search. Gene. Genome. Genomics 3 1–14. [Google Scholar]
- Lamb, J. C., A. Kato and J. A. Birchler, 2005. Sequences associated with A chromosome centromeres are present throughout the maize B chromosome. Chromosoma 113 337–349. [DOI] [PubMed] [Google Scholar]
- Lamb, J. C., N. C. Riddle, Y. Cheng, J. Theuri and J. A. Birchler, 2007. Localization and transcription of a retrotransposon-derived element on the maize B chromosome. Chromosome Res. 15 383–398. [DOI] [PubMed] [Google Scholar]
- Langdon, T., C. Seago, R. N. Jones, H. Ougham, H. Thomas et al., 2000. De novo evolution of satellite DNA on the rye B chromosome. Genetics 154 869–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado, P., and J. Rozas, 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lopez-Leon, M. D., J. Cabrero, J. P. M. Camacho, M. I. Cano and J. L. Santos, 1992. A widespread B chromosome polymorphism maintained without apparent drive. Evolution 46 529–539. [DOI] [PubMed] [Google Scholar]
- Lopez-Leon, M., J. Cabrero, M. Pardo, E. Viseras, J. Camacho et al., 1993. a Generating high variability of B chromosomes in Eyprepocnemis plorans (grasshopper). Heredity 71 352–362. [Google Scholar]
- Lopez-Leon, M., J. Cabrero, M. Pardo, E. Viseras and J. Camacho, 1993. b Paternity displacement in the grasshopper Eyprepocnemis plorans. Heredity 71 539–545. [Google Scholar]
- López-León, M. D., N. Neves, T. Schwarzacher, J. S. Heslop-Harrison, G. M. Hewitt et al., 1994. Possible origin of a B chromosome deduced from its DNA composition using double FISH technique. Chromosome Res. 2 87–92. [DOI] [PubMed] [Google Scholar]
- López-León, M. D., J. Cabrero, V. V. Dzyubenko, A. G. Bugrov, T. V. Karamysheva et al., 2008. Differences in ribosomal DNA distribution on A and B chromosomes between eastern and western populations of the grasshopper Eyprepocnemis plorans plorans. Cytogenet. Genome Res. 121 260–265. [DOI] [PubMed] [Google Scholar]
- Manrique-Poyato, I., 2010. Spatial and temporal dynamics of the B chromosomes of the grasshopper Eyprepocnemis plorans. Ph.D. Thesis, University of Granada, Spain (in Spanish).
- McAllister, B. F., and J. H. Werren, 1997. Hybrid origin of a B chromosome (PSR) in the parasitic wasp Nasonia vitripennis. Chromosoma 106 243–253. [DOI] [PubMed] [Google Scholar]
- McVean, G. T., 1995. Fractious chromosomes: hybrid disruption and the origin of selfish genetic elements. Bioessays 17 579–582. [DOI] [PubMed] [Google Scholar]
- Muñoz, E., F. Perfectti, A. Martin-Alganza and J. P. M. Camacho, 1998. Parallel effects of a B chromosome and a mite that decrease female fitness in the grasshopper Eyprepocnemis plorans. Proc. Roy. Soc. Ser. B 265 1903–1909. [Google Scholar]
- Östergren, G., 1945. Parasitic nature of extra fragment chromosomes. Botaniska Notiser 2 157–163. [Google Scholar]
- Oyama, R. K., S. M. Volz and S. S. Renner, 2009. A sex-linked SCAR marker in Bryonia dioica (Cucurbitaceae), a dioecious species with XY sex-determination and homomorphic sex chromosomes. J. Evol. Biol. 22 214–224. [DOI] [PubMed] [Google Scholar]
- Paran, I., and R. W. Michelmore, 1993. Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor. Appl. Genet. 85 985–993. [DOI] [PubMed] [Google Scholar]
