Abstract
The carotenoid-binding protein (CBP) of the domesticated silkworm, Bombyx mori, a major determinant of cocoon color, is likely to have been substantially influenced by domestication of this species. We analyzed the structure of the CBP gene in multiple strains of B. mori, in multiple individuals of the wild silkworm, B. mandarina (the putative wild ancestor of B. mori), and in a number of other lepidopterans. We found the CBP gene copy number in genomic DNA to vary widely among B. mori strains, ranging from 1 to 20. The copies of CBP are of several types, based on the presence of a retrotransposon or partial deletion of the coding sequence. In contrast to B. mori, B. mandarina was found to possess a single copy of CBP without the retrotransposon insertion, regardless of habitat. Several other lepidopterans were found to contain sequences homologous to CBP, revealing that this gene is evolutionarily conserved in the lepidopteran lineage. Thus, domestication can generate significant diversity of gene copy number and structure over a relatively short evolutionary time.
DOMESTICATION represents a relatively recent evolutionary event, occurring over the past 13,000 years after the Neolithic revolution (Diamond 1997; Purugganan and Fuller 2009). This process frequently leads to the improvement of economically important traits and the diversification of morphological traits in domesticated species compared to their wild ancestors. Elucidating the molecular genetic basis of the domestication process can provide insights into the early and short-term mechanisms of evolutionary changes and reveal how humans have performed one of the most influential technological innovations in history.
The silkworm, Bombyx mori, which was domesticated for sericulture, is the only domesticated insect that is completely dependent on humans for its survival and reproduction (Goldsmith 2009). Genetic analysis suggests that B. mori originated from a Chinese population of the wild silkworm B. mandarina (Arunkumar et al. 2006; Li et al. 2010). More than 1000 inbred B. mori strains maintained worldwide exhibit phenotypic diversity in morphology (Banno et al. 2010). In particular, cocoon color varies substantially, with various shades of white, yellow, pink, and green silk. The variation in B. mori cocoon color is apparently more diversified than that in B. mandarina, which generally produces pale yellow to green cocoons.
Recently, a critical gene (locus) of B. mori for yellow cocoon color, Yellow blood (Y), located on chromosome 2, was identified (Sakudoh and Tsuchida 2009). Y encodes the carotenoid-binding protein (CBP) (Tabunoki et al. 2002), which likely functions as a cytosolic transporter of carotenoids, a group of yellowish hydrophobic pigments. In B. mori strains harboring the dominant allele of Y (the Y allele), CBP facilitates the extensive transport of dietary carotenoids from the midgut lumen to the middle silk gland via hemolymph, resulting in yellow cocoons. By contrast, in B. mori strains that are homozygous for the recessive allele of Y (the +Y allele), CBP expression is completely absent (Tabunoki et al. 2002; Tsuchida et al. 2004). These recessive strains are characterized by a significant decrease in carotenoid transport and a consequent loss of carotenoids in their cocoons, which appear white when other pigments, such as flavonoids, do not accumulate. These observations suggest that CBP can be used as a tool to dissect the molecular evolutionary mechanisms underlying morphogenetic change during domestication.
In the course of our previous characterization of Y, the genomic structure of CBP was analyzed in four B. mori strains (Sakudoh et al. 2005, 2007). Our results revealed that the Y allele contains duplicate copies of CBP, one of which carried a full-length nonlong terminal repeat (non-LTR) retrotransposon, termed CATS. In contrast, the +Y allele contains only one copy of CBP, which contained a partial genomic deletion of the coding sequence and a truncated CATS sequence. To investigate the impact of the domestication process on CBP, the presence and structure of the gene were further examined in multiple B. mori strains, multiple B. mandarina individuals from several geographical regions, and species from several other lepidopteran genera.
MATERIALS AND METHODS
Insects:
The B. mori strains used in this study were obtained in Japan from the silkworm stock center of Kyushu University, Fukuoka, University of Ryukyu, Okinawa, the National Institute of Agrobiological Sciences, Ibaraki, and the National Institute of Infectious Diseases, Tokyo. Japanese and Taiwanese B. mandarina individuals were obtained from laboratory stock by sib breeding. Samia cynthia ricini, Sa. c. pryeri, Antheraea pernyi, and A. yamamai were provided by Zenta Kajiura (Shinshu University, Nagano, Japan). Helicoverpa armigera armigera was provided by Dr. Hidetoshi Iwano (Nihon University, Kanagawa, Japan).
Western blotting analysis of CBP expression:
The midgut and the middle silk gland from last-instar larvae of each species were dissected and frozen at −80° prior to use. Western blotting analysis using rabbit anti-CBP antibody (Tabunoki et al. 2002) was performed as described previously (Tsuchida et al. 2004).
Determination of CBP cDNA sequences:
The cDNA sequence of CBP from B. mori was determined after amplification by RT–PCR of the gene using the middle silk gland from one individual of strain N4 as a template with primer-43 (5′-GATCCCAAAAGCGATGTGTAGCTCCGTG-3′) and primer-30 (5′-CTTAATCCGGCCGATGGGTGAACATTGG-3′). The products were subcloned into pTA2 (Toyobo, Osaka, Japan) for sequencing.
The cDNA sequence of CBP from B. mandarina was determined after amplification by RT–PCR, using the midgut of a Japanese individual as the template, with primer-CBP-pBac-XbaI-1 (5′-ATGCTCTAGAGAAACCCTAAGCTCTTGAAGTG-3′) and primer-CBP-pBac-XbaI-2 (5′-ATGCTCTAGAGGCCGATGGGTGAACATTGG-3′). The product was subcloned into pTA2 for sequencing. The nucleotide sequences obtained from two independent subclones were identical, indicating that the identified nucleotide differences in the CBP genes of B. mori and B. mandarina were not a result of PCR-generated errors.
Homology-based searches of GenBank retrieved the CBP gene of Spodoptera frugiperda from a midgut cDNA library (accession no. DV076780). A pair of degenerate PCR primers, primer-CBP-dege-5 (5′-DSNSTNGCNAAYTGGATGAA-3′) and primer-CBP-dege-4 (5′-NARDATNGTNGGRTTCCA-3′) was then designed from the conserved regions between the putative protein sequences of B. mori and S. frugiperda CBP and used to amplify CBP from S. c. ricini, S. c. pryeri, A. pernyi, A. yamama, and H. a. armigera using larval midgut as the template for RT–PCR. Primer-CBP-dege-5 is complementary to exon B2 of B. mori CBP, which is not present in BmStart1, an alternative-spliced isoform that contains putative transmembrane helices (Sakudoh et al. 2005). The sequences amplified by these degenerate primers should, therefore, represent a portion of the authentic CBP cDNA rather than BmStart1.
