Abstract
The characterization of a new class of pyrrole–imidazole hairpin polyamides with β-amino-γ-turn units for recognition of the DNA minor groove is reported. A library of eight hairpins containing (R)-and (S)-3,4-diaminobutyric acid (β-amino-γ-turn) has been synthesized, and the impact of the molecules on DNA-duplex stabilization was studied for comparison with the parent γ-aminobutyric acid (γ-turn) and standard (R)-2,4-diaminobutyric acid (α-amino-γ-turn)-linked eight-ring polyamides. For some, but not all, sequence compositions, melting temperature analyses have revealed that both enantiomeric forms of the β-amino-γ-turn increase the DNA-binding affinity of polyamides relative to the (R)-α-amino-γ-turn. The (R)- β-amine residue may be an attractive alternative for constructing hairpin polyamide conjugates. Biological assays have shown that (R)- β-amino-γ-turn hairpins are able to inhibit androgen receptor-mediated gene expression in cell culture similar to hairpins bearing the standard (R)-α-amino-γ-turn, from which we infer they are cell-permeable.
Introduction
The ability to modulate the expression of eukaryotic gene networks by small molecules is a challenge in the field of chemical biology. Hairpin pyrrole–imidazole polyamides are a class of programmable small molecules that bind to the minor groove of DNA with affinities similar to transcription factors and have been shown to inhibit gene expression in living cells by interfering with transcription factor/DNA interfaces.1 The DNA sequence specificity of polyamides arise from interactions of pairs of the aromatic amino acids N-methylpyrrole (Py), N-methylimidazole (Im), and N-methylhydroxypyrrole (Hp) with the edges of the Watson–Crick base pairs.2 The generality of the polyamide pairing rules has been demonstrated by numerous studies3 and applications of polyamide conjugates include DNA alkylations,4 DNA-templated ligations,5 sequence-specific DNA intercalators,6 fluorescent DNA paints,7 DNA nanoarchitectures,8 and transcription factor mimics.9 Efforts have been made to improve the DNA-binding properties of hairpin polyamides with modified turn units.10 Substitution of γ-aminobutyric acid (γ-turn) by (R)-2,4-diaminobutyric acid (α-amino-γ-turn) increases the DNA-binding affinity by ~15-fold.10b,11 In contrast, hairpins containing the opposite enantiomer, (S)-α-amino-γ-turn, result in diminished binding affinities. This decrease is most likely caused by an unfavorable steric clash of the amine residue with the DNA minor groove.10b Sugiyama and co-workers have introduced polyamides containing the α-hydroxy-γ-turn.10c These hairpins provide discrimination for A • T/T • A base pairs at the turn position, although a ~70-fold reduced DNA-binding affinity relative to analogue (R)- α-amino-γ-turn-linked polyamides has been observed.
Here we introduce a new class of hairpin polyamides which are linked by 3,4-diaminobutyric acid which results in a β -amine residue at the turn unit (β-amino-γ-turn) (Figure 1). DNA-binding affinities of four different eight-ring polyamide core sequences (with incrementally increasing Im/Py pairs) have been investigated and were compared to analogue hairpins bearing the parent γ-turn and the standard (R)-α-amino-γ-turn. We show that, for certain series of hairpin polyamides, both enantiomers of the β-amino-γ-turn are able to increase the relative DNA-binding affinity. However, this is sequence context dependent. Biological assays revealed that hairpin polyamides bearing the (R)-β-amino-γ-turn are able to inhibit specific gene expression in cell culture, which is taken as evidence for cell permeability.
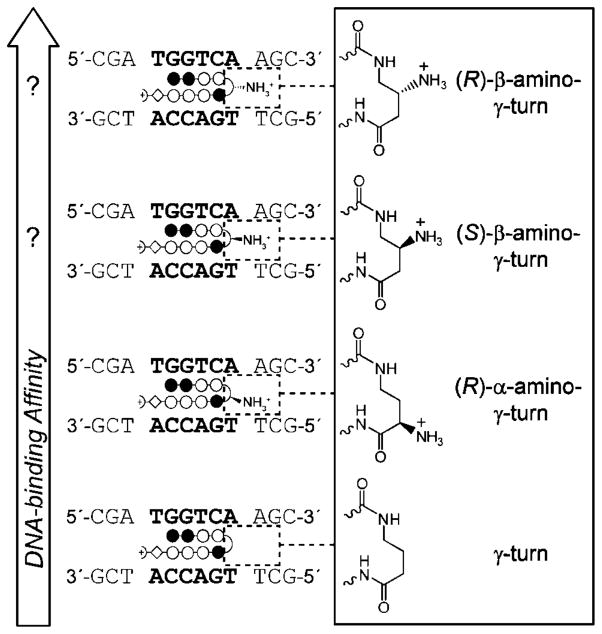
Figure 1.
Schematic representation of hairpin polyamides with increased DNA-binding affinity caused by different γ-turn units. Hairpin polyamides targeted to DNA sequence 5′-TGGTCA-3′ are shown as ball-and-stick models. Ball-and-stick representation legend: black and white circles represent N-methylimidazole and N-methylpyrrole units, respectively, half-circles represent γ-aminobutyric acid, white diamonds represent β-alanine units, and half-circles containing a cross represent 3-(dimethylamino)-1-propylamine (Dp) as tail.
