Prediction of physical protein-protein interactions represents a key challenge in computational systems biology. This study provides a proof-of-principle that high-throughput in silico protein docking results can be used to predict interaction partners.
Keywords: interactome, protein docking, protein–protein interaction
Abstract
Deciphering the whole network of protein interactions for a given proteome (‘interactome') is the goal of many experimental and computational efforts in Systems Biology. Separately the prediction of the structure of protein complexes by docking methods is a well-established scientific area. To date, docking programs have not been used to predict interaction partners. We provide a proof of principle for such an approach. Using a set of protein complexes representing known interactors in their unbound form, we show that a standard docking program can distinguish the true interactors from a background of 922 non-redundant potential interactors. We additionally show that true interactions can be distinguished from non-likely interacting proteins within the same structural family. Our approach may be put in the context of the proposed ‘funnel-energy model'; the docking algorithm may not find the native complex, but it distinguishes binding partners because of the higher probability of favourable models compared with a collection of non-binders. The potential exists to develop this proof of principle into new approaches for predicting interaction partners and reconstructing biological networks.
Introduction
The thousands of proteins expressed in cells perform many of their functions through interactions with other proteins. Protein–protein interactions are therefore essential to understanding the function of biological systems, and their characterisation has become an important task for both experimental and computational approaches in systems biology. Experimental methods include yeast two-hybrid systems (Uetz et al, 2000; Ito et al, 2001), mass spectrometry (Ewing et al, 2007) and protein chips, among others (reviewed in Piehler, 2005).
Computational methods are based on simple sequence and genomic features intuitively related to interactions and functional relationships (Skrabanek et al, 2008), including colocalisation or gene neighbourhood methods (Ramani et al, 2008), analysis of gene fusion (Enright et al, 1999), phylogenetic profiling (Pellegrini et al, 1999) and co-evolution (Burger and van Nimwegen, 2008; Juan et al, 2008; Pazos and Valencia, 2008). Such methods can be used on a large scale to predict whole interactomes (Juan et al, 2008) but their applicability has limitations. Protein structure information is often incorporated with interaction data (reviewed in Lee et al, 2009); however, this structural information has rarely been used for the large-scale prediction of protein interaction partners, which is surprising given that it is the structural features of two proteins that determine if they interact. Aloy and Russell (2002) developed a method that mapped homologues of interacting proteins onto the structure of complexes and assessed if the homologues were likely to fit together and therefore interact.
Protein docking lies at the other extreme of the prediction of interaction partners. It is generally applied to individual pairs of proteins that are known to interact, to model the three-dimensional structure of the complexes they form. Docking programs usually comprise two standard steps: generation of thousands of alternative poses to sample all possible interaction modes, followed by scoring these poses using a ‘pseudo-energy' function. Approximately correct solutions are generated by the first step, but scoring functions often fail to rank them properly (Lensink et al, 2007).
The potential to use protein docking algorithms to infer protein–protein interactions has often been discussed. Docking algorithms aim to compute the structure of protein complexes, so in principle, they should be able to discriminate between pairs of proteins that interact and those that do not. However, given their poor performance in ranking correct conformations, it has generally been regarded to be beyond the scope of current protein docking algorithms to detect interaction partners (Russell et al, 2004; Aloy and Russell, 2006; Gray, 2006). We have challenged this preconception, by assessing the potential of a general docking method to detect interaction partners. We report a high-throughput docking experiment, which shows that current docking algorithms can be used to distinguish between native interacting and non-interacting proteins.
To date, only one other high-throughput docking experiment has been reported (Mosca et al, 2009). Mosca et al (2009) used protein–protein docking, in a conventional manner to expand the structural coverage of the Saccharomyces cerevisiae interactome. Their interest was therefore in identifying accurate models of the complex structure of pairs of proteins that are known to interact. In this study, protein docking is used to address a different problem, namely, predicting which pairs of proteins interact sampling from a large set of alternative possibilities.
Fifty-six proteins in their unbound form from a well-established non-redundant benchmark set of protein complexes (Mintseris et al, 2005; the ‘benchmark set' in this study) were docked with a set of 922 monomeric non-redundant (at superfamily level) structures, which provide a background of docking score distributions for pairs of proteins that are unlikely to interact. The background set contains full-length proteins (further details in Supplementary Figures S1–S4 and Supplementary Tables SI, SII). By comparing the docking of a protein with its known interactor to those of this large background of potentially non-interacting proteins, we test whether the docking scores can be used to identify the known interactor from the background and therefore be used for the prediction of interaction partners (Supplementary Figure S16). In a further example, three of the interactors are tested against proteins of the same homologous family (SCOP superfamily; Murzin et al (1995)) to test the possibility of distinguishing true interactions from similar but not likely binding proteins.
