Abstract
The GABAA receptor is a multisubunit protein that transduces the binding of a neurotransmitter at an intersubunit interface into the opening of a central ion channel. The structural components that mediate the steps involved in this action are poorly defined. A large amount of work has focused on clarifying the specific functions and interactions of residues believed to surround the GABA binding pocket. Here, we explored two charged residues (β2Asp163 and α1Arg120), which have been suggested by homology models to participate in a salt-bridge interaction. When mutated to alanine, both single mutants, as well as the double mutant, increase EC50-GABA, decrease the GABA binding rate, and accelerate deactivation and GABA unbinding rates. Double-mutant cycle analysis demonstrates that the effects of each alanine mutation on the GABA binding rate were additive and independent. In contrast, a significant coupling energy was found during an analysis of deactivation time constants. Using kinetic modeling, we further demonstrated that the GABA unbinding rates, in particular, are strongly coupled. These data suggest that β2Asp163 and α1Arg120 form a state-dependent salt bridge, interacting when GABA is bound to the receptor but not when the receptor is in the unbound state.
Introduction
The GABAA receptor is a member of the Cys-loop family of ligand-gated ion channels (LGICs), which also includes nicotinic acetylcholine, serotonin (5-HT3), and glycine receptors. The Cys-loop LGICs mediate fast synaptic transmission throughout the central and peripheral nervous systems as the binding of a neurotransmitter to the extracellular domain of a receptor leads to the opening of an intrinsic ion pore. The GABAA receptor contains a chloride channel and is the major type of inhibitory neurotransmitter receptor in the mammalian central nervous system. It is the target of sedatives, anxiolytics, antiepileptics, and general anesthetics.
The GABAA receptor, along with all Cys-loop LGICs, is a pentameric protein, constructed of homologous subunits. These subunits are arranged pseudosymmetrically around a central ion-conducting pore. The most abundant complex found in the brain consists of two α1 subunits, two β2 subunits, and one γ2 subunit (Benke et al., 1994; McKernan and Whiting, 1996). It is arranged counterclockwise as βαγβα (Baumann et al., 2002). The neurotransmitter GABA is believed to bind at the extracellular interface of the β and α subunits. This idea is supported by a plethora of studies that have used site-directed mutagenesis, photoaffinity labeling, and the substituted cysteine accessibility method (SCAM) to identify more than 20 amino acids from the α and β subunits that seem to play a role in GABA binding (Kash et al., 2004). Specific roles for a handful of these residues have been proposed. For example, β2Tyr97 may form a cation-π bond with the amino group of GABA (Padgett et al., 2007), but by and large it is unclear whether a given residue implicated in GABA binding directly interacts with GABA, stabilizes the structure of the binding pocket, or is involved in transducing binding into channel gating.
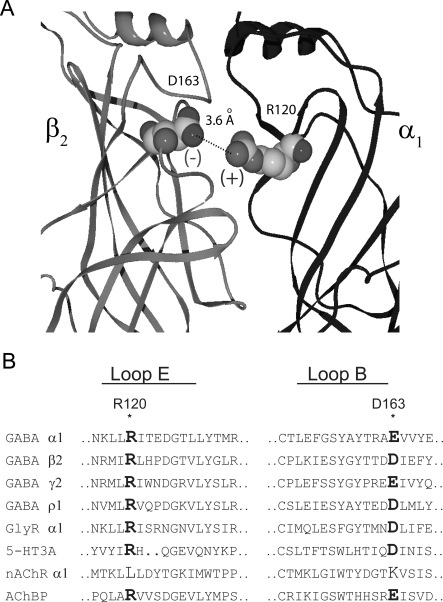
Homology models (Cromer et al., 2002; O'Mara et al., 2005) derived from the crystallized structure of the molluscan acetylcholine binding protein (Brejc et al., 2001) have been used to generally position the residues in the binding pocket and can further inform us as to the role of an individual residue. However, interpretations drawn from these models must be experimentally verified. One interpretation drawn from homology models is that an arginine from the α subunit (α1Arg120) forms an intersubunit salt bridge with an aspartate from the β subunit (β2Asp163), and that this salt bridge is conserved at every subunit interface (Cromer et al., 2002) (Fig. 1). These two residues are highly conserved among LGIC subunits, and experimental evidence for a corresponding salt-bridge interaction has been presented for the γ/β intersubunit interface of the GABAA receptor (Goldschen-Ohm et al., 2010). Mutagenesis studies show the GABA concentration-response curve is sensitive to changes at either α1Arg120 or β2Asp163 (Westh-Hansen et al., 1999; Newell et al., 2004; Kloda and Czajkowski, 2007). In addition, SCAM studies demonstrated that both residues are accessible to modification by sulfhydryl reactive reagents, indicating that they are present at the aqueous surface (Newell et al., 2004; Kloda and Czajkowski, 2007).