Phylogenetic analysis:
Phylogenetic trees were constructed via the neighbor-joining method using ClustalX2 (Larkin et al. 2007). The authentic CBP cDNA sequence of B. mori was obtained from GenBank (accession no. AB263201).
Quantification of CBP copy numbers in the silkworm genome by quantitative PCR:
The general principle for quantifying copy number by quantitative PCR is described in Andersson and Hughes (2009). Genomic DNA was extracted from larval tissues using standard procedures and stored in Tris-EDTA (TE) buffer at 4°. The concentration of DNA in each sample was then determined by quantitative PCR with the following primer pairs for each exon (gene) sequence: A4, primer-139 (5′-CGTAACGAGACGTTAGACCAAATATTGC-3′) and primer-140 (5′-CTTATCACAGCTAACACCACTATGTCG-3′); A7, primer-81 (5′-TACTGGACCCTCAGCCCGGTGTGAATTC-3′) and primer-16 (5′-TCAGTATGACCTGCTCCCCGAACCTGCG-3′); B1, Primer-43 and Primer-54 (5′-GGCCAACCAGAGTTGTTAGCCTTGTATC-3′); C4, primer-143 (5′-GGGACACAATAAAGTCGGTTGCTGG-3′) and primer-144 (5′-CACAACACATCAACCATTGGAACAGG-3′); Y-b, primer-145 (5′-GATCAGTAGCGCCCTAGCGAACTGGATG-3′) and primer-146 (5′-CACAGGCATGTGCGCCTTCACTTTCCGC-3′), and primer-149 (5′-ACCACGACCAAGGAAGAATTCGATCCTG-3′) and primer-152 (5′-CGTCGTATTTTCGTGGCGCTTGGACTC-3′); +Y-a, primer-1 (5′-ATGGCCGACTCTACGTCGAAAAGCG-3′) and primer-40 (5′-TGCAGATGTCAGCAGTCAAACCATCCGC-3′), and primer-151 (5′-GAAACCCTAAGCTCTTGAAGTG-3′) and primer-154 (5′-TGATAGTCATCATCCTGTCCTC-3′); dib, primer-dib-1 (5′-GAAAGATGATGAGATAACCGCTGACG-3′) and primer-dib-2 (5′-CCAATCGATACCGGATTAAGTCTCAAAC-3′); Cameo2, primer-Cameo2-21 (5′-TCCTTACCGTTACCAGGAGCATAG-3′) and primer-Cameo2-20 (5′-GCGGTTATAACGTCAATGGTTGTG-3′); and OR2, primer-OR2-1 (5′-GCTGGATGTCATTTTCTGTTCGTGG-3′) and primer-OR2-2 (5′-AACTCGGAACAACTCAGCCGTATTG-3′). OR2 is a single-copy gene found on chromosome 16 according to the B. mori genome database (International Silkworm Genome Consortium 2008; Shimomura et al. 2009; Duan et al. 2010) with a highly-conserved sequence (Krieger et al. 2003) and function (Nakagawa et al. 2005) among insects, and is generally considered to be essential for chemosensory behavior (Larsson et al. 2004). LightCycler FastStartDNA MasterPLUS SYBR Green I (Roche, Mannheim, Germany) and LightCycler ST300 (Roche) were used for PCRs.
To calculate the copy number of exons A4, A7, B1, and C4, dib, and Cameo2, their concentrations in each strain (individual) were normalized by the concentration of OR2 and then further divided by the corresponding value for strain p50 (also known as Daizo or Dazao). Thus, for example, the copy number of exon A4 is represented by the following mathematical formula:
 |
The strain p50 was taken as a standard for determining gene copy number because it was used for constructing the B. mori genome database (International Silkworm Genome Consortium 2008), and each of these sequences was identified as a single copy.
The copy number of Y-b is given by
 |
The copy number for Y-b in strain N4 was estimated as three because the copy numbers of that strain for exons A4, A7, B1, and C4 were found to be about four in this study and previous results from Southern blotting showed a significantly stronger band intensity for Y-b than that for Y-a in strain N4 (Sakudoh et al. 2007). Southern analysis of supporting information, Figure S1 also supported our estimation that there is one copy of Y-a and three copies of Y-b in strain N4.
The copy number of +Y-a is given by
 |
The copy number for +Y-a in strain w1-pnd was estimated as one because the copy numbers for exons A4, A7, B1, and C4 in that strain were proven in this study to be nearly one.
The copy number values obtained using the method described can vary due to differential PCR efficiency caused by unidentified nucleotide mutations in specific copies, including point mutations located in the primer annealing sites. Copy number calculations can also vary depending on the standard gene used for normalization. For example, the use of Cameo2 as the standard in place of OR2 led to a decrease of ∼25% in the calculated copy number values for strain c11. However, Southern blot analyses from this and previous (Sakudoh et al. 2005, 2007) studies clearly validated the overall approach and results, as detailed in results.
Genotyping of CBP by genomic PCR:
For CBP genotyping, the following primers were used in addition to several primers described above: primer-18 (5′-GCCTTCAACTTTCCTTGACTCCACGACG-3′), primer-33 (5′-GGTAGACTCCACACTCACACAG-3′), primer-150 (5′-CATGCTCTCGTTAGCCTGACTCTTGTAC-3′), primer-153 (5′-GCACGGCCTTTTGTATTGCACAAAGACG-3′), and primer-156 (5′-AGCGTGATGATGCGCCAAGCGTTC-3′).
Construction of the genomic fosmid library of B.mandarina:
Genomic DNA of B. mandarina was prepared from the posterior silk glands of female fifth-instar larvae of strain UT-Sakado. This strain originated from a wild population in Sakado, Saitama, Japan, and had been inbred at the University of Tokyo for >20 generations before DNA preparation. Digested and linearized DNAs were inserted into the vector pKS150 as described previously (Shizuya et al. 1992; Kim et al. 2003). A total of 147,456 independent clones were stocked. The average size of the clones was ∼40 kbp.
Sequencing of fosmid ends:
Primers used for sequencing were primer_Fos_T7+21 (5′-TTATCGATGATAAGCGGTCAAA-3′) and primer_Fos_T3 (5′-TATCACGAGGCCCTTTCGTCT-3′). A total of 153,216 reads from 76,608 independent clones were performed. Given that the genome size of the silkworm is ∼530 Mbp (Gage 1974), the clones represent more than five times the coverage of the whole genome. Consequently, a total of 161,554,214 nucleotide letters, which is estimated to be >30% of the whole genome sequence by simple summation, were obtained and used for the BLAST system.