This large-scale experiment generated over one billion complex models, requiring the use of supercomputing facilities (see Materials and methods). The energy distributions of the models generated by docking the individual complex components with this library of proteins were compared with the models generated from the docking of the benchmark complexes. We demonstrate that the distribution of scores obtained by the docking of the known interactors is distinguishable from the distributions of scores obtained from the models of the individual complex components docked with the structures in the background set.
Results
Benchmark and background docking score distributions
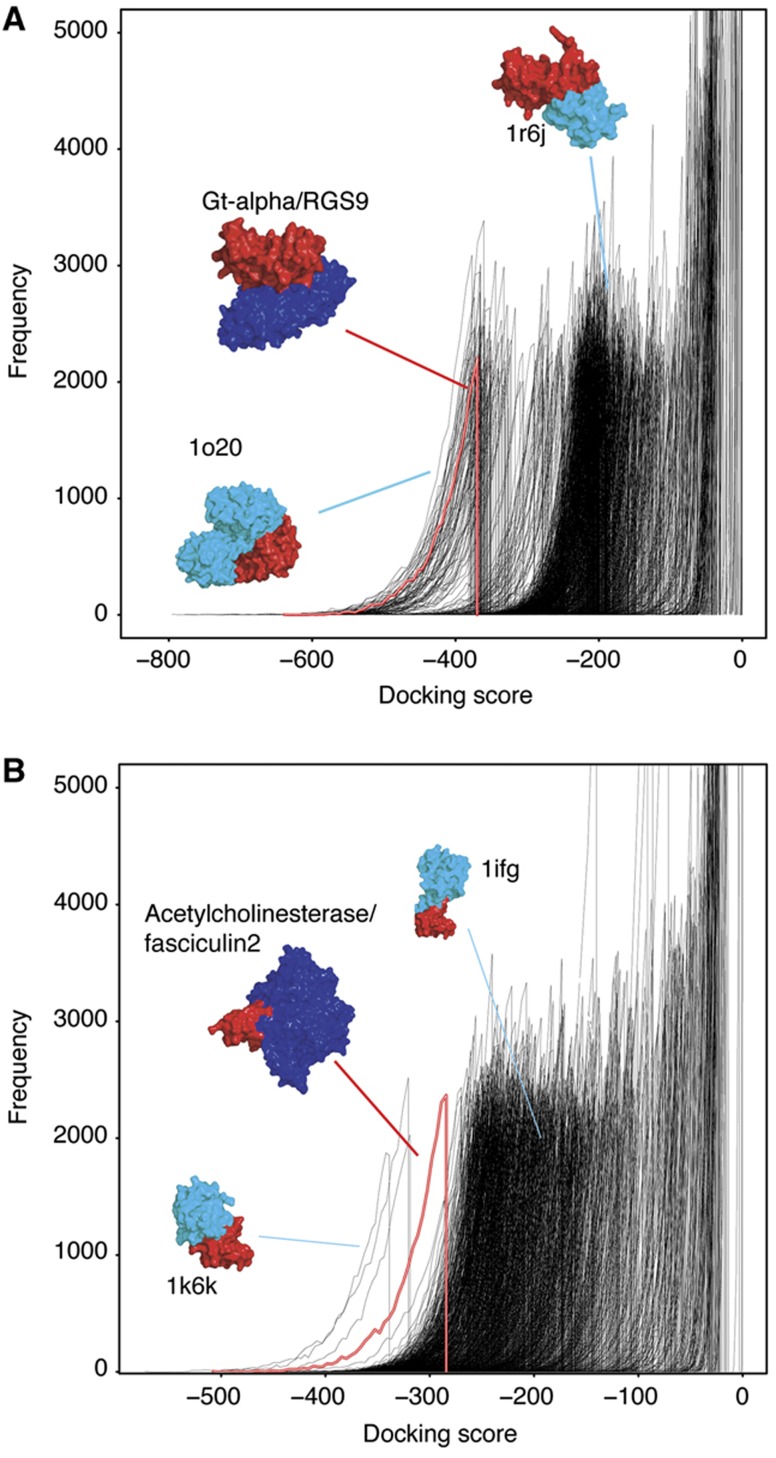
Our high-throughput approach is designed to assess if it is possible to predict which pairs of proteins interact using a rigid-body docking approach (Halperin et al, 2002), using only the geometric complementarity of the two molecular surfaces to score the docking poses. Docking was performed using the docking program HEX (Ritchie and Kemp, 2000). We compared the models generated for known interactors with those for a large background set of different structures that are unlikely to interact with either of the partners. For each of the 56 unbound proteins, the 20 000 best scoring poses with the known interactor and background structures (18.44 M models per benchmark set protein) were retained for comparison. The docking score distributions for the models of the benchmark complexes were plotted with those for the 922 proteins in the background set (Figure 1 and Supplementary Figure S5). In Figure 1A, the benchmark complex of Gt-α docked with RGS9 has a docking score distribution (red distribution) at lower (better) scores than the distributions for RGS9 docked with the background set (black distributions).