Fig. 1.
Homology models of the GABAA receptor depict a putative salt bridge between β2Asp163 and α1Arg120. A, side view of the extracellular domain at a single β/α interface. The nearest charged atoms of α1Arg120 and β2Asp163 are 3.6 Å apart. B, sequences of various human GABAA receptor, glycine receptor (GlyR), 5-HT3A receptor, and nicotinic acetylcholine receptor (nAChR) subunits aligned with the acetylcholine binding protein (AChBP) sequence, demonstrate the conservation of the residues mutated in this study.
We explored this postulated interaction between α1Arg120 and β2Asp163 by characterizing macroscopic parameters (EC50-GABA, deactivation, and desensitization) and microscopic parameters (GABA binding and unbinding rates) for alanine mutations at each residue. These results were subjected to double-mutant cycle analysis. It is noteworthy that mutation of both residues affected the GABA binding rate (kon-GABA), independently. However, when analyzing the deactivation time constants or the GABA unbinding rates, the two residues seem to be coupled. These results suggest that α1Arg120 and β2Asp163 form a state-dependent salt bridge.
Materials and Methods
cDNA Constructs and Mutagenesis.
Human α1, β2, and γ2S subunits inserted into the pcDNA 3.1 vector were used. A β2 variant (β2-GKER) that has been developed experimentally was used to rescue expression with α1R120A. β2-GKER has four amino acids of β2 replaced with the aligned residues found on the β3 subunit: D171G, N173K, T179E, and K180R (Taylor et al., 1999; Bollan et al., 2003). These residues are located proximal to the cell membrane and are on the non-GABA binding face of the subunit. This subunit has been shown to assemble more efficiently (Bollan et al., 2003), and we have found no differences in kinetics, EC50-GABA, or amplitude between β2- and β2-GKER-containing receptors. For all of the results presented here α1β2-GKERγ2S receptors were used as the control. Mutant α1 and β2 subunits were created using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA), and double-stranded sequencing of the entire coding region was conducted to verify fidelity.
Cell Culture, Transfection, and Labeling.
Human embryonic kidney (HEK)-293 cells were cultured in Eagle minimum essential medium with Earle's salts (Mediatech, Herndon, VA) supplemented with 10% newborn calf serum (Thermo Fisher Scientific, Waltham, MA) and penicillin-streptomycin-glutamine (Mediatech) in a 37°C incubator under a 5% CO2 atmosphere. Cells were plated onto 35-mm dishes coated with poly-l-lysine and were transfected 18 to 24 h later using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and the following amounts of cDNA: 250 ng of enhanced green fluorescent protein, 1 μg of α1 (wild type or R120A), 1 μg of β2-GKER (wild type or D163A), and 3 μg of γ2S. Cells were recorded from 48 to 96 h after transfection.
Electrophysiology.