Results and Discussion
Thermal Stabilization of DNA Duplexes by Hairpin Polyamides
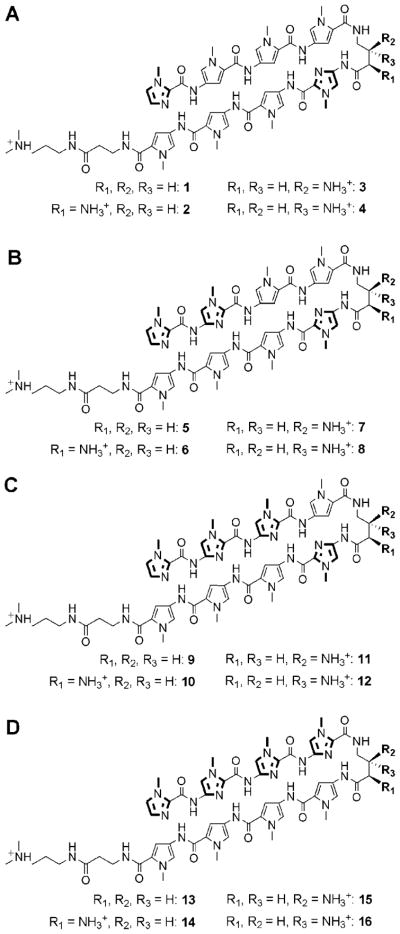
Hairpin polyamides 1–16 were synthesized with different Im/Py and Py/Py compositions targeted to the four DNA sequences with increasing G/C content 5′-TGTTCA-3′, 5′-TGGTCA-3′, 5′-TGGGCA-3′, and 5′-TGGGGA-3′ (Figure 2). The energetics of DNA-binding properties of polyamides are typically characterized by quantitative DNase I footprint titrations.12 These measurements provide precise information regarding the affinity and specificity of DNA/polyamide complexes. Unfortunately, quantitative footprinting experiments revealed similar equilibrium association constants (Ka values ~ 2 × 1010 M−1) for hairpins 1–8, reaching an upper limit of the standard procedure12,13 (see Supporting Information).
Figure 2.
Chemical structures for hairpins 1–16 targeted to DNA sequences: (A) 5′-TGTTCA-3′, (B) 5′-TGGTCA-3′, (C) 5′-TGGGCA-3′, and (D) 5′-TGGGGA-3′.
Prior results have shown that the increase in melting temperature (ΔTm) of DNA duplexes bound by hairpin polyamides correlates with DNA-binding affinity and can be utilized to detect single base pair mismatched DNA/polyamide complexes.14 Accordingly, we have used melting temperature analysis for dissecting differences in DNA affinities of hairpin polyamides. Spectroscopic analyses were performed on 12mer DNA duplexes containing the appropriate match sequence in the absence and presence of polyamides in order to derive the desired ΔTm values (Figure 3). Table 1 shows that all hairpins provided an increase in melting temperature, relative to the individual DNA duplexes, confirming the formation of DNA/polyamide complexes. As expected, spectroscopic analysis with (R)-α-amino-γ-turn hairpins revealed stronger stabilizations than the parent γ-turn analogues; for example, achiral polyamide 1 targeted to DNA sequence 5′-TGTTCA-3′ resulted in a ΔTm value of 15.9 °C, while chiral hairpin (R)-α-2 led to a 3.6 °C higher melting temperature (ΔTm = 19.5 °C). Remarkably, melting temperature analyses in the presence of β-amino-γ-turn hairpins (S)-β-3 (ΔTm = 20.9 °C) and (R)-β-4 (ΔTm = 22.2 °C) revealed higher ΔTm values compared to those for the α-series hairpin (R)-α-2 (ΔTm = 19.5 °C). The same trend was observed for hairpins 5–8 targeted to DNA sequence 5′-TGGTCA-3′ (Table 1, Figure 2). First, it is noteworthy that both the (R)- and (S)-β-amino-γ-turn generated higher melting temperatures than the standard (R)-α-amino-γ-turn. Second, the enhancement (relative to achiral hairpins) observed for the (R)-β-series is almost twice that of the (R)-α-series targeted to DNA sequences 5′-TGTTCA-3′ and 5′-TGGTCA-3′; for example, polyamide (R)-β-8 provided a ΔΔTm value of 6.9 °C, while the α-series (R)-α-6 led to a ΔΔTm value of 3.5 °C relative to achiral hairpin 5. Interestingly, by further increasing the amounts of Im/Py pairs in the polyamides, significantly less DNA duplex stabilizations have been observed. For example, achiral polyamide 9 and chiral hairpin (R)-α-10 targeted to DNA sequence 5′-TGGGCA-3′ yielded ΔTm values of 8.6 and 13.2 °C, while the β-series (S)-β-11 and (R)-β-12 led to ΔTm values of 13.3 and 13.6 °C, respectively (Table 1). Even lower melting temperatures were observed for hairpins 13–16 designed to bind DNA sequence 5′-TGGGGA-3′. Both β-amino-γ-turns, as in (S)-β-15 (ΔTm = 6.7) and (R)-β-16 (ΔTm = 6.8), resulted in significantly lower ΔTm values than the α-series analogue (R)-α-14 (ΔTm = 9.1). These results imply that the impact of polyamide turn units on DNA-duplex stabilization is sequence context dependent.
Figure 3.
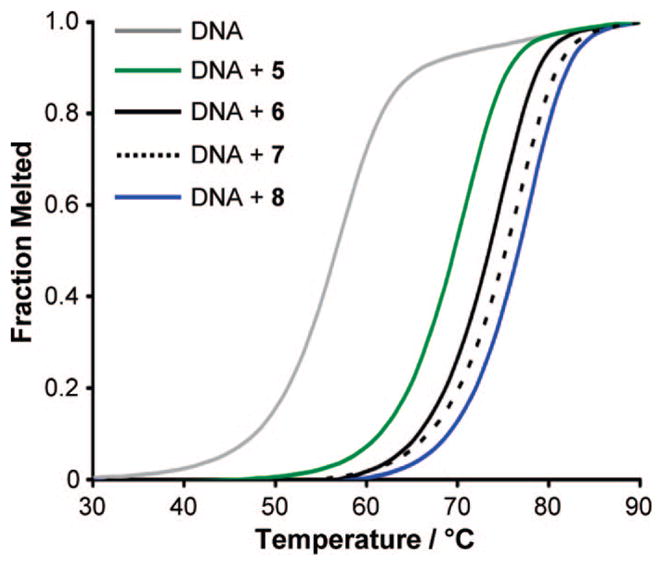
Normalized UV denaturation profiles of 12mer DNA duplex 5′-CGATGGTCAAGC-3′/5′-GCTTGACCATCG-3′ in the absence and presence of hairpin polyamides 5–8.