Figure 1.
Docking score distributions for benchmark complexes. The docking score distribution for a benchmark complex (red and white line) and one of the components docked with the background set (black lines) are shown for (A) Gt-α docked with RGS9 and (B) acetylcholinesterase complexed with fasciculin2. The native benchmark complexes are shown (red/blue structure) and selected models for the benchmark component docked with the background set (red/cyan).
The statistical significance of the docking score distributions for the 56 known interactors compared with the background set was assessed using the Wilcoxon rank-sum test (Wilcoxon, 1945). The distribution of the known interactors was compared with each of the individual background distributions and the percentage of background distributions of the known interactor is significantly less than (i.e., at better scores) was recorded. At the 1% significance level, 14 of the known interactors' score distributions are at significantly better scores than 99% of the background distributions, with the score distributions of 25 known interactors being significantly better than 90% of background pairs and 36 examples being significantly better than 80% (Table I). There are, however, some benchmark complexes whose score distributions cannot be distinguished from the background pairs, particularly eight examples where the known interactor's distribution is significantly better than less than 50% of the background distributions (e.g., Supplementary Figure S5, complex 2SNI).
Table 1. Statistical significance of benchmark complex docking score distributions.
| % Background better than | All (%) | Enzyme/ inhibitors (%) | Others (%) |
|---|---|---|---|
| The percentage of the background docking score distributions that the benchmark complex score distribution is significantly less than is shown. Cumulative counts and percentages are displayed. Statistical significance was tested using Wilcoxon rank-sum test at a 1% significance level. The results are also split for the two complex categories in the docking benchmark (enzyme/inhibitor and others). | |||
| 100 | 3 (5) | 1 (3) | 2 (7) |
| >99 | 14 (25) | 5 (17) | 10 (37) |
| >95 | 19 (34) | 6 (21) | 13 (48) |
| >90 | 25 (45) | 7 (24) | 18 (67) |
| >85 | 31 (55) | 10 (34) | 21 (78) |
| >80 | 36 (64) | 14 (48) | 22 (81) |
| >70 | 40 (71) | 17 (59) | 23 (85) |
| >60 | 46 (82) | 22 (76) | 24 (89) |
| >50 | 48 (85) | 23 (79) | 25 (93) |
| <50 | 56 (100) | 29 (100) | 27 (100) |
We also applied receiver operating characteristic (ROC) analysis to assess the global ability of the method in separating the 56 pairs representing real interactions (positives) from the 51 632 negative pairs (see Materials and methods). The AUC parameter of the ROC analysis is 0.80 (in a 0.0–1.0 scale), indicating that the discriminative power of the method is clearly better than the random value of 0.5 (Supplementary Figure S12).
Docking algorithms tend to perform better for enzyme/inhibitor complexes, as this kind of interaction is highly based on shape complementarity (Vajda, 2005). To assess the effect of this on our results, we evaluated the performance separately for enzyme inhibitor and other types of complexes. Overall, the general tendency to discriminate the right interactions can also be observed in the 29 available enzyme/inhibitor complexes, with a slight tendency to be worse (15 of poor discrimination out of 29, Table I).
If it is assumed that proteins from different species are less likely to interact, then as the background set contains structures from many different species, many of the dockings performed are between proteins that will not interact in natural conditions. Species-specific background sets (i.e., only docking proteins from the same species) were generated and the experiment repeated. This experiment is limited to 32 of the 56 interactors, for which there were sufficient structures from the same species. Differences in performance are mainly small compared with the results with the full background set (See Supplementary Table SIV). This demonstrates that it is valid to use a multispecies background set.
We compared the performance (from here on performance refers to how distinguishable the benchmark distribution is from the background distributions) of the benchmark dockings to the amount of conformational change between the unbound and bound forms of the protein, which is often used to indicate how difficult the proteins are to dock. Interestingly, there is no correlation between the conformational change and the performance of the benchmark docking (Supplementary Figure S8). Additionally, we found that good shape complementarity is identified for all of the benchmark complexes, as their score distributions are generally in the range of −800 to −300 (Supplementary Figure S6). So even for complexes where the rigid body docking is not able to accurately predict the correct interface of the complex, areas of shape complementarity are still identified. This contrasts with the background dockings, which show a much greater range of score distributions, and many have high scores (close to zero) where no shape complementarity is identified. This is also demonstrated in the ROC curve (Supplementary Figure S12).
We also observed no correlation between performance of complexes in our experiment and the size of the proteins in the benchmark complex (Supplementary Figure S7) or their affinity (described by their dissociation equilibrium constants; Kastritis and Bonvin (2010); Supplementary Figure S9). The results are also invariant to the number of docking models that are retained when 10 000, 5000 or only 1000 models are considered (Supplementary Figure S10). Comparable results were obtained for four of the complexes when the number of poses retained was increased to 100 000 (Supplementary Figure S10).