All recordings for this study were collected from outside-out patches excised from HEK-293 cells (−60 mV). Recordings were made using borosilicate glass pipettes filled with 140 mM KCl, 10 mM EGTA, 2 mM MgATP, 20 mM phosphocreatine, and 10 mM HEPES, pH 7.3. Rapid solution exchange was accomplished by using a multibarreled flow pipe array (Vitrodynamics, Rockaway, NJ) mounted on a piezoelectric bimorph (Vernitron, Bedford, OH). A computer-controlled constant current source drove the bimorph to move solution interfaces over the patch with 10 to 90% exchange times of ∼200 μs, as measured by the liquid junction current at the open pipette tip after each experiment. GABAA receptor agonists and antagonists were dissolved in the perfusion solution, which contained 145 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, and 4 mM glucose, pH 7.4. For extracellular solutions that contained >30 mM GABA, the concentration of NaCl was reduced to 95 mM, and a combination of sucrose and GABA was added to compensate for the reduced osmolarity. The pipette solution was adjusted in conjunction, reducing the KCl concentration to 90 mM, and adding 50 mM potassium gluconate to maintain a constant Cl− driving force. GABA and 6-imino-3-(4-methoxyphenyl)-1(6H)-pyridazinebutanoic acid hydrobromide (SR-95531) were obtained from Sigma-Aldrich (St. Louis, MO). Currents were low-pass-filtered at 2 to 5 kHz with a four-pole Bessel filter and digitized at a rate no less than twice the filter frequency. Data were collected at 20 kHz using an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA) and an ITC-1600 digitizer (InstruTECH Corporation, Port Washington, NY), controlled by Axograph X software (Axograph Scientific, Sydney, Australia). Macroscopic current ensembles were collected with 15-s intervals between consecutive solution applications. Curve fitting was also performed using Axograph X software. Deactivation and desensitization phases were fit with biexponential functions. For deactivation, the time of GABA removal was set to zero, and the region of deactivation was fit with the equation Y = A1 · e−t/τ1 + A2 · e−t/τ2. For desensitization, the onset of desensitization was set to zero, and region of desensitization was fit with the equation Y = A1 · e−t/τ1 + A2 · e−t/τ2 + C. A weighted time constant (τw) was also calculated for each analysis. τw = (A1/(A1 + A2)) · τ1 + (A2/(A1 + A2)) · τ2. Figures of raw data represent ensemble averages of 10 to 30 traces that have been decimated. In all cases significant differences were tested using one-way ANOVA with a post hoc Dunnett's test, at a significance level of p < 0.05 (Prism 4; GraphPad Software, Inc., San Diego, CA).
Antagonist Unbinding Experiments.
Outside-out patches were pre-equilibrated in SR-95531 for 750 ms, and then rapidly switched to a solution containing saturating GABA. The evoked current was shaped by the convolution of the time course of antagonist unbinding and the wave form of the control current (evoked with no pre-equilibration in antagonist). Home-written Matlab (The Mathworks Inc., Natick, MA) routines were used for the deconvolution of the time course of antagonist unbinding from this evoked current (Jones et al., 2001). The time course of antagonist unbinding was fit with an exponential function, yielding koff-SR and the percentage of receptors occupied by antagonist at equilibrium. The experiment was repeated several times, pre-equilibrating in different concentrations of SR-95531. Kd-SR was determined by plotting the antagonist occupancy versus concentration.
Measuring the Microscopic Binding Rate of GABA.
The microscopic binding rate of GABA (kon-GABA) was measured using “race” experiments (Jones et al., 1998). In a race experiment, agonist (having an unknown binding rate) is coapplied with an antagonist that has a previously determined binding rate (kon-ant; see Antagonist Unbinding Experiments). The amplitude of the current evoked by coapplication of agonist and antagonist is compared with the amplitude of current evoked by agonist alone, and this ratio (Iag-ant/Iag-only) is called Irace · Irace depends only on the relative concentrations and binding rates of agonist and antagonist. The only unknown is the agonist binding rate, kon-agonist, which can be solved for using the following equation: kon-agonist = ([antagonist] · kon-ant)/([agonist] · (1/Irace − 1)).
Mutant Cycle Analysis.
Mutant cycle analysis was performed on EC50 values, deactivation rates, and binding rates. ΔΔG′o was calculated as R · T ln(kmutant/kwild-type), where R is the ideal gas constant (1.987 calories/mol) and T is the absolute temperature (296 K). Although EC50 and deactivation rates do not provide true kinetic rate constants, comparison of macroscopic parameters have been previously used to support side-chain interactions and establish coupling coefficients (Kash et al., 2003; Price et al., 2007; Gleitsman et al., 2008). If two mutations have independent effects ΔΔG′o(3) = ΔΔG′o(1) + ΔΔG′o(2). For our evaluation we set a coupling energy [ΔΔG′ocoupling = (ΔΔG′o(1) + ΔΔG′o(2)) − ΔΔG′o(3))] of 0.5 kcal/mol as the cutoff for nonadditivity.
Nonstationary Variance Analysis.
Nonstationary variance analysis (Sigworth, 1980) was performed on responses to repeated 3-ms pulses of saturating GABA. As described previously (Wagner et al., 2004; Goldschen-Ohm et al., 2010), mean current (I) and variance (σ2) of the repeated pulses were calculated at each time point, mean current was divided into 100 equally sized bins, and the variances in each bin were averaged. The binned variance was plotted versus current and fit with the equation: σ2 = (i · I) − (I2 · N) − 1, where i is the single-channel current and N is the number of channels. Conductance was calculated by dividing I by the holding potential of −60 mv. Variance resulting from slow drift (i.e., rundown or run-up) was corrected by local linear fitting of the drift, calculating the variance caused by this trend at each point, and subtracting the drift variance (scaled by the square current amplitude) from the total variance before fitting. This method yields accurate estimates of i and N when tested on simulated data with drift (Wagner et al., 2004).