Determination of CBP genomic sequences:
To sequence the full length of B. mandarina CBP, a fosmid clone containing CBP was isolated by searching the BLAST system described above. Four positive clones were found by a BLAST search using the repeat-masked sequences of the CBP gene from strain Kinshu X Showa (Sakudoh et al. 2005) and strain p50 (International Silkworm Genome Consortium 2008; Shimomura et al. 2009; Duan et al. 2010). A 11,009-bp length of one positive clone (#188_e09) was then sequenced by primer walking.
The Y-a and Y-b sequences for single-nucleotide polymorphism (SNP) analysis were amplified by PCR using primer-1 and primer-18 and primer-1 and primer-146, respectively, except for the Y-a sequence of Japanese B. mandarina from the genomic fosmid library. The resulting products were subcloned. The sequences from the annealing site of primer-1 in exon B2 to the insertion site of CATS were then determined. One subclone was determined for each sequence unless otherwise noted.
Southern blot analysis:
The procedures used for Southern blotting are described previously (Sakudoh et al. 2005). DIG-labeled probes for exons A4, B1 and B2, and dib were amplified from cDNA-containing vectors by PCR using the primer pairs: primer-139 and primer-140, primer-43 and primer-54, primer-147 (5′-TACCCGCAATCAAGCTTATAAAAACAC-3′) and primer-158 (5′-ATTTTTGGCGCGCTTTTCGACGTAG-3′), and primer-dib-1 and primer-dib-2, respectively.
Data deposition:
The CBP sequences reported in this article have been deposited in GenBank under accession nos. AB570176 (CBP cDNA sequence from Japanese B. mandarina), AB570177 (CBP cDNA sequence from A. pernyi), AB570178 (CBP cDNA sequence from A. yamamai), AB570179 (CBP cDNA sequence from S. c. pryeri), AB570180 (CBP cDNA sequence from S. c. ricini), AB570181 (CBP cDNA sequence from H. armigera), AB570182 (CBP genomic DNA sequence from Japanese B. mandarina), AB570183 (Y-a sequence from strain c11), AB570184 (Y-b sequence from strain c11), AB570185 (Y-a sequence from strain e09), AB570186 (Y-b sequence from strain e09), AB570187 (Y-a sequence from strain N4), AB570188 (Y-b sequence from strain N4), AB570189 (Y-a sequence from strain Nistari), AB570190 (Y-b sequence from strain Nistari), AB570191 and AB570192 (Y-a sequence from Chinese B. mandarina), AB570193 (Y-a sequence from Taiwanese B. mandarina), and AB570194 (Y-a sequence from Korean B. mandarina).
RESULTS
B.mandarina and several other lepidopteran insects possess CBP:
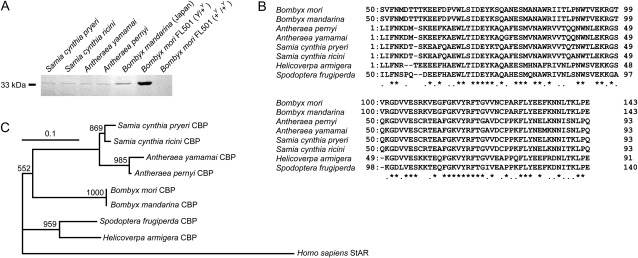
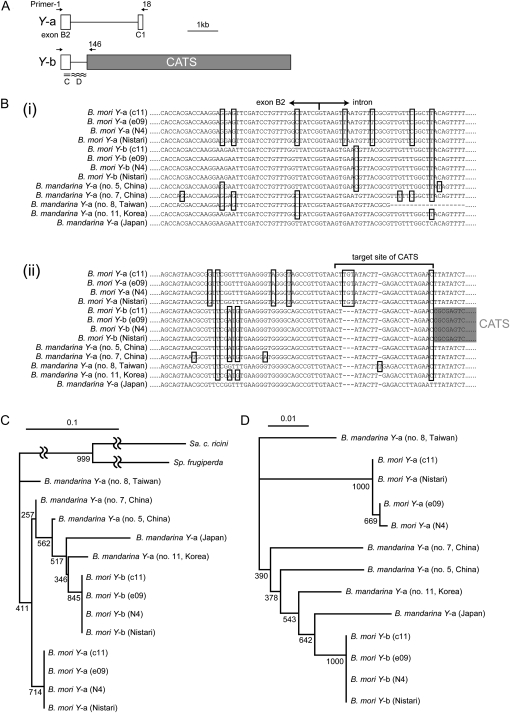
To assess the presence of CBP in lepidopterans other than B. mori, immunoreactivity against an anti B. mori CBP antibody in B. mandarina and several other lepidopteran insects was explored by Western analysis. A band of ∼33 kDa, similar to that observed in the sample from B. mori individuals carrying the Y allele, was present in the midgut of all the species tested (Figure 1A). The coding regions of CBP from B. mandarina and several other lepidopterans, which are homologous, possibly orthologous, to the B. mori sequence, were then fully or partially sequenced (Figure 1B). The CBP of B. mandarina from Japan was almost identical to that of B. mori, with no insertions/deletions in the complete coding sequence, 97.4% identity in the nucleotide sequence, and only one nonsynonymous mutation resulting in a change from glutamine to histidine at residue 240. The phylogenetic tree of lepidopterans based on these CBP sequences (Figure 1C) was consistent with trees generated using other gene sequences (Regier et al. 2009). Therefore, CBP has likely been evolutionarily conserved and vertically transferred in the lepidopteran lineage. The existence of CBP in S. frugiperda (fall armyworm) and H. armigera (cotton bollworm), which do not produce cocoons, was not unexpected because lepidopteran larvae utilize carotenoids for functions other than cocoon coloration, including larval integumental coloration (Landrum et al. 2009).
Figure 1.—
Presence of CBP in lepidopteran insects, including B. mandarina, the putative wild ancestor of B. mori. (A) Western blot analysis of larval midguts from lepidopterans using an anti-B. mori CBP antibody. Weak bands were observed in the lepidopteran species other than B. mori. (B) Alignment of the partial predicted amino acid sequences of CBP from lepidopterans. (C) A neighbor-joining tree generated using the amino acid sequences of CBP from lepidopterans. Human StAR, one of the closest homologs to CBP in humans (Alpy and Tomasetto 2006; Bhosale et al. 2009), was chosen as the outgroup. B. mori and B. mandarina share an identical position in this tree. Bootstrap values were based on 1000 replicates.