The background set is representative at the SCOP superfamily level. To investigate if the results are sensitive to the size of the background set and to identify if smaller background sets could be used in future research, we simulated the results obtained when the background set is reduced to 50, 100, 300 or 500 structures. The results suggest that a minimum size of ∼500 is appropriate for the background set (Supplementary Figure S11).
Description of the distributions
The docking score distributions are divided into three populations. The first contains protein pairs in which the docking program failed to find shape complementarity between them. These distributions are narrow and have negligible docking scores. Distributions in the second population have intermediate scores and are broader than the first population. Finally, the third population exhibits the best scores with the broadest distributions. In the latter, the docking program has identified significant shape complementarity between the structures, resulting in a large number of poses with good scores. Such distributions may be indicative of potential interactors.
Overall, the 922 background score distributions are observed in two patterns, which may be useful when identifying potential interactors. In the first pattern, many distributions have a negligible score and most of the remaining distributions are in the medium scoring population. The results for acetylchoinesterase docked with fasciculin2 and Gt-α docked with RGS9 follow this pattern (Figure 1). The second pattern has few distributions in the negligible scoring population, with the majority present in the medium scoring population (Supplementary Figure S5, complex 2MTA). In two-thirds of the complexes where the real interactors are clearly distinguishable from the background dockings, the score distributions exhibit the first pattern. The opposite is true for those interactors whose models were indistinguishable from the background set, with the majority of these distributions showing populations belonging to the second pattern. Most proteases (particularly Subtilisin) in the test set exhibit the second pattern of distributions. This may be due to the broad substrate specificity of such enzymes (Supplementary information).
Investigation of the interactors' docking models
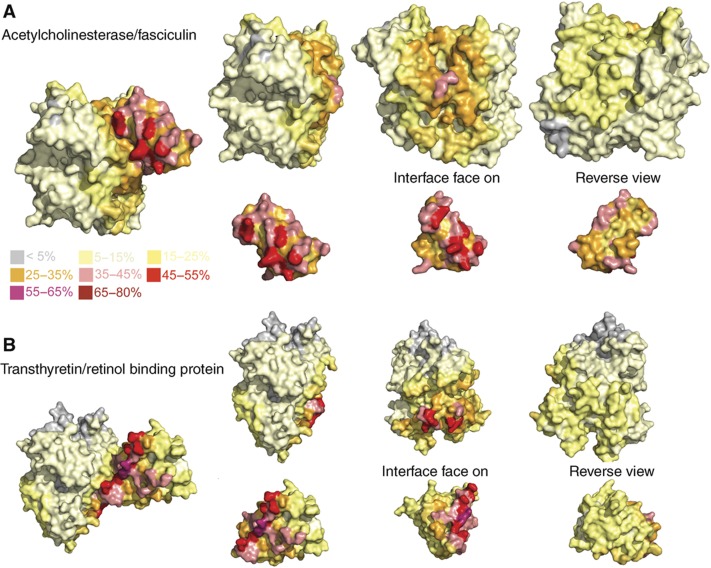
The docking models generated for each of the benchmark complexes were further investigated by considering how frequently each residue is part of the predicted interface. Heat map style figures were visually assessed (see Materials and methods) for their agreement with the native binding site (Figure 2 and Supplementary Figure S13). For all of the complexes, both components had either one or two areas on the surface that were present in a greater proportion of the predicted interfaces. These patches suggest that HEX identifies one or two areas of shape complementarity, which result in good scoring models. For example, for acetlycholinesterase (Figure 2) one patch contains many residues present in up to 45% of models and the other patch has only a few residues present in up to 35% of models. For 64% of complex components, a patch on the heat map overlaps with the native binding site (Supplementary Table SIII). For example, heat maps demonstrating the range of results obtained are shown in Figure 2 and Supplementary Figure S13.
Figure 2.
Heat map of the docking model putative binding sites. The Heat map shows how often each residue is present in the binding site modelled by HEX for (A) Acetylcholinesterase/fasciculin2 and (B) transthyretin/retinol binding protein. The unbound structures are shown and they have been aligned with their equivalent component in the native complex. The colour scheme as shown in the key indicates the percentage of HEX poses that a residue formed part of the putative interface.