Table 1.
Melting Temperatures of DNA/Polyamide Complexes for A • T and T • A Base Pairs at the Turn Position of Hairpin Polyamidesa
| Polyamides | A • T | T • A | ||
|---|---|---|---|---|
| 5′-CGA
TGTTCA
AGC-3′ |
5′-CGA
TGTTCT
AGC-3′ |
|||
| Tm/°C | ΔTm/°C | Tm/°C | ΔTm/°C | |
| — | 54.0 (±0.2) | — | n.d. | |
|
|
69.9 (±0.3) | 15.9 | n.d. | |
|
|
73.5 (±0.2) | 19.5 | n.d. | |
|
|
74.9 (±0.2) | 20.9 | n.d. | |
|
|
76.2 (±0.2) | 22.2 | n.d. | |
|
| ||||
| 5′-CGA
TGGTCA
AGC-3′ |
5′-CGA
TGGTCT
AGC-3′ |
|||
| — | 57.2 (±0.1) | — | 55.8 (±0.1) | — |
|
|
70.6 (±0.2) | 13.4 | 69.0 (±0.3) | 13.2 |
|
|
74.1 (±0.3) | 16.9 | 72.9 (±0.2) | 17.1 |
|
|
76.1 (±0.2) | 18.9 | 73.2 (±0.1) | 17.4 |
|
|
77.5 (±0.3) | 20.3 | 74.2 (±0.1) | 18.4 |
|
| ||||
| 5′-CGA
TGGGCA
AGC-3′ |
5′-CGA
TGGGCT
AGC-3′ |
|||
| — | 60.2 (±0.2) | — | 59.8 (±0.3) | — |
|
|
68.8 (±0.2) | 8.6 | 67.4 (±0.3) | 7.6 |
|
|
73.4 (±0.2) | 13.2 | 72.0 (±0.1) | 12.2 |
|
|
73.5 (±0.1) | 13.3 | 70.5 (±0.3) | 10.7 |
|
|
73.8 (±0.1) | 13.6 | 71.3 (±0.3) | 11.5 |
|
| ||||
| 5′-CGA
TGGGGA
AGC-3′ |
5′-CGA
TGGGGT
AGC-3′ |
|||
| — | 57.5 (±0.1) | — | 57.9 (±0.1) | — |
|
|
60.9 (±0.1) | 3.4 | 61.4 (±0.3) | 3.5 |
|
|
66.6 (±0.1) | 9.1 | 67.0 (±0.1) | 9.1 |
|
|
64.2 (±0.1) | 6.7 | 64.2 (±0.1) | 6.3 |
|
|
64.3 (±0.3) | 6.8 | 64.4 (±0.3) | 6.5 |
All values reported are derived from at least three melting temperature experiments with standard deviations indicated in parentheses (n.d. = not determined). ΔTm values are given as Tm(DNA/polyamide) −Tm(DNA).
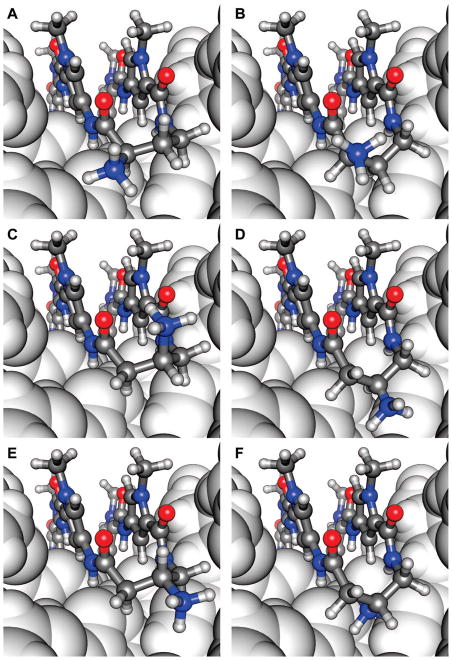
The general increase in DNA-binding affinity for polyamides containing the (R)-α-substitued γ-turn, relative to achiral hairpins, is most likely caused by a superposition of favorable noncovalent interactions of the positively charged substituent and conformational preferences of the turn unit.10b The (R)-α-amino-γ-turn can exist in two different conformations, one orienting the α-ammonium in a pseudoequatorial position (Figure 4A), which directs the substituent toward the wall of the minor groove with the potential of steric interactions. The alternate conformation places the α-amine residue in a pseudo-axial position out of the minor groove, orienting the β-methylene to the floor of the double helix (Figure 4B). Modeling of the (S)-β-amino-γ-turn conformations suggests that the β-ammonium in a pseudoaxial position is directed out of the minor groove (Figure 4C) relieving the potential steric interactions with the wall in comparison to the α-series. In contrast, the (S)-β-amine in a pseudoequatorial orientation is following the curvature of the minor groove (Figure 4D). The possibility for favorable noncovalent interactions should exist in both conformations without the detriment of steric interactions. As shown in Figure 4E, the pseudoequatorial β-amine residue of the (R)-β-amino-γ-turn is well accommodated in the DNA minor groove, while the pseudoaxial position should result in a steric clash of the substituent with the groove floor (Figure 4F). Previous results have shown that polyamides constructed with several continuous Im/Py pairs are overcurved with respect to the DNA minor groove, significantly influencing the DNA-binding affinity and sequence specificity.15 We assume that this curvature affects the alignment of the turn units in the DNA minor groove. This is supported by the observation that the presence of fewer continuous Im’s improves the DNA affinity of β-amino-γ-turns while diminishing the affinity for α-amino-γ-turns, and vice versa. However, illustrative modeling is not sufficient to explain the sequence context dependence of chiral hairpin polyamides, highlighting the pressing need for high-resolution structural studies.
Figure 4.
Illustrative models of different turn conformations for hairpin polyamides containing the (R)-α-amino-γ-turn (A and B), (S)-β-amino-γ-turn (C and D), and the (R)-β-amino-γ-turn (E and F) bound to the minor groove of DNA (dark gray = carbons, white = hydrogen, blue = nitrogen, red = oxygen).