Large copy number variations for CBP in the B. mori genome revealed by quantitative PCR:
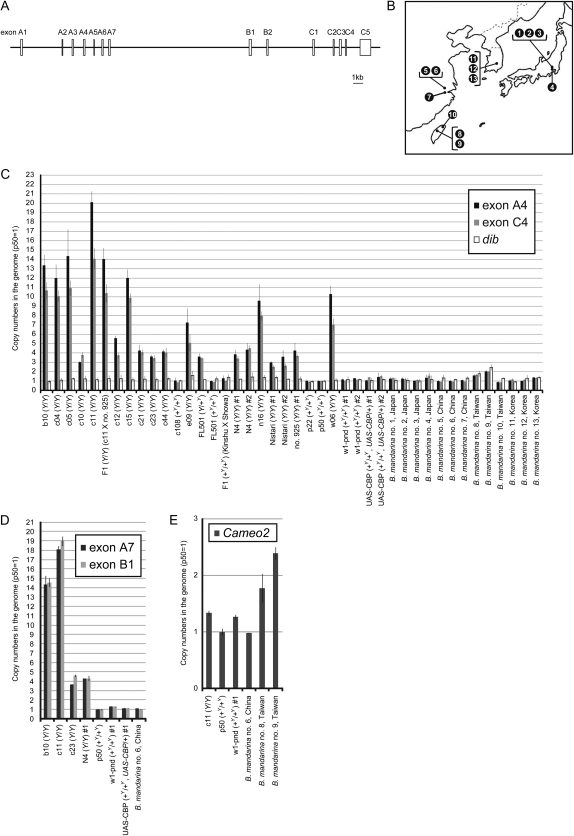
To investigate the alterations in CBP structure that occurred during silkworm domestication, we first analyzed the copy numbers of CBP in B. mori and B. mandarina genomes using quantitative PCR. For this analysis, primer pairs were designed to amplify several exons, including A4 and A7, both of which are contained in an alternative spliced isoform of CBP that contains putative transmembrane helices (known as BmStart1; Sakudoh et al. 2005) (Figure 2A). Multiple B. mori strains and multiple B. mandarina individuals, obtained from several places in East Asia (Figure 2B), the natural habitat of this insect, were analyzed. To our surprise, the copy number of CBP in B. mori strains carrying the Y allele varied extensively, ranging from 3 to 20 (Figure 2, C and D). This variation was not limited to a specific exon, suggesting that the entire gene, which spans 35 kbp in the B. mori genome, at least partially behaves as a single unit for multiple duplication events. In contrast to these strains, the copy number of CBP in B. mori strains carrying the +Y allele homozygously was determined to be ∼1. A transgenic line containing the UAS-CBP transgene (Sakudoh et al. 2007), which includes the complete sequence of exons C1–4, was calculated to contain 1.40 or 1.52 copies of exon C4. This is consistent with the presence of the homozygous native +Y allele and the heterozygous UAS-CBP transgene, supporting the validity of our copy number determination. For the B. mandarina individuals, the CBP copy number was found to be ∼1, with the exception of two Taiwanese individuals that were calculated to have copy numbers of ∼2. Our calculation also showed that the two Taiwanese individuals had approximately two copies of dib (Niwa et al. 2005) and Cameo2 (Sakudoh et al. 2010) (Figure 2, C and E), the control nuclear genes used for copy number quantification. Therefore, we concluded that B. mandarina most likely had only one copy of CBP, and the apparent duplication in the Taiwanese individuals may be due to an unexplained 50% decrease in the copy number of OR2 (Krieger et al. 2003; Sakurai et al. 2004), which was used for the normalization of all copy numbers in this study (for details of the normalization process, see materials and methods).
Figure 2.—
Amplification of the copy number of CBP in B. mori strains containing the Y allele. (A) Schematic exon/intron structure of CBP, including its alternatively spliced isoform BmStart1. Authentic CBP is composed of seven exons, namely exons B1–2 and C1–5, while BmStart1 is composed of 12 exons, including exons A1–7 and C1–5. Although this structure was derived from the strain Kinshu X Showa, which contains the +Y allele (Sakudoh et al. 2005), the sequence of the CATS retrotransposon was intentionally left out of this figure for the purpose of clarity. Furthermore, although this structure was generated by connecting partial nucleotide sequences, Southern blot analysis confirmed the overall structural integrity (Sakudoh et al. 2005). (B) Geographical distribution of B. mandarina individuals used in this study: 1–3, Sakado, Saitama, Japan; 4, Fuchu, Tokyo, Japan; 5 and 6, Huzhou, Zhejiang, China; 7, Hangzhou, Zhejiang, China; 8 and 9, Taichung, Taiwan; 10, Taipei, Taiwan; and 11–13, Daegu, South Korea. (C–E) Copy numbers of each exon or gene in multiple B. mori strains and B. mandarina individuals calculated by quantitative PCR. Error bars represent mean ± standard deviation (SD), n = 5 for strain p50, while n = 3 for all other strains. dib and Cameo2 were used as internal standards. For B. mori, the genotype of the Y gene is shown in parentheses. Two or more individuals of strains N4, w1-pnd, UAS-CBP, Nistari, and 925 were analyzed in this study.
Genotyping of CBP in B. mori and B. mandarina by genomic PCR:
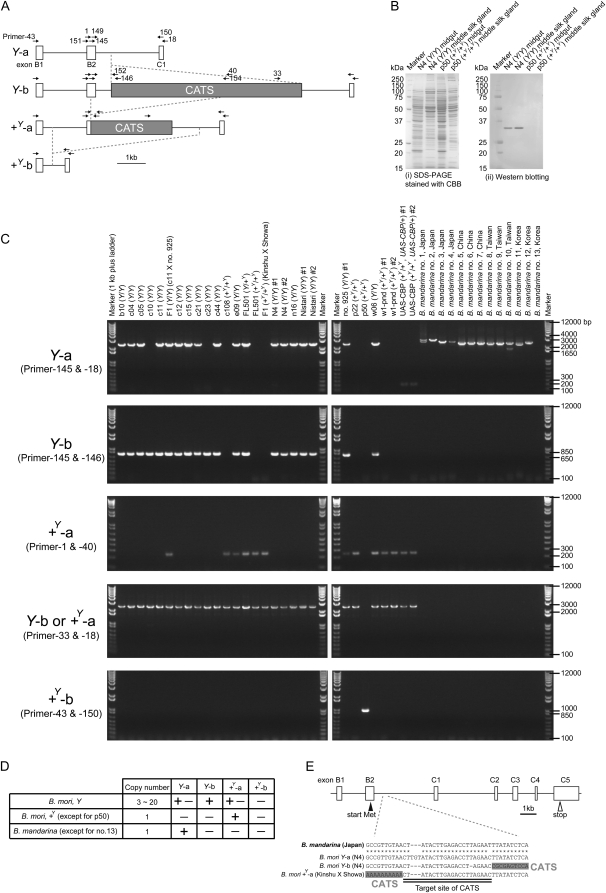
The structure of CBP in each strain and individual was assessed next. As described in the Introduction, three types of genomic sequences have been identified for the CATS insertion of CBP (Figure 3A) (Sakudoh et al. 2007), including: a Y-a, which lacks the CATS retrotransposon; a Y-b, which contains an insertion of the full-length CATS; and a +Y-a, which contains a truncated version of CATS together with a 169-bp deletion that lies within the coding sequence of CBP at the 3′ end of exon 2. In addition to these sequences, in the silkworm genome database constructed with strain p50 (International Silkworm Genome Consortium 2008; Shimomura et al. 2009; Duan et al. 2010), another version of CBP lacking both exon 2 and CATS was found on chromosome 2 (Figure 3A). Strain p50, which produces a cocoon lacking carotenoids, does not express CBP (Figure 3B), indicating that CBP is not produced from its CBP sequence as +Y-a (Sakudoh et al. 2007). The novel CBP sequence identified in strain p50 was, therefore, termed a +Y-b sequence. PCR primers were designed on the exons and CATS to examine their genomic structure in yellow and white cocoon strains of B. mori and in B. mandarina (Figure 3A).