Distinguishing interactors within the same superfamily
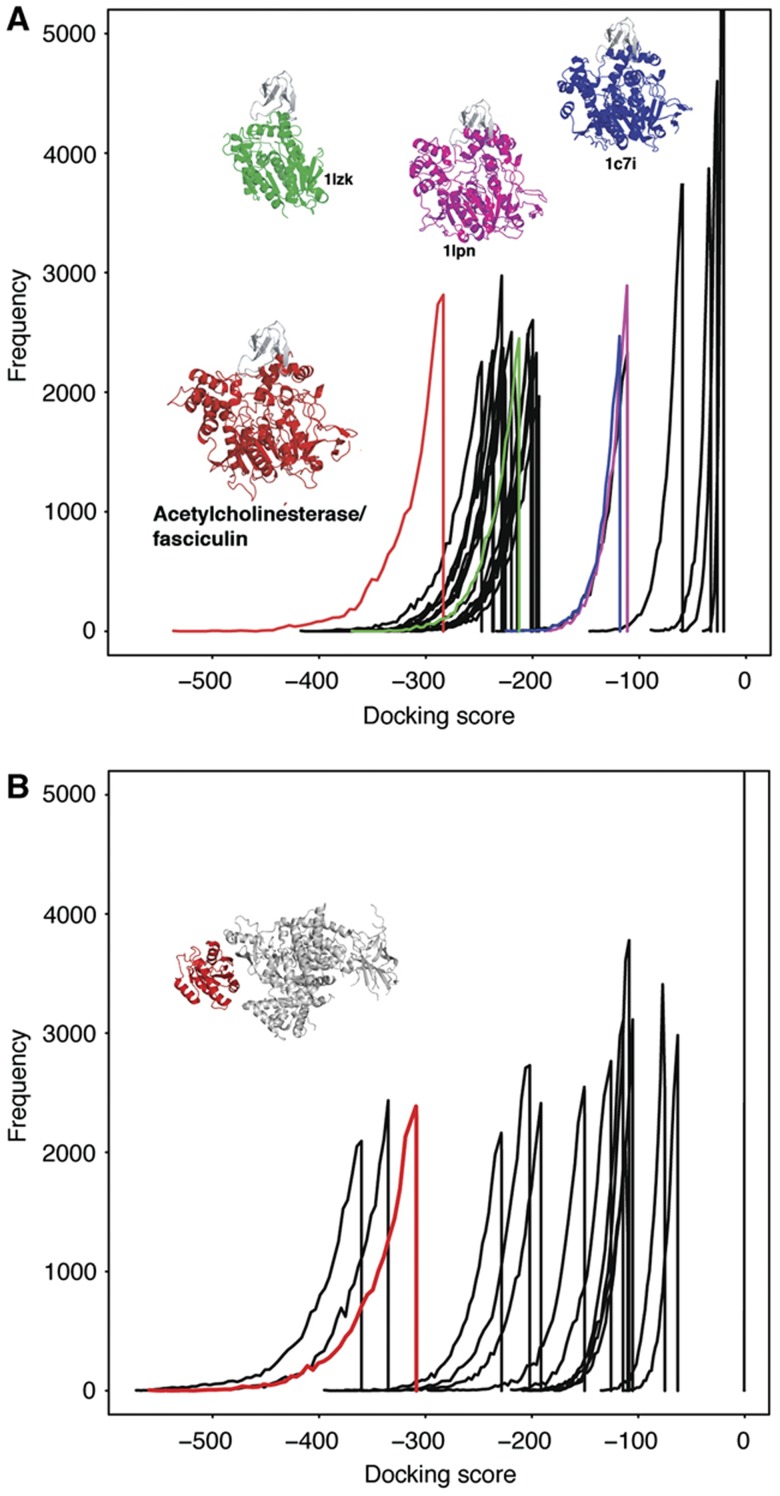
Our initial experiment compared the docking results of known interactors with those for a representative set of diverse structures from different protein folds. To indicate if our approach can be used to distinguish interactors within a more homogeneous set of proteins belonging to the same superfamily, an experiment was performed on three of the benchmark complexes; acetylcholinesterase and fasciculin2, Ras GTPase/PIP3 kinase and Rac GTPase/p67 Phox. For acetylcholinesterase and fasciculin2, 23 representative structures from different protein families belonging to the α/β-hydrolases superfamily (the acetylcholinesterase superfamily) were docked with fasciculin2 (see Materials and methods). The score distribution of the benchmark complex is distinct from the 23 representatives from the same superfamily (Figure 3). Further, we did not find any relationship between overall structural similarity with the real interactor and position of the score distribution, meaning that similar structures had score distributions indistinguishable from the background distributions (Supplementary Figure S14).
Figure 3.
Docking for a single superfamily. (A) Docking score distributions for the acetylcholinesterase and fasciculin2 complex (red) and for fasciculin2 docked to members of the α/β-hydrolases superfamily. The docking scores for the three most structurally similar proteins to acetylcholinesterase are shown in different colours with their structures. The score distributions for the remaining structures are shown in black. (B) Docking score distributions for Ras GTPase docked with PIP3 kinase (red line), and PIP3 kinase docked with other structures from the same SCOP superfamily as Ras GTPase. The native form of the GTPase/PIP3 kinase complex is shown.