Sequence Specificity at the Turn Position
Hairpin polyamides containing the γ-turn have been shown to possess an equal preference for A • T/T • A over G • C/C • G base pairs at the turn position, presumably for steric reasons.16 Sugiyama’s α-hydroxy-γ-turns have been demonstrated to discriminate A • T versus T • A at the turn position.10c In order to study the sequence specificity for polyamides 5–16, we performed melting temperature analyses in the presence of DNA duplexes bearing a T • A base pair at the turn position. Experiments involving hairpins 1–4 have been omitted due to the palindromic core sequence specified by the polyamides. As shown in Table 1, most γ-turn and (R)-α-amino-γ-turn hairpins provided similar ΔTm values for T • A and A • T base pairs. In contrast, significantly lower thermal stabilizations for T • A over A • T base pairs were observed for β-amino-γ-turn-linked polyamides targeting DNA sequences 5′-TGGTCA-3′ (5–8) and 5′-TGGGCA-3′ (9–12). Even more diminished duplex stabilizations were observed in presence of C • G and G • C base pairs (see Supporting Information). These observations suggest that polyamides containing β-amino-γ-turns prefer A • T > T • A ≫ C • G > G • C base pairs at the turn position. However, sequence specificity studies by thermal denaturation measurements require binding enthalpies (ΔHb) of DNA/polyamide complexes in order to determine equilibrium association constants.14a One could also imagine using six-ring hairpin polyamides with lower DNA-binding affinities in order to discriminate sequence specificities at the turn position by quantitative DNase I footprint titration methods.
Acetylated Chiral Hairpin Polyamides
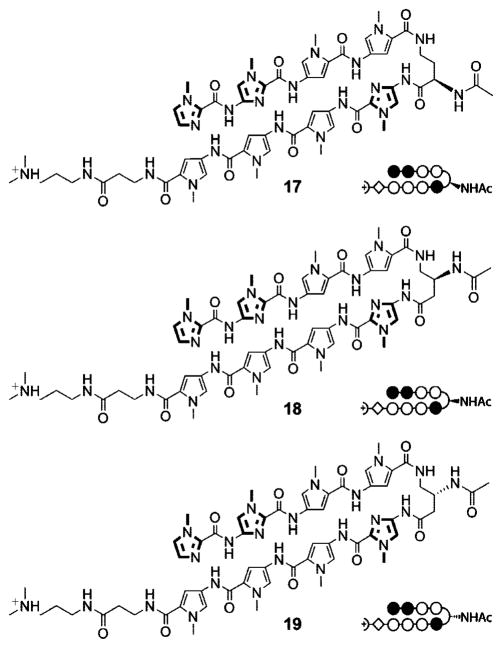
Several approaches have been reported wherein the (R)-α-amino-γ-turn was utilized as a position for synthetic modifications of hairpin polyamides.4,5,17 It has been shown that acetylation of the (R)-α-amine in six-ring hairpin polyamides results in ~15-fold reduced DNA-binding affinity.10b To study the tolerance of synthetic modifications for eight-ring polyamides containing the β-amino-γ-turns, we examined acetylated hairpins 17–19 by melting temperature analysis (Figure 5). Indeed, hairpin 17 containing the acetylated (R)-α-amino-γ-turn yielded a markedly lower ΔTm value (12.8 °C) than non-acetylated analogue 6 (ΔTm = 16.9 °C, Table 2). Even more pronounced was the decrease in DNA duplex stabilization for acetylated (S)-β-amino-γ-turn hairpin 18 leading to a ΔTm value of 11.7 °C. Remarkably, the opposite enantiomer (R)-β-19 resulted in significantly less destabilization (ΔTm = 17.8 °C). All hairpins lose the positive charge at the turn unit by acetylation. This implies that the cationic state of the amine residue is not the only contribution impacting the energetics of the DNA/polyamide complexes, as evidenced by the differences in melting temperatures between hairpins 17–19. Increased steric demands of the acetylated substituents may also be responsible for differing binding affinities, due to the restricting DNA minor groove and alternate conformations of the γ-turn units (Figure 4).
Figure 5.
Chemical structures and ball-and-stick models of acetylated hairpin polyamides 17–19 targeted to DNA sequence 5′-TGGTCA-3′.
Table 2.
Melting Temperatures for DNA Complexes Containing Nonacetylated and Acetylated Hairpin Polyamides Targeted to DNA Sequence 5′-TGGTCA-3′a
| DNA sequence =
5′-CGA TGGTCA AGC-3′ | ||
|---|---|---|
| Polyamides | Tm/°C | ΔTm/°C |
| — | 57.2 (±0.2) | — |
|
|
74.1 (±0.3) | 16.9 |
|
|
76.1 (±0.2) | 18.9 |
|
|
77.5 (±0.3) | 20.3 |
|
| ||
|
|
70.0 (±0.1) | 12.8 |
|
|
68.9 (±0.2) | 11.7 |
|
|
75.0 (±0.1) | 17.8 |
All values reported are derived from at least three melting temperature experiments with standard deviations indicated in parentheses. ΔTm values are given as Tm(DNA/polyamide) – Tm(DNA).
Biological Assay for Cell Permeability
Hairpin polyamide conjugates bearing- the standard (R)-α-amino-γ-turn have been shown to modulate the expression of certain gene pathways in living cells by interfering with transcription factor/DNA interfaces.1 Recently, a hairpin designed to bind DNA sequence 5′-AGAACA-3′, found in the androgen response element (ARE), has been demonstrated to inhibit androgen receptor-mediated expression of prostate specific antigen (PSA) in LNCaP cells (Figure 6).1b We utilized this cell culture transcription assay to investigate the cell permeability of (R)-β-amino-γ-turn hairpins because small structural changes within polyamides can influence nuclear uptake properties.18 Hairpin polyamide 21 was examined in comparison to the previously used (R)-α-amino-γ-turn hairpin 20 (Figure 7A). Chiral polyamides 22 and 23, designed to target different DNA sequences, have been used as controls. Melting temperature analyses for polyamide conjugates 20–23 confirmed the results obtained for hairpins 1–4, revealing highest DNA-duplex stabilizations for (R)-β-amino- γ-turn hairpins (see Supporting Information). The induction of PSA mRNA by dihydrotestosterone (DHT) in LNCaP cells for matched and mismatched polyamides 20–23 was measured by quantitative real-time RT-PCR. As shown in Figure 7B, hairpin 21 provided significant inhibition of AR-mediated expression of PSA mRNA, KLK2, FKBP5, and TMPRSS2 mRNA (see Supporting Information) which supports cell-permeable properties for (R)-β-amino-γ-turn hairpins.