Figure 3.—
Genotyping of CBP in multiple B. mori strains and B. mandarina individuals. (A) Schematic genomic structures of several versions of the CBP gene, identified either in a previous study (Y-a, Y-b, and +Y-a) (Sakudoh et al. 2007) or from the silkworm genome database generated from B. mori strain p50 (+Y-b) (International Silkworm Genome Consortium 2008). The annealing sites for the primers used in C and Figure 4 are indicated. (B) Western blot analysis showing that CBP protein was absent from strain p50, indicating that the protein was not produced from the +Y-b sequence, likely due to the complete absence of exon B2. (C) Genotyping of CBP by genomic PCR. The lower band of ∼200 bp for Y-a in strain UAS-CBP is likely from the UAS-CBP transgene (Sakudoh et al. 2007). The structure of CBP in B. mandarina individual number 13 remains undetermined. (D) Provisional summary of the presence/absence (+/−) of each CBP sequence in B. mori and B. mandarina. (E) Schematic genomic structure of CBP, as assessed by analysis of one clone from a Japanese B. mandarina genomic fosmid library. A single target site for the CATS retrotransposon was observed in the B. mandarina sequence between exons B2 and C1, suggesting that the insertion of CATS had not occurred.
Our genomic PCR results are presented in Figure 3C and Figure S2 and summarized in Figure 3D. Y-a was present in almost all B. mori strains carrying the Y allele, in no B. mori strain homozygous for the +Y allele, and in all but one of the B. mandarina individuals. Polymorphisms in the length of Y-a were observed in B. mandarina. Y-b was present in all B. mori strains carrying the Y allele, but was not found in the other strains or individuals analyzed. +Y-a was present in a few B. mori strains carrying the Y allele, in all B. mori strains homozygous for the +Y allele, except strain p50, and in none of the B. mandarina individuals. +Y-b was present only in strain p50. The detection of only a single type in the B. mori strains homozygous for the +Y allele and in the B. mandarina individuals is consistent with our copy number quantification results (Figure 2, C and D). The data from the present study suggest that strain p50, which was used to construct the B. mori genome database, could be considered an outlier strain for CBP.
To confirm the existence of Y-a in B. mandarina, a fosmid clone containing CBP was isolated by exploring our constructed BLAST search system of the end sequences of a Japanese B. mandarina genomic fosmid library (available at the Web site: http://pistil.ab.a.u-tokyo.ac.jp/wild/). The sequence of the fosmid clone was then determined by primer walking. The sequence did not contain the CATS insertion and exhibited a single target site for CATS retrotransposition (Figure 3E). This is similar to B. mori Y-a, supporting our classification of the B. mandarina CBP as Y-a type sequence.
Most copies of CBP in the Y allele of B. mori are of type Y-b:
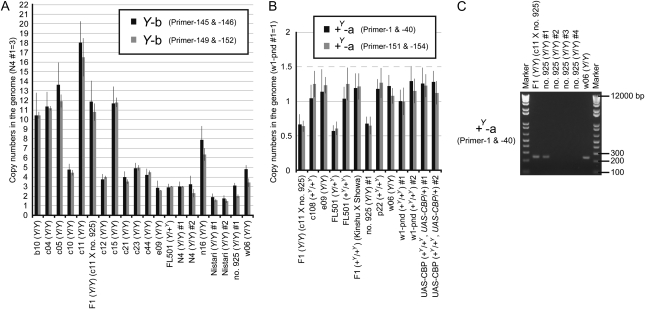
The copy numbers for Y-b and +Y-a in the B. mori strains were examined by quantitative PCR. The copy number for Y-b varied between strains (Figure 4A), with a pattern of variation similar to that of the exons of CBP (Figure 2, C and D). Most copies of CBP in the B. mori Y allele are, therefore, likely to be of the Y-b. The copy number for +Y-a was found to be ∼1 or 0.5 (Figure 4B). The calculated copy number for +Y-a in strain 925 was ∼0.5, which was curious because this strain should be homozygous for the Y gene, and an F1 hybrid between strain c11, which did not contain +Y-a, and strain 925 also carried ∼0.5 copy of +Y-a. Consequently, we examined several individuals of strain 925, which revealed that some individuals did not have +Y-a (Figure 4C). Therefore, these data suggested that strain 925 could be in the process of gaining or losing +Y-a. The copy number of Y-a unfortunately has not yet been determined due to the difficulty in designing suitable PCR primers for this sequence alone, while it may be expected to be a few or 1 because the copy number of Y-b approximated that of exons of CBP in the B. mori Y allele.
Figure 4.—
Copy numbers of types Y-b and +Y-a in B. mori strains. (A and B) The copy number of each CBP sequence in diverse B. mori strains was calculated by quantitative PCR (mean, SD; n = 4 for primer-145 and -146 and primer-1 and -40; n = 3 for primer-149 and -152 and primer-151 and -154). The primers used were the same as those used to generate the data in Figure 3C and Figure S2B. Strains for which no bands for Y-b and +Y-a were detected in Figure 3C were also confirmed by quantitative PCR to have a value of zero. (C) Genotyping of the +Y-a sequence by genomic PCR in several individuals from strain 925. The 925 individuals in which bands corresponding to +Y-a were not detected were found to be zero by quantitative PCR.