Ras GTPase and Rac GTPase both belong to the P-loop containing nucleoside triphosphate hydrolases SCOP superfamily. Fourteen structures belonging to different families within this superfamily were docked with both PIP3 kinase and p67 Phox. For both complexes, the known interactors have docking score distributions that are distinct from the majority of the representatives from the same superfamily (Figure 3B and Supplementary Figure S15). The results for all three complexes suggest that our approach may be able to distinguish between the real interactors and decoys that have similar structures to the interactor (See Supplementary information for further analysis).
Discussion
This high-throughput docking experiment proves the principle that a current docking algorithm can be used to distinguish between interacting and non-interacting proteins. We have shown that the score distribution of docking conformations can be used to distinguish known interactors among a diverse background of non-interacting proteins. We have also illustrated the feasibility of this strategy to identify known interactors from a set of more similar proteins belonging to the same superfamily.
The results presented here suggest that while a simple docking algorithm may not accurately find the native complex, it can distinguish between binding and non-binding partners, based on their score distributions. This may indicate that although protein surface morphology is not enough to find the native interface (otherwise protein docking programs will always find the right conformation of protein complexes); it at least contains sufficient information to identify a ‘bona fide' interactor when a general binding profile is compared with those of many non-interacting proteins. This observation may be put in the context of the proposed concept of the funnel-like intermolecular energy landscape in protein–protein interactions (McCammon, 1998; Tsai et al, 1999). This theory states that the assembly of two proteins is initiated by the formation of nonspecific encounter complexes (Blundell and Fernandez-Recio, 2006), followed by rearrangements of them driven by stronger and more specific interactions. Such hypothesis implies that not only the ‘final' binding surface contains information for interacting with the partner but also other parts of the surface. Hence, despite the limitations of the docking algorithm, it nevertheless is able to distinguish binding partners, because they will be present in a larger number of favourable poses compared with a collection of non-binders. Indeed, binding surfaces seem to be similar to general surfaces (Levy, 2010).
In the search for an interpretation of physical principles behind the results, we did not find any obvious feature common to those complexes with distinct profiles. It is tempting to think that the results correspond instead to a complex mixture of the characteristics of the proteins (i.e., protein size, protein shape, size of the binding interaction and number, organisation and structure of potential binding sites) together with characteristics of their binding partners and the characteristics necessary to avoid nonspecific binding with other proteins.
The protocol used here can be extended to a predictive method with the development of thresholds and rules for the statistical comparison of the docking score distributions between pairs of proteins. This approach could further be used for the analysis of large data sets by docking sets of protein structures to discover their potential binding partners in the growing catalogue of proteins of known structure. Furthermore, it is possible to think of an extension of this methodology to provide predictions for all known structures. Recent advances in the docking protocol used have shown 45-fold speed improvements, which makes such experiments feasible (Ritchie and Venkatraman, 2010). As this docking-based methodology for predicting interaction partners is complementary to all other existing methods, based on sequence or genome information, the increase in the number of known protein structures will make it possible to combine them in a systematic way. This would extend our knowledge on the interactomes of organisms and consequently improve our capacity to perform systematic studies on them.
Materials and methods
Generating the background set of structures
The background set of structures was designed to be non-redundant at the SCOP (Murzin et al, 1995) superfamily level and includes an example of each superfamily for which a monomeric (based on PDB biological unit data) structure was available. Where several X-ray structures were available, the highest resolution structure was chosen. Where only NMR models were available, the PDB file was selected arbitrarily and the first model from the ensemble retained. Using SCOP version 1.75, this resulted in 922 structures in the background set, which contain 988 different SCOP superfamilies. The full list of pdb codes and analysis of the characteristics of these structures is provided in Supplementary information (Supplementary Table SV). Although the SCOP domain classification was used to select a set of structurally non-redundant proteins, the full-length proteins containing these domains (and not the isolated domains) were used to perform the docking experiments.
Selecting the benchmark complexes
The docking Benchmark 2.0 (Mintseris et al, 2005) is a widely accepted data set of complexes used for benchmarking docking-based studies. These complexes were selected based on a number of criteria. Proteins were only included if they were characterised in an unbound form, such that no other chain was found in the PDB file. This resulted in a test set of 56 proteins representing 42 different benchmark complexes (see Supplementary information for further details). The unbound structures were used and their orientations randomised prior to docking.
Docking the benchmark complexes
The Benchmark complexes were docked using HEX (Ritchie and Kemp, 2000; version 4.5), with the unbound forms of the complex used as input structures, after randomisation of their orientations. HEX was run using a shape complementarity scoring function (i.e., electrostatics turned off). A full table of parameters used for HEX is shown in Supplementary Table SVI. The top 20 000 scoring models were retained for analysis. In additional analyses, the top 1 000 000, 10 000, 5000 and 1000 models were considered (Supplementary information).