Figure 6.
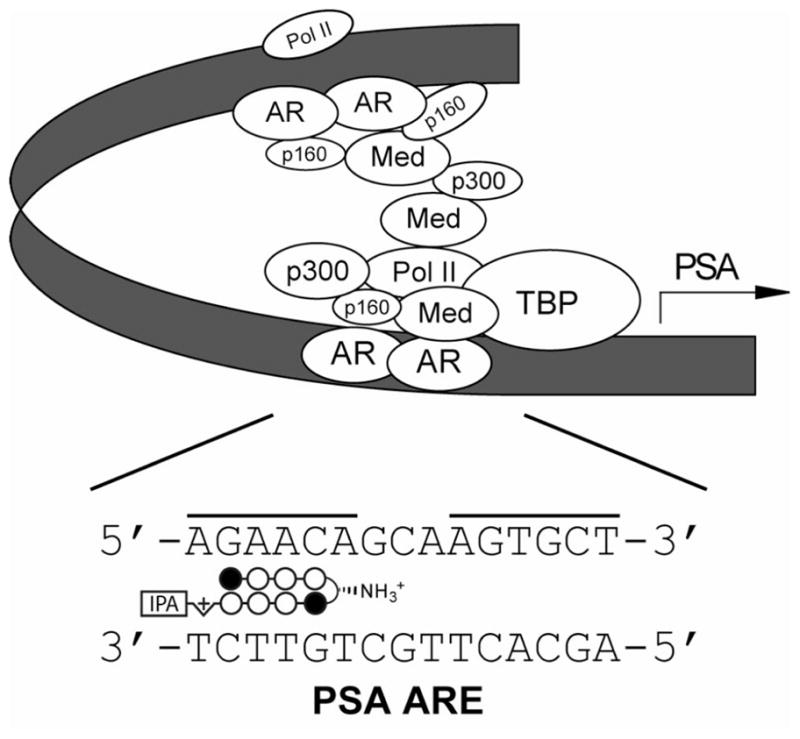
Schematic representation of the androgen receptor (AR)-mediated transcription complex with the androgen response element (ARE).
Figure 7.
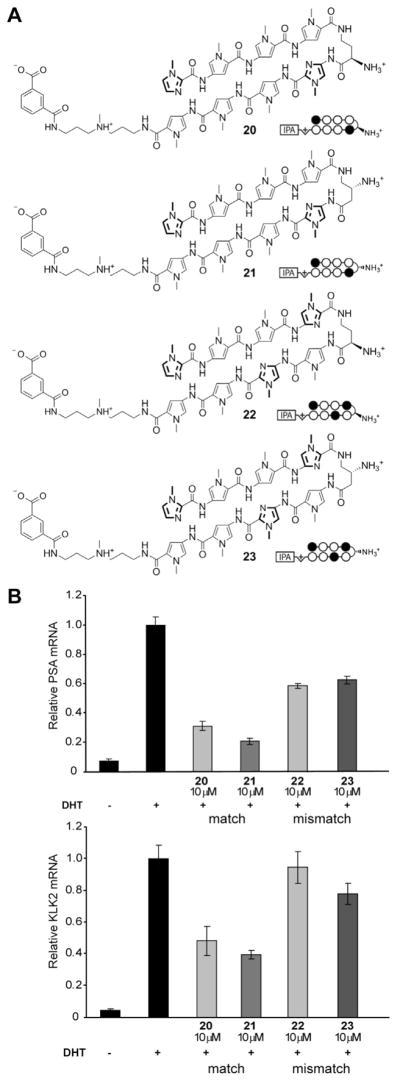
(A) Chemical structures and ball-and-stick models of matched and mismatched polyamides 20–23, targeted to DNA sequence 5′-AGAACA-3′. (B) Inhibition of DHT-induced PSA and KLK2 expression by 20–23 measured by quantitative real-time RT-PCR.
Conclusions
Herein we have introduced (R)- and (S)-β-amino-γ-turn hairpin polyamides. Eight new polyamides targeting different DNA-binding motifs have been synthesized, and their impact on DNA duplex stabilization in relation to hairpins containing the parent γ-turn and the standard (R)-α-amino-γ-turn was investigated. It was found that changing the turn unit from the (R)-α-amino-γ-turn to either enantiomeric forms of the β-amino-γ-turn increases the relative DNA-binding affinity of polyamides targeted to 5′-TGTTCA-3′ and 5′-TGGTCA-3′ but not to 5′-TGGGCA-3′ and 5′-TGGGGA-3′ sequences, rendering the impact of β-amino-substituted γ-turns sequence context dependent. Acetylation of the (S)-β-amino-γ-turn has been demonstrated to significantly impact the DNA-binding affinity but has minimal effect for the (R)-β-amino-γ-turn, which makes the (R)-β-amino residue attractive for synthetic modifications and conjugate design. Upper limits presented by DNase I footprinting titrations of high affinity binders rendered melting temperature analysis a more practical choice for dissecting improvements by structural changes of new turn units in hairpin polyamides. Due to the strong thermal stabilizations, reported for eight-ring hairpin polyamides 1–8 targeted to 5′-TGTTCA-3′ and 5′-TGGTCA-3′ sequences, it is not unreasonable to estimate that the DNA-binding equilibrium association constants are markedly higher than Ka = 2 × 1010 M−1. Biological experiments have demonstrated that (R)-β-amino-γ-turn hairpins possess biological activity to inhibit AR-mediated gene expression within a human cancer cell line and may have similar uptake properties as polyamides bearing the standard (R)-α-amino-γ-turn. Ongoing work is focused on the use of the next generation hairpins in biological investigations as well as turn unit sequence specificity and high-resolution crystallographic studies for DNA/chiral hairpin polyamide complexes. These efforts will be reported in due course.