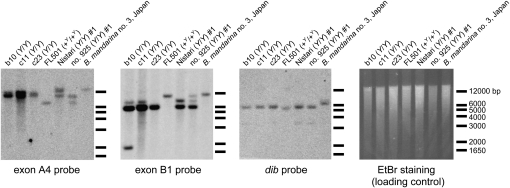
Southern blotting confirms the structural diversification of CBP in B. mori:
To support the observations of structural diversity in the B. mori CBP gene, Southern blot analysis was performed (Figure 5). Variations in signal intensity were largely consistent with our CBP copy number results (Figure 2, C and D). For a probe specific to exon B1, the migration of the bands for the Y-a, Y-b, and +Y-a alleles of B. mori was predicted from available sequence data to be 8200 bp, 5890 bp, and 10,320 bp, respectively (Sakudoh et al. 2007). As expected, the strongest signal in each strain carrying the Y allele was observed at the predicted size for Y-b (Figure 5). The sizes of the bands observed for the exon A4 probe, which should not to be affected by the CATS insertion, were diversified among analyzed strains. Divergent CBP sequences, therefore, likely exist in addition to those in Figure 3A.
Figure 5.—
Confirmation of copy number variation and structural differences in Bombyx CBP by Southern blot analysis. Genomic DNA was fully digested with EcoRV. For the exon B1 probe, the lengths of the bands for Y-a, Y-b, and +Y-a were predicted to be 8200, 5890, and 10,320 bp, respectively, on the basis of available sequences (Sakudoh et al. 2007). A dib probe that hybridized to dib was used as an internal standard.
Comparison of B. mandarina and B.mori Y-a and Y-b genes based on SNPs:
To investigate the evolutionary changes that occurred in CBP during silkworm domestication, Y-a and Y-b were partially sequenced from several B. mori strains and B. mandarina individuals (Figure 6, A and B). Phylogenetic trees were then generated on the basis of SNPs present in the third codon positions of exon B2 (Figure 6C) and in the intron located next to the 3′ end of exon B2 (Figure 6D). The trees suggest that B. mori Y-b is closer to B. mandarina Y-a, especially in Japan and Korea, than to B. mori Y-a. The presence of a CATS target site duplication (Figure 3E) indicates that Y-b was generated from Y-a. Therefore, B. mori Y-a and Y-b were likely derived from different populations of B. mandarina, and B. mori Y-b may have emerged by a CATS insertion into the Y-a sequence, which is close to Japanese and Korean B. mandarina Y-a.
Figure 6.—
Phylogenetic analysis based on SNPs in B. mori and B. mandarina Y-a and Y-b. (A) Schematic representation of the region of genomic sequence used in our phylogenetic analysis. The double underline is for C and the double wavy underline is for D. The sequences of Y-a and Y-b were determined after amplification by PCR using the primer pairs, primer-1 and primer-18 and primer-1 and primer-146, respectively, except for the Y-a sequence of Japanese B. mandarina from a genomic fosmid library. (B) Representative portions of the alignment of the genomic sequences near the 3′ end of exon B2 (i) and the target site of CATS (ii). Positions in which the sequences were not identical to Japanese B. mandarina Y-a are boxed. The Y-a and Y-b sequences of B. mori were nearly or completely identical among the strains analyzed. The sequences of the CATS target site were not included in this phylogenetic analysis. (C and D) Neighbor-joining trees showing each version of CBP from B. mori and B. mandarina, based on an analysis of SNPs at the third codon positions in exon B2 (C) and in the intron located next to the 3′ end of exon B2 (D). Bootstrap values were based on 1000 replicates.
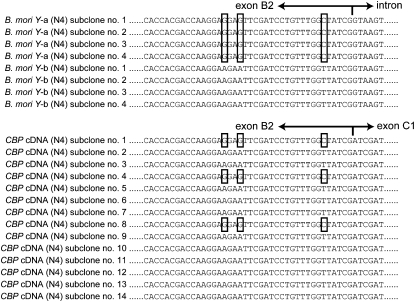
Both Y-a and Y-b of B. mori produced CBP mRNA:
Taking advantage of the SNP identified between Y-a and Y-b from B. mori strain N4, we assessed whether these sequences produced functional CBP mRNA. Fourteen independent clones of CBP cDNA from the middle silk gland of strain N4 were sequenced. The clones exhibited no nonsynonymous or frameshift mutations in the coding sequence. SNPs revealed that 3 clones could be classified as from Y-a, while 11 could be classified as from Y-b (Figure 7A). This result suggests that both Y-a and Y-b produced functional CBP mRNA. Multiple duplications of CBP would, therefore, explain the observed differences in CBP protein expression between B. mandarina and B. mori carrying the Y allele (Figure 1A).
Figure 7.—
Potential contributions of both Y-a and Y-b to mRNA transcription of CBP. A representative alignment of the CBP genomic DNA and cDNA sequences from strain N4. SNPs in Y-a are boxed.
DISCUSSION
We demonstrated the presence of CBP in several lepidopteran insects. Furthermore, we identified large copy number variations and retrotransposon-associated structural differences in CBP from B. mori, which were absent from B. mandarina. Silkworm domestication, which is a relatively recent evolutionary event, may have generated a large number of alterations and diversification in the structure of an evolutionarily conserved morphogenetic gene.
Genome-wide scanning studies (Scherer et al. 2007) have recently provided other examples of an increase in copy number variation in domesticated species, including yeast (Liti et al. 2009) and maize (Springer et al. 2009). Copy number variation may, therefore, be a common phenomenon in domesticated species. However, with the exception of a few studies (Parker et al. 2009; Blackman et al. 2010; Wang et al. 2010), the extent to which these variations are related to morphological phenotypes remains to be elucidated.
On the basis of the present data, we developed the following possible scenario that could explain the emergence of CBP variants due to silkworm domestication. At a relatively early stage of domestication, B. mori obtained two Y-a alleles from two different populations of B. mandarina. The CATS retrotransposon was then inserted into one of the alleles, evidenced by the target site duplication (Figure 3E), resulting in the emergence of Y-b. A deletion in Y-b would have occurred subsequently, resulting in the emergence of +Y-a. The +Y-b sequence could have been produced through deletions in Y-a, Y-b, or +Y-a. The copy numbers of CBP could have increased or decreased by means of unequal crossovers or deletions, resulting in the current state of diversity. Compared to Y-a, +Y-a, and +Y-b, Y-b tended to increase by unknown mechanisms. All copies of CBP are likely located on chromosome 2, where Y has been used as a stable marker in studies utilizing many yellow cocoon strains, without reported exception. Further elucidation of SNPs through an analysis of additional copies of CBP in more strains would elucidate more details and test the validity of this hypothetical process.