Generating the background set models
The unbound components from the docking benchmark set were docked with each of the structures from the background set using HEX as described above. Overall, this experiment generated over 1 billion complex models. Docking runs were performed using the facilities of MareNostrum at the Barcelona Supercomputing Centre. Distributions of the docking scores are plotted as explained in Supplementary Figure S17.
ROC analysis
The ROC analysis (Fawcett, 2006) is used to evaluate the performance of a binary classifier separating two populations (positive and negative cases). This analysis can be applied to classifiers (prediction methods), which associate a numerical score to each case. Ideally, the classifier would tend to associate high scores to the positive cases and low scores to the negative ones (or the other way around). All the possible pairs between chains within the benchmark set and the background set were considered together for the ROC analysis. The score associated with each pair was the average value of the corresponding distribution of docking scores. These scores were converted to ranks in order to compare between different cases within the benchmark set. Intuitively, these ranks represent the positions of the energy distributions, such as the ones represented in Figure 1. Good ranks (distributions comparatively shifted to better scores) are expected to be associated with real interactors (as discussed qualitatively in results). The final list contains 56 positives and 51 632 negatives.
Generating the interactor heat maps
The putative binding sites from the docked models of the benchmark complexes were assessed by calculating the percentage of models that each individual residue formed as a part of the binding site. Residues with non-hydrogen atoms within 5 Å of non-hydrogen atoms on residues in the other interactor were classed as part of the putative binding site. The results were mapped onto the structures, generating heat maps indicating the locations where HEX had preferentially docked the interactors. The heat maps were visually inspected to assess their overlap with the native binding site.
Areas (patches) on the proteins present in a large percentage of the docking models were identified. Where there was more than one patch, the patch present in the greatest number of models was ranked as the first patch and the other as the second patch. The overlap of these patches with the interaction site was visually assessed. Where most of the interaction site was a part of a patch, they were classed as being in agreement. In some examples, patches and the interaction sites were classed as having a partial overlap, such that a significant part of the binding site and patch overlapped; it was generally required that the overlap occur on the area of highest intensity on the patch. Finally, the patch and interaction site were classed as non-overlapping, if there was a very small overlap or none at all between them.
Statistical testing
The statistical difference between the score distributions for the benchmark complex and the background set were assessed using the Wilcoxon rank-sum test (Wilcoxon, 1945). The Wilcoxon rank-sum tests compared the ranked set of 20 000 docking scores from the benchmark complex and the individual background set dockings, and determines if the two sets of scores are significantly different. For each benchmark complex, a one-sided test was performed to assess if the benchmark complex score distribution was significantly less than each of the background score distributions. An individual test was performed against each member of the background set. The number of background distributions that the benchmark distribution is significantly less than (at P<0.01) was recorded and expressed as a percentage of the total number of structures in the background set (922).
Superfamily examples
For the acetylcholinesterase/fasciculin2 complex, structures belonging to the same superfamily (c.69.1-α/β-hydrolases) as acetlycholinesterase (pdb: 1J06) were identified. Representative structures were chosen from the 32 different families, such that it was required that the chain was isolated in the pdb file (i.e., monomeric). This resulted in a final set of 23 structures: 1j1i, 1s8o, 1a8s, 1pja, 1lpn, 1cvl, 1bu8, 1c7i, 1scq, 1lns, 1imj, 1m33, 1q0z, 1tqh, 1lzk, 2axe, 1uxo, 1dqy, 1vz2, 1ac5, 1mu0, 1d07 and 1ggv. Each of the 23 structures was docked with the unbound form of fasciculin2 using the same HEX settings as for the benchmark complexes. Dalilite (Holm and Park, 2000) was used to align each of the structures to the acetylcholinesterase to identify how similar the structures are.
For Ras GTPase and Rac GTPase, 14 monomeric proteins belonging to different families within the P-loop containing nucleoside triphosphate hydrolases SCOP superfamily were identified and docked with the unbound forms of Phox67 and PIP3 kinase. The 14 pdb structures are as follows: 1ls1, 1ls6, 1ye8, 1rif, 1rz3, 1yks, 1lv7, 1kgd, 1r2q, 1htw, 1c4o, 2iyv, 1f9v and 1oxx.
Supplementary Material
Main content of the supplementary information excluding Table SV which is uploaded separately
Acknowledgments
MNW was supported by a British Council Research Exchange award. GF was awarded a Marie Curie Action European Reintegration Grant (MERG-CT-2007-200179). This work was funded by the BioSapiens Network of Excellence (grant number LSHG-CT-2003-503265) and in part by the BIO2006-15318 project from the Spanish Ministry for Education and Science. Computer support was provided by the Barcelona Supercomputer Centre.