Experimental Section
General
Chemicals and solvents were purchased from Sigma-Aldrich and were used without further purification. Boc-γ-Abu-OH was purchased from Novabiochem. (R)-2,4-Fmoc-Dbu(Boc)-OH and Boc-β-Ala-PAM resin were purchased from Peptides International. (R)-3,4-Cbz-Dbu(Boc)-OH and (S)-3,4-Cbz-Dbu(Boc)-OH were purchased from Senn Chemicals AG. All DNA oligomers were purchased HPLC purified from Integrated DNA Technologies. Water (18 MΩ) was purified using a Millipore MilliQ purification system. The pH of buffers was adjusted using a Beckman 340 pH/temp meter. Analytical HPLC was performed on a Beckman Gold system equipped with a diode array detector using a Phenomenex Gemini column (5 μm particle size, C18 110A, 250 × 4.6 mm, 5 μm). Preparative HPLC was performed on a Beckman Gold system equipped with a single-wavelength detector monitoring at 310 nm using a Phenomenex Gemini column (5 μm particle size, C18 110A, 250 × 21.2 mm, 5 μm). For both analytical and preparative HPLC, solvent A was 0.1% (v/v) aqueous trifluoroacetic acid (TFA) and solvent B was acetonitrile. Solvent gradients were adjusted as needed. Polyamide concentrations were measured in 0.1% (v/v) aqueous TFA on a Hewlett-Packard diode array spectrophotometer “Model 8452 A” and were determined by using an extinction coefficient of 69200 M−1 • cm−1 at λmax near 310 nm. Matrix-assisted, LASER desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) was performed on an Applied Biosystems Voyager DR Pro spectrometer using α-cyano-4-hydroxycinnamic acid as matrix.
Synthesis of Polyamides
Polyamide monomers and oligomers were synthesized as described previously.19 All β-amino-γ-turn hairpins were synthesized by performing the following procedure: the polyamide was cleaved from the resin with 3-(dimethylamino)-1-propylamine, purified by preparative HPLC, and characterized by MALDI-TOF MS, UV–vis spectroscopy, and analytical HPLC. A 500 nmol fraction of the Cbz-protected hairpin polyamide was dissolved in a 9:1 mixture (500 μL) of TFA and trifluoromethane-sulfonic acid (TFMSA). After 5 min reaction time, the solution was flash-frozen by liquid N2 and overlaid with N,N′-dimethylformamide (1 mL). The thawed solution was diluted with 20% aqueous acetonitrile (8 mL), purified by preparative HPLC, and characterized by MALDI-TOF MS, UV–vis spectroscopy, and analytical HPLC. Acetylated polyamides 17–19 were synthesized by performing the following procedure: A 500 nmol fraction of the polyamide was dissolved in N,N′-dimethylformamide (900 μL) and a 9:1 mixture of pyridine/acetic anhydride (100 μL) was added. After 5 min reaction time, the solution was diluted with 10% aqueous TFA (8 mL), purified by preparative HPLC, and characterized by MALDI-TOF MS, UV–vis spectroscopy, and analytical HPLC. Polyamide conjugates 20–23 were synthesized as described previously.20 Polyamide 1: MALDI-TOF [M + H]+ calcd for C58H72N21O10+ = 1222.6, observed = 1222.7. Polyamide 2: MALDI-TOF [M + H]+ calcd for C58H73N22O10+= 1237.6, observed = 1237.8. Polyamide 3: MALDI-TOF [M + H]+ calcd for C58H73N22O10+= 1237.6, observed = 1237.8. Polyamide 4: MALDI-TOF [M + H]+ calcd for C58H73N22O10+= 1237.6, observed = 1237.8. Polyamide 5: MALDI-TOF [M + H]+ calcd for C57H71N22O10+= 1223.6, observed = 1223.5. Polyamide 6: MALDI-TOF [M + H]+ calcd for C57H72N23O10+= 1238.6, observed = 1238.6. Polyamide 7: MALDI-TOF [M + H]+ calcd for C57H72N23O10+= 1238.6, observed = 1238.5. Polyamide 8: MALDI-TOF [M + H]+ calcd for C57H72N23O10+= 1238.6, observed = 1238.5. Polyamide 9: MALDI-TOF [M + H]+ calcd for C56H70N23O10+= 1224.6, observed = 1224.8. Polyamide 10: MALDI-TOF [M + H]+ calcd for C56H71N24O10+= 1239.6, observed = 1239.6. Polyamide 11: MALDI-TOF [M + H]+ calcd for C56H71N24O10+= 1239.6, observed = 1239.5. Polyamide 12: MALDI-TOF [M + H]+ calcd for C56H71N24O10+= 1239.6, observed = 1239.6. Polyamide 13: MALDI-TOF [M + H]+ calcd for C56H70N23O10+= 1224.6, observed = 1224.6. Polyamide 14: MALDI-TOF [M + H]+ calcd for C56H71N24O10+= 1239.6, observed = 1239.7. Polyamide 15: MALDI-TOF [M + H]+ calcd for C56H71N24O10+= 1239.6, observed = 1239.4. Polyamide 16: MALDI-TOF [M + H]+ calcd for C56H71N24O10+= 1239.6, observed = 1239.5. Polyamide 17: MALDI-TOF [M + H]+ calcd for C59H74N23O11+= 1280.6, observed = 1280.6. Polyamide 18: MALDI-TOF [M + H]+ calcd for C59H74N23O11+= 1280.6, observed = 1280.7. Polyamide 19: MALDI-TOF [M + H]+ calcd for C59H74N23O11+= 1280.6, observed = 1280.6. Polyamide 20: MALDI-TOF [M + H]+ calcd for C65H77N22O12+= 1357.7, observed = 1357.6. Polyamide 21: MALDI-TOF [M + H]+ calcd for C65H77N22O12+= 1357.6, observed = 1357.7. Polyamide 22: MALDI-TOF [M + H]+ calcd for C64H76N23O12+= 1358.6, observed = 1358.6. Polyamide 23: MALDI-TOF [M + H]+ calcd for C64H76N23O12+=1358.6, observed =1358.6.