Recently, Xia et al. (2009) reported the comparative whole genome sequencing of 29 B. mori strains and 11 Chinese B. mandarina individuals by 1.50 billion short reads. The 29 B. mori strains included three of the strains used in our study: c108, N4, and p50. From an analysis of ∼16 million SNPs, Xia et al. (2009) concluded that B. mori was clearly genetically differentiated from B. mandarina. At the same time, on the basis of the high level of conservation of genetic variability, the authors estimated that a large number of B. mandarina individuals were used for domestication (i.e., the population bottleneck during silkworm domestication might not have been severe.) Therefore, gene flow limited to B. mori could have occurred for many genes during silkworm domestication. Our genomic-PCR results (Figure 3C and Figure S2B) suggest that the CBP genes containing CATS, Y-b, or +Y-a sequence, may be examples of such gene flow.
Were there human demands for cocoon color which drove the structural diversification of CBP during domestication? With the exception of thin silks, such as organza, most silk fibers used in modern textile production lack the majority of the sericin layer, which is the major site of pigmentation in colored cocoons and is largely removed during the refining process. Thus, most silk fibers from colored cocoons are nearly white after refinement. Therefore, cocoon color is not thought to be of crucial economic importance, at least in the modern age with refining technology. A hypothetical explanation for a probable driving force for genomic diversification in this area may be that cocoon colors were chosen by silk farmers who promoted them simply as a matter of local fashion or historical trend. Indeed, a Japanese editorial at the turn of the 20th century argued that Japanese sericulture should review the need to use yellow cocoon strains, despite their popularity (Houchi-shinbun in Japan; January 14, 1918). We speculate that a tendency of human nature to prefer unambiguous or pure colors may have caused the diversification of CBP structure from a duplicated allele to a null allele, resulting in the production of both deep yellow and clear white B. mori cocoons. On the other hand, it is noteworthy that cocoon color is also affected by genes other than CBP, such as Cameo2 (Sakudoh et al. 2010) or UGT10286 (Daimon et al. 2010). To properly evaluate the contribution of the changes of CBP structure to cocoon color, elucidation of the genetic diversity and domestication history of other cocoon color genes would be useful.
Our results illustrate the presence of multiple duplicated copies of a morphogenetic gene containing high nucleotide identity. Although genes can arise de novo primarily from ancestrally noncoding DNA (Levine et al. 2006; Begun et al. 2007; Chen et al. 2007), gene duplication represents a major driving force in the production of new genes and phenotypic novelties during biological evolution (Ohno 1970). Such a process is also occasionally tightly linked to human disease and medical therapeutic effects (Mouches et al. 1986; Johansson et al. 1993; Carvalho et al. 2010). However, the molecular evolutionary mechanisms underlying the emergence, maintenance, and evolution of gene duplication are just beginning to be understood (Innan and Kondrashov 2010). In particular, knowledge regarding the early stages in the evolution of duplicated genes is limited (Moore and Purugganan 2003). The CBP of the silkworm is an excellent resource for studying these early evolutionary stages, as multiple duplications of the gene are likely to have occurred relatively recently. Notably, although significant diversity has already been identified, the B. mori strains analyzed in this study represent only a small fraction of the >1000 strains maintained in sericultural and genetic stock centers. In future studies, we plan to further investigate the details of the domestication process using CBP with additional strains of B. mori to gain insight into how and when each copy of CBP was generated and how evolutionary constraints have affected the fates of its many duplicated copies.
Acknowledgments
We thank Zenta Kajiura, Hidetoshi Iwano, Yumiko Nakajima, and Toshiki Tamura for providing some of the insects and their genomic DNAs used in this study. The article has been improved thanks to comments of Marian Goldsmith and Michael Kanost. This work was supported by the Kieikai Research Foundation (Japan), the Futaba Electronics Memorial Foundation (Japan), the Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science and the National Bioresource Project (Silkworm) of the Ministry of Education, Culture, Sports, Science, and Technology (Japan).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.124982/DC1.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. AB570176–AB570194.
References
- Alpy, F., and C. Tomasetto, 2006. MLN64 and MENTHO, two mediators of endosomal cholesterol transport. Biochem. Soc. Trans. 34 343–345. [DOI] [PubMed] [Google Scholar]
- Andersson, D. I., and D. Hughes, 2009. Gene amplification and adaptive evolution in bacteria. Annu. Rev. Genet. 43 167–195. [DOI] [PubMed] [Google Scholar]
- Arunkumar, K. P., M. Metta and J. Nagaraju, 2006. Molecular phylogeny of silkmoths reveals the origin of domesticated silkmoth, Bombyx mori from Chinese Bombyx mandarina and paternal inheritance of Antheraea proylei mitochondrial DNA. Mol. Phylogenet. Evol. 40 419–427. [DOI] [PubMed] [Google Scholar]
- Banno, Y., T. Shimada, Z. Kajiura and H. Sezutsu, 2010. The silkworm-an attractive BioResource supplied by Japan. Exp. Anim. 59 139–146. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., H. A. Lindfors, A. D. Kern and C. D. Jones, 2007. Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics 176 1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhosale, P., B. Li, M. Sharifzadeh, W. Gellermann, J. M. Frederick et al., 2009. Purification and partial characterization of a lutein-binding protein from human retina. Biochemistry 48 4798–4807. [DOI] [PubMed] [Google Scholar]
- Blackman, B. K., J. L. Strasburg, A. R. Raduski, S. D. Michaels and L. H. Rieseberg, 2010. The role of recently derived FT paralogs in sunflower domestication. Curr. Biol. 20 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, C. M., F. Zhang and J. R. Lupski, 2010. Evolution in health and medicine Sackler colloquium: Genomic disorders: a window into human gene and genome evolution. Proc. Natl. Acad. Sci. USA 107(Suppl 1): 1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. T., H. C. Cheng, D. A. Barbash and H. P. Yang, 2007. Evolution of hydra, a recently evolved testis-expressed gene with nine alternative first exons in Drosophila melanogaster. PLoS Genet. 3 e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daimon, T., C. Hirayama, M. Kanai, Y. Ruike, Y. Meng et al., 2010. The silkworm Green b locus encodes a quercetin 5-O-glucosyltransferase that produces green cocoons with UV-shielding properties. Proc. Natl. Acad. Sci. USA 107 11471–11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond, J., 1997. Guns, Germs, and Streel: The Fates of Human Societies. Norton, New York.
- Duan, J., R. Li, D. Cheng, W. Fan, X. Zha et al., 2010. SilkDB v2.0: a platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res. 38 D453–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage, L. P., 1974. The Bombyx mori genome: analysis by DNA reassociation kinetics. Chromosoma 45 27–42. [DOI] [PubMed] [Google Scholar]
- Goldsmith, M. R., 2009. Recent progress in silkworm genetics and genomics, pp. 25–48 in Molecular Biology and Genetics of the Lepidoptera, edited by M. R. Goldsmith and F. Marec. CRC Press, Boca Raton, FL.