- Pasquali, M., F. Dematheis, M. L. Gullino and A. Garibaldi, 2007. Identification of race 1 of Fusarium oxysporum f. sp. lactucae on lettuce by inter-retrotransposon sequence-characterized amplified region technique. Phytopathology. 97 987–996. [DOI] [PubMed] [Google Scholar]
- Perfectti, F., and J. H. Werren, 2001. The interspecific origin of B chromosomes: experimental evidence. Evolution 55 1069–1073. [DOI] [PubMed] [Google Scholar]
- Perfectti, F., J. M. Corral, J. A. Mesa, J. Cabrero, M. Bakkali et al., 2004. Rapid suppression of drive for a parasitic B chromosome. Cytogenet. Genome Res. 106 338–343. [DOI] [PubMed] [Google Scholar]
- Riera, L., E. Petitpierre, C. Juan, J. Cabrero and J. P. M. Camacho, 2004. Evolutionary dynamics of a B chromosome invasion in island populations of the grasshopper Eyprepocnemis plorans. J. Evol. Biol. 17 716–719. [DOI] [PubMed] [Google Scholar]
- Stark, E. A., I. Connerton, S. T. Bennett, S. R. Barnes, J. S. Parker et al., 1996. Molecular analysis of the structure of the maize B-chromosome. Chromosome Res. 4 15–23. [DOI] [PubMed] [Google Scholar]
- Teruel, M., 2009. Origen, expresión y efectos fenotípicos de un parásito genómico. Ph.D. thesis. Universidad de Granada, Granada, Spain.
- Teruel, M., J. Cabrero, F. Perfectti, M. J. Acosta, A. Sánchez et al., 2009. Microdissection and chromosome painting of X and B chromosomes in the grasshopper Eyprepocnemis plorans. Cytogenet. Genome Res. 125 286–291. [DOI] [PubMed] [Google Scholar]
- Tosta, V. C., T. M. Fernandes-Salomão, M. G. Tavares, S. G. Pompolo, E. G. Barros et al., 2004. A RAPD marker associated with B chromosomes in Partamona helleri (Hymenoptera, Apidae). Cytogenet. Genome Res. 106 279–283. [DOI] [PubMed] [Google Scholar]
- Tosta, V. C., M. G. Tavares, T. M. Fernandes-Salomão, E. G. Barros, L. A. O. Campos et al., 2007. Development of a SCAR marker for the analysis of B chromosome presence in Partamona helleri (Hymenoptera, Apidae). Cytogenet. Genome Res. 116 127–129. [DOI] [PubMed] [Google Scholar]
- Troutt, A. B., M. G. McHeyzer-Williams, B. Pulendran and G. J. Nossal, 1992. Ligation-anchored PCR: a simple amplification technique with single-sided specificity. Proc. Natl. Acad. Sci. USA 89 9823–9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler, K. S., and B. T. Wakimoto, 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29 577–605. [DOI] [PubMed] [Google Scholar]
- Whitaker, V. M., J. M. Bradeen, T. Debener, A. Biber and S. C. Hokanson, 2010. Rdr3, a novel locus conferring black spot disease resistance in tetraploid rose: genetic analysis, LRR profiling, and SCAR marker development. Theor. Appl. Genet. 120 573–585. [DOI] [PubMed] [Google Scholar]
- Wilson, E. B., 1907. The supernumerary chromosomes of Hemiptera. Science 26 870–871. [Google Scholar]
- Yau, F. C., K. Wong, J. Wang, P. P. But and P. Shaw, 2002. Generation of a sequence characterized amplified region probe for authentication of crocodilian species. J. Exp. Zool. 294 382–386. [DOI] [PubMed] [Google Scholar]
- Zurita, S., J. Cabrero, M. D. López-León and J. P. M. Camacho, 1998. Polymorphism regeneration for a neutralized selfish B chromosome. Evolution 52 274–277. [DOI] [PubMed] [Google Scholar]