UV Absorption Spectrophotometry
Melting temperature analysis was performed on a Varian Cary 100 spectrophotometer equipped with a thermocontrolled cell holder possessing a cell path length of 1 cm. The buffer for the spectroscopic measurements was chosen to match as closely as possible the conditions of DNase I footprinting experiments. We used 10 mM sodium cacodylate since the temperature dependence of Tris-HCl makes it poorly suited for melting temperature analyses.14a A degassed aqueous solution of 10 mM sodium cacodylate, 10 mM KCl, 10 mM MgCl2, and 5 mM CaCl2 at pH 7.0 was used as analysis buffer. DNA duplexes and hairpin polyamides were mixed in 1:1 stoichiometry to a final concentration of 2 μM for each experiment. Prior to analysis, samples were heated to 90 °C and cooled to a starting temperature of 25 °C with a heating rate of 5 °C/min for each ramp. Denaturation profiles were recorded at λ = 260 nm from 25 to 90 °C with a heating rate of 0.5 °C/min. The reported melting temperatures were defined as the maximum of the first derivative of the denaturation profile.
Molecular Modeling
DNA/polyamide models are based on coordinates derived from NMR structure studies using standard bond length and angles.3c The molecular graphics images are non-minimized and have been created by introducing ammonium residues to the appropriate position of the turn unit using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081).21
Measurement of Androgen-Induced PSA mRNA
Experiments were performed as described previously.1b
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (GM27681). C.D. is grateful to the Alexander von Humboldt foundation for a postdoctoral research fellowship. M.E.F. is grateful for a predoctoral NIH NRSA training grant. D.M.C. is grateful to the Kanel Foundation for a predoctoral fellowship.
Footnotes
Supporting Information Available: Further experimental procedures and methods, DNase I footprint titration details, plasmid sequences, equilibrium association constants, melting temperatures, and further biological studies. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Olenyuk BZ, Zhang GJ, Klco JM, Nickols NG, Kaelin WG, Dervan PB. Proc Natl Acad Sci USA. 2004;101:16768–16773. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nickols NG, Dervan PB. Proc Natl Acad Sci USA. 2007;104:10418–10423. doi: 10.1073/pnas.0704217104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Nickols NG, Jacobs CS, Farkas ME, Dervan PB. ACS Chem Biol. 2007;2:561–571. doi: 10.1021/cb700110z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Dervan PB. Bioorg Med Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]; (b) Dervan PB, Edelson BS. Curr Opin Struct Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 3.(a) Kielkopf CL, White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB, Rees DC. Science. 1998;282:111–115. doi: 10.1126/science.282.5386.111. [DOI] [PubMed] [Google Scholar]; (b) Kielkopf CL, Baird EE, Dervan PD, Rees DC. Nat Struct Biol. 1998;5:104–109. doi: 10.1038/nsb0298-104. [DOI] [PubMed] [Google Scholar]; (c) Zhang Q, Dwyer TJ, Tsui V, Case DA, Cho JH, Dervan PB, Wemmer DE. J Am Chem Soc. 2004;126:7958–7966. doi: 10.1021/ja0373622. [DOI] [PubMed] [Google Scholar]; (d) Puckett JW, Muzikar KA, Tietjen J, Warren CL, Ansari AZ, Dervan PB. J Am Chem Soc. 2007;129:12310–12319. doi: 10.1021/ja0744899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Wurtz NR, Dervan PB. Chem Biol. 2000;7:153–161. doi: 10.1016/s1074-5521(00)00085-5. [DOI] [PubMed] [Google Scholar]; (b) Sasaki S, Bando T, Minoshima M, Shimizu T, Shinohara K, Takaoka T, Sugiyama H. J Am Chem Soc. 2006;128:12162–12168. doi: 10.1021/ja0626584. [DOI] [PubMed] [Google Scholar]; (c) Tsai SM, Farkas ME, Chou CJ, Gottesfeld JM, Dervan PB. Nucleic Acids Res. 2007;35:307–316. doi: 10.1093/nar/gkl1025. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Minoshima M, Bando T, Sasaki S, Shinohara K, Shimizu T, Fujimoto J, Sugiyama H. J Am Chem Soc. 2007;129:5384–5390. doi: 10.1021/ja065235a. [DOI] [PubMed] [Google Scholar]
- 5.Poulin-Kerstien AT, Dervan PB. J Am Chem Soc. 2003;125:15811–15821. doi: 10.1021/ja030494a. [DOI] [PubMed] [Google Scholar]
- 6.(a) Fechter EJ, Dervan PB. J Am Chem Soc. 2003;125:8476–8485. doi: 10.1021/ja030125e. [DOI] [PubMed] [Google Scholar]; (b) Fechter EJ, Olenyuk B, Dervan PB. Angew Chem, Int Ed. 2004;43:3591–3594. doi: 10.1002/anie.200454231. [DOI] [PubMed] [Google Scholar]; (c) Fechter EJ, Olenyuk B, Dervan PB. J Am Chem Soc. 2005;127:16685–16691. doi: 10.1021/ja054650k. [DOI] [PubMed] [Google Scholar]