- Innan, H., and F. Kondrashov, 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11 97–108. [DOI] [PubMed] [Google Scholar]
- International Silkworm Genome Consortium, 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect. Biochem. Mol. Biol. 38 1036–1045. [DOI] [PubMed] [Google Scholar]
- Johansson, I., E. Lundqvist, L. Bertilsson, M. L. Dahl, F. Sjoqvist et al., 1993. Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquine. Proc. Natl. Acad. Sci. USA 90 11825–11829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C. G., A. Fujiyama and N. Saitou, 2003. Construction of a gorilla fosmid library and its PCR screening system. Genomics 82 571–574. [DOI] [PubMed] [Google Scholar]
- Krieger, J., O. Klink, C. Mohl, K. Raming and H. Breer, 2003. A candidate olfactory receptor subtype highly conserved across different insect orders. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 189 519–526. [DOI] [PubMed] [Google Scholar]
- Landrum, J. T., D. Callejas and F. Alvarez-Calderon, 2009. Specific accumulation of lutein within the epidermis of butterfly larvae, pp. 525–535 in Carotenoids: Physical, Chemical, and Biological Functions and Properties, edited by J. T. Landrum. CRC Press, Boca Raton, FL.
- Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan et al., 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. [DOI] [PubMed] [Google Scholar]
- Larsson, M. C., A. I. Domingos, W. D. Jones, M. E. Chiappe, H. Amrein et al., 2004. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43 703–714. [DOI] [PubMed] [Google Scholar]
- Levine, M. T., C. D. Jones, A. D. Kern, H. A. Lindfors and D. J. Begun, 2006. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc. Natl. Acad. Sci. USA 103 9935–9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D., Y. Guo, H. Shao, L. C. Tellier, J. Wang et al., 2010. Genetic diversity, molecular phylogeny and selection evidence of the silkworm mitochondria implicated by complete resequencing of 41 genomes. BMC Evol. Biol. 10 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti, G., D. M. Carter, A. M. Moses, J. Warringer, L. Parts et al., 2009. Population genomics of domestic and wild yeasts. Nature 458 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R. C., and M. D. Purugganan, 2003. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 100 15682–15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouches, C., N. Pasteur, J. B. Berge, O. Hyrien, M. Raymond et al., 1986. Amplification of an esterase gene is responsible for insecticide resistance in a California Culex mosquito. Science 233 778–780. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., T. Sakurai, T. Nishioka and K. Touhara, 2005. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307 1638–1642. [DOI] [PubMed] [Google Scholar]
- Niwa, R., T. Sakudoh, T. Namiki, K. Saida, Y. Fujimoto et al., 2005. The ecdysteroidogenic P450 Cyp302a1/disembodied from the silkworm, Bombyx mori, is transcriptionally regulated by prothoracicotropic hormone. Insect Mol. Biol. 14 563–571. [DOI] [PubMed] [Google Scholar]
- Ohno, S., 1970. Evolution by Gene Duplication. Springer-Verlag, New York.
- Parker, H. G., B. M. VonHoldt, P. Quignon, E. H. Margulies, S. Shao et al., 2009. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science 325 995–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan, M. D., and D. Q. Fuller, 2009. The nature of selection during plant domestication. Nature 457 843–848. [DOI] [PubMed] [Google Scholar]
- Regier, J. C., A. Zwick, M. P. Cummings, A. Y. Kawahara, S. Cho et al., 2009. Toward reconstructing the evolution of advanced moths and butterflies (Lepidoptera: Ditrysia): an initial molecular study. BMC Evol. Biol. 9 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakudoh, T., and K. Tsuchida, 2009. Transport of carotenoids by a carotenoid-binding protein in the silkworm, pp. 511–523 in Carotenoids: Physical, Chemical, and Biological Functions and Properties, edited by J. T. Landrum. CRC Press, Boca Raton, FL.
- Sakudoh, T., K. Tsuchida and H. Kataoka, 2005. BmStart1, a novel carotenoid-binding protein isoform from Bombyx mori, is orthologous to MLN64, a mammalian cholesterol transporter. Biochem. Biophys. Res. Commun. 336 1125–1135. [DOI] [PubMed] [Google Scholar]
- Sakudoh, T., H. Sezutsu, T. Nakashima, I. Kobayashi, H. Fujimoto et al., 2007. Carotenoid silk coloration is controlled by a carotenoid-binding protein, a product of the Yellow blood gene. Proc. Natl. Acad. Sci. USA 104 8941–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakudoh, T., T. Iizuka, J. Narukawa, H. Sezutsu, I. Kobayashi et al., 2010. A CD36-related transmembrane protein is coordinated with an intracellular lipid-binding protein in selective carotenoid transport for cocoon coloration. J. Biol. Chem. 285 7739–7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai, T., T. Nakagawa, H. Mitsuno, H. Mori, Y. Endo et al., 2004. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. USA 101 16653–16658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer, S. W., C. Lee, E. Birney, D. M. Altshuler, E. E. Eichler et al., 2007. Challenges and standards in integrating surveys of structural variation. Nat. Genet. 39 S7–S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura, M., H. Minami, Y. Suetsugu, H. Ohyanagi, C. Satoh et al., 2009. KAIKObase: an integrated silkworm genome database and data mining tool. BMC Genomics 10 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizuya, H., B. Birren, U. J. Kim, V. Mancino, T. Slepak et al., 1992. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. USA 89 8794–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, N. M., K. Ying, Y. Fu, T. Ji, C. T. Yeh et al., 2009. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5 e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabunoki, H., H. Sugiyama, Y. Tanaka, H. Fujii, Y. Banno et al., 2002. Isolation, characterization, and cDNA sequence of a carotenoid binding protein from the silk gland of Bombyx mori larvae. J. Biol. Chem. 277 32133–32140. [DOI] [PubMed] [Google Scholar]
- Tsuchida, K., Z. E. Jouni, J. Gardetto, Y. Kobayashi, H. Tabunoki et al., 2004. Characterization of the carotenoid-binding protein of the Y-gene dominant mutants of Bombyx mori. J. Insect Physiol. 50 363–372. [DOI] [PubMed] [Google Scholar]
- Wang, E., X. Xun, L. Zhang, H. Zhang, L. Lin et al., 2010. Duplication and independent selection of cell-wall invertase genes GIF1 and OsCIN1 during rice evolution and domestication. BMC Evol. Biol. 10 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Q., Y. Guo, Z. Zhang, D. Li, Z. Xuan et al., 2009. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science 326 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]