Author contributions: MNW designed and performed experiments, analysed data and wrote the paper. CP designed and performed experiments and wrote the paper. GF, FP and AV designed experiments and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aloy P, Russell RB (2002) Interrogating protein interaction networks through structural biology. Proc Natl Acad Sci USA 99: 5896–5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloy P, Russell RB (2006) Structural systems biology: modelling protein interactions. Nat Rev Mol Cell Biol 7: 188–197 [DOI] [PubMed] [Google Scholar]
- Blundell TL, Fernandez-Recio J (2006) Cell biology: brief encounters bolster contacts. Nature 444: 279–280 [DOI] [PubMed] [Google Scholar]
- Burger L, van Nimwegen E (2008) Accurate prediction of protein-protein interactions from sequence alignments using a Bayesian method. Mol Syst Biol 4: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA (1999) Protein interaction maps for complete genomes based on gene fusion events. Nature 402: 86–90 [DOI] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y et al. (2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 3: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett T (2006) An introduction to ROC analysis. Pattern Recogn Lett 27: 861–874 [Google Scholar]
- Gray JJ (2006) High-resolution protein-protein docking. Curr Opin Struct Biol 16: 183–193 [DOI] [PubMed] [Google Scholar]
- Halperin I, Ma B, Wolfson H, Nussinov R (2002) Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins 47: 409–443 [DOI] [PubMed] [Google Scholar]
- Holm L, Park J (2000) DaliLite workbench for protein structure comparison. Bioinformatics 16: 566–567 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan D, Pazos F, Valencia A (2008) High-confidence prediction of global interactomes based on genome-wide coevolutionary networks. Proc Natl Acad Sci USA 105: 934–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastritis PL, Bonvin AMJJ (2010) Are scoring functions in protein-protein docking ready to predict interactomes? Clues from a novel binding affinity benchmark. J Proteome Res 9: 2216–2225 [DOI] [PubMed] [Google Scholar]
- Lee S, Brown A, Pitt WR, Higueruelo AP, Gong S, Bickerton GR, Schreyer A, Tanramluk D, Baylay A, Blundell TL (2009) Structural interactomics: informatics approaches to aid the interpretation of genetic variation and the development of novel therapeutics. Mol BioSyst 5: 1456–1472 [DOI] [PubMed] [Google Scholar]
- Lensink MF, Mendez R, Wodak SJ (2007) Docking and scoring protein complexes: CAPRI 3rd edition. Proteins 69: 704–718 [DOI] [PubMed] [Google Scholar]
- Levy ED (2010) A simple definition of structural regions in proteins and its use in analyzing interface evolution. J Mol Biol 403: 660–670 [DOI] [PubMed] [Google Scholar]
- McCammon JA (1998) Theory of biomolecular recognition. Curr Opin Struct Biol 8: 245–249 [DOI] [PubMed] [Google Scholar]
- Mintseris J, Wiehe K, Pierce B, Anderson R, Chen R, Janin J, Weng Z (2005) Protein-protein docking benchmark 2.0: an update. Proteins 60: 214–216 [DOI] [PubMed] [Google Scholar]
- Mosca R, Pons C, Fernandez-Recio J, Aloy P (2009) Pushing structural information into the yeast interactome by high-throughput protein docking experiments. PLoS Comput Biol 5: e1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247: 536–540 [DOI] [PubMed] [Google Scholar]
- Pazos F, Valencia A (2008) Protein co-evolution, co-adaptation and interactions. Embo J 27: 2648–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO (1999) Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci USA 96: 4285–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehler J (2005) New methodologies for measuring protein interactions in vivo and in vitro. Curr Opin Struct Biol 15: 4–14 [DOI] [PubMed] [Google Scholar]
- Ramani AK, Li Z, Hart GT, Carlson MW, Boutz DR, Marcotte EM (2008) A map of human protein interactions derived from co-expression of human mRNAs and their orthologs. Mol Syst Biol 4: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie DW, Kemp GJ (2000) Protein docking using spherical polar Fourier correlations. Proteins 39: 178–194 [PubMed] [Google Scholar]
- Ritchie DW, Venkatraman V (2010) Ultra-fast FFT protein docking on graphics processors. Bioinformatics 26: 2398–2405 [DOI] [PubMed] [Google Scholar]
- Russell RB, Alber F, Aloy P, Davis FP, Korkin D, Pichaud M, Topf M, Sali A (2004) A structural perspective on protein-protein interactions. Curr Opin Struct Biol 14: 313–324 [DOI] [PubMed] [Google Scholar]
- Skrabanek L, Saini HK, Bader GD, Enright AJ (2008) Computational prediction of protein-protein interactions. Mol Biotechnol 38: 1–17 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Kumar S, Ma B, Nussinov R (1999) Folding funnels, binding funnels, and protein function. Protein Sci 8: 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Po, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- Vajda S (2005) Classification of protein complexes based on docking difficulty. Proteins 60: 176–180 [DOI] [PubMed] [Google Scholar]
- Wilcoxon F (1945) Individual comparisons by ranking methods. Biometrics Bull 1: 80–83 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Main content of the supplementary information excluding Table SV which is uploaded separately