- 7.(a) Rucker VC, Foister S, Melander C, Dervan PB. J Am Chem Soc. 2003;125:1195–1202. doi: 10.1021/ja021011q. [DOI] [PubMed] [Google Scholar]; (b) Chenoweth DM, Viger A, Dervan PB. J Am Chem Soc. 2007;129:2216–2217. doi: 10.1021/ja0682576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Cohen JD, Sadowski JP, Dervan PB. Angew Chem, Int Ed. 2007;46:7956–7959. doi: 10.1002/anie.200702767. [DOI] [PubMed] [Google Scholar]; (b) Schmidt TL, Nandi CK, Rasched G, Parui PP, Brutschy B, Famulok M, Heckel A. Angew Chem, Int Ed. 2007;46:4382–4384. doi: 10.1002/anie.200700469. [DOI] [PubMed] [Google Scholar]; (c) Cohen JD, Sadowski JP, Dervan PB. J Am Chem Soc. 2008;130:402–403. doi: 10.1021/ja0772400. [DOI] [PubMed] [Google Scholar]
- 9.(a) Arndt HD, Hauschild KE, Sullivan DP, Lake K, Dervan PB, Ansari AZ. J Am Chem Soc. 2003;125:13322–13323. doi: 10.1021/ja0371395. [DOI] [PubMed] [Google Scholar]; (b) Kwonj Y, Arndt HD, Qian M, Choi Y, Kawazoe Y, Dervan PB, Uesugi M. J Am Chem Soc. 2004;126:15940–15941. doi: 10.1021/ja0445140. [DOI] [PubMed] [Google Scholar]; (c) Hauschild KE, Metzler RE, Arndt HD, Moretti R, Raffaelle M, Dervan PB, Ansari AZ. Proc Natl Acad Sci USA. 2005;102:5008–5013. doi: 10.1073/pnas.0501289102. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Stafford RL, Arndt HD, Brezinski ML, Ansari AZ, Dervan PB. J Am Chem Soc. 2007;129:2660–2668. doi: 10.1021/ja067971k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Stafford RL, Dervan PB. J Am Chem Soc. 2007;129:14026–14033. doi: 10.1021/ja075247b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Xiao X, Yu P, Lim HS, Sikder D, Kodadek T. Angew Chem, Int Ed. 2007;46:2865–2868. doi: 10.1002/anie.200604485. [DOI] [PubMed] [Google Scholar]
- 10.(a) Mrksich M, Parks ME, Dervan PB. J Am Chem Soc. 1994;116:7983–7988. [Google Scholar]; (b) Herman DM, Baird EE, Dervan PB. J Am Chem Soc. 1998;120:1382–1391. [Google Scholar]; (c) Zhang W, Minoshima M, Sugiyama H. J Am Chem Soc. 2006;128:14905–14912. doi: 10.1021/ja064369l. [DOI] [PubMed] [Google Scholar]; (d) Farkas ME, Tsai SM, Dervan PB. Bioorg Med Chem. 2007;15:6927–6936. doi: 10.1016/j.bmc.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu CF, Phillips JW, Trauger JW, Farkas ME, Belitsky JM, Heckel A, Olenyuk BZ, Puckett JW, Wang CCC, Dervan PB. Tetrahedron. 2007:6146–6151. doi: 10.1016/j.tet.2007.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trauger JW, Dervan PB. Methods Enzymol. 2001;340:450–466. doi: 10.1016/s0076-6879(01)40436-8. [DOI] [PubMed] [Google Scholar]
- 13.(a) For quantitative footprinting experiments, the DNA concentrations of equilibrium mixtures should be at least 10-fold less than the total association constant of the DNA/ligand complex in order to ensure the approximation [ligand]free = [ligand]total for numerical analysis. However, the concentration of the labeled DNA fragment specified by the standard DNA/polyamide footprinting protocol is ~5 pM.12 Consequently, DNA association constants become compressed and hence unreliable for comparison studies at Ka values ≥ 2 × 1010 M−1. Brenowitz M, Senear DF, Shea MA, Ackers GK. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9.Senear DF, Dalma-Weiszhausz DD, Brenowitz M. Electrophoresis. 1993;14:704–712. doi: 10.1002/elps.11501401112.
- 14.(a) Pilch DS, Poklar N, Gelfand CA, Law SM, Breslauer KJ, Baird EE, Dervan PB. Proc Natl Acad Sci USA. 1996;93:8306–8311. doi: 10.1073/pnas.93.16.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Pilch DS, Poklar N, Baird EE, Dervan PB, Breslauer KJ. Biochemistry. 1999;38:2143–2151. doi: 10.1021/bi982628g. [DOI] [PubMed] [Google Scholar]
- 15.(a) Turner JM, Swalley SE, Baird EE, Dervan PB. J Am Chem Soc. 1998;120:6219–6226. [Google Scholar]; (b) Floreancig PE, Swalley SE, Trauger JW, Dervan PB. J Am Chem Soc. 2000;122:6342–6350. [Google Scholar]
- 16.Swalley SE, Baird EE, Dervan PB. J Am Chem Soc. 1999;121:1113–1120. [Google Scholar]
- 17.(a) Weyermann P, Dervan PB. J Am Chem Soc. 2002;124:6872–6878. doi: 10.1021/ja020258k. [DOI] [PubMed] [Google Scholar]; (b) Herman DM, Baird EE, Dervan PB. Chem—Eur J. 1999;5:975–983. [Google Scholar]; (c) Wang CCC, Dervan PB. J Am Chem Soc. 2001;123:8657–8661. doi: 10.1021/ja010392p. [DOI] [PubMed] [Google Scholar]; (d) Edayathumangalam RS, Weyermann P, Gottesfeld JM, Dervan PB, Luger K. Proc Natl Acad Sci USA. 2004;101:6864–6869. doi: 10.1073/pnas.0401743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Best TP, Edelson BS, Nickols NG, Dervan PB. Proc Natl Acad Sci USA. 2003;100:12063–12068. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Edelson BS, Best TP, Olenyuk B, Nickols NG, Doss RM, Foister S, Heckel A, Dervan PB. Nucleic Acids Res. 2004;32:2802–2818. doi: 10.1093/nar/gkh609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baird EE, Dervan PB. J Am Chem Soc. 1996;118:6141–6146. [Google Scholar]
- 20.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Nucleic Acids Res. 2007;35:363–370. doi: 10.1093/nar/gkl1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.