Kinetic Modeling.
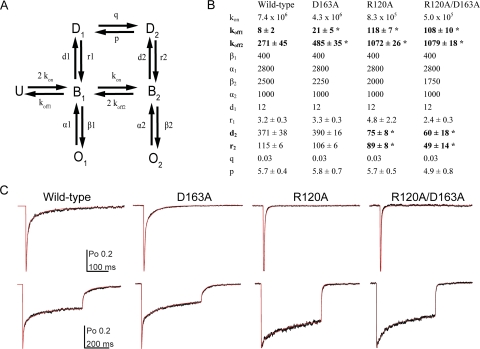
Kinetic modeling was performed with home-written software using the Q-matrix method (Colquhoun and Hawkes, 1995a,b). We used a simplified model of GABAA receptor behavior (Fig. 6) that had been described previously (Jones et al., 1998; Wagner et al., 2004; Goldschen-Ohm et al., 2010). Although we considered more complex variations of this model that included additional desensitized and open states, the simplified model was equally suitable to recapitulate our data. During optimization the rate constant kon and Po-max were constrained to the value obtained from experiments in this study; all other parameters were initially set to values reported by Goldschen-Ohm et al. (2010) and were unconstrained. Current responses from 3- and 500-ms pulses of saturating GABA were simultaneously fit for each patch. After initial optimization, only koff, r1, d2, r2, and p were left unconstrained, and fits were repeated. Optimization used a simplex algorithm to minimize the amplitude-weighted sum of squared errors between actual and simulated currents. In all cases, significant differences in transition rates were tested using a two-tailed unpaired Student's t test at a significance level of p < 0.05 (Prism 4).
Fig. 6.
Kinetic modeling demonstrates that the effects of R120A, D163A, and R120A/D163A can be similarly explained by faster unbinding rates, while differential changes in desensitization occur. A, the seven-state Markov model used to simulate GABA responses (U, unbound; B, bound; O, open; D, desensitized) is shown. B, rate constants used to simultaneously simulate responses to short and long pulses of saturating GABA for wild-type, D163A, R120A, and R120A/D163A are shown. The units are s−1 except for GABA binding steps, which are M−1 · s−1. Only koff1, koff2, r1, d2, r2, and p (in bold) are reported as ± S.E. because they were allowed to vary while the model was optimized. C, current responses (black traces) and simulated responses (red traces) to both short and long pulses of GABA are displayed for each receptor type. *, significant differences (P < 0.05) between control and mutant transition rates were calculated using a two-tailed unpaired Student's t test.
Results
In homology models β2Asp163 and α1Arg120 are juxtaposed across the “top” of the GABA binding pocket. To further explore the roles of these residues in receptor function and determine whether they functionally interact, we replaced each wild-type side chain with the methyl group of alanine. Alanine replacement serves to remove electrostatic or hydrogen bonding interactions inherent to the wild-type side chains and minimizes potentially confounding interactions from the introduced side chain. Receptors containing the α1R120A mutation, the β2D163A mutation, or both mutations were characterized.
β2-GKER Rescues α1R120A Expression and γ2 Rescues β2Asp163A Expression.
HEK-293 cells transfected with wild-type β2 and α1R120A display neither GABA-evoked (100 mM) nor propofol-evoked (300 μM) current. The propofol binding site is distinct from the GABA binding site where α1Arg120 is located. Therefore it is likely that the α1Arg120A mutation negatively affects functional expression of receptors in HEK-293 cells. To rescue functional expression, we used the β2-GKER construct. β2-GKER contains four point mutations where a given residue is replaced with its counterpart from the β3 subunit. This construct has been shown to rescue expression of another binding site mutant, α1R67A (Bollan et al., 2003), and has subsequently been used in our laboratory to restore functional expression to several other mutant constructs (D. A. Wagner, unpublished results).
Transfections with wild-type α1 and β2D163A also failed to display GABA- or propofol-evoked currents. In an attempt to rescue expression, the β2D163A mutation was recreated in the β2-GKER background, but this construct failed to give current. Ultimately, cotransfection with the γ2 subunit was necessary to obtain robust GABA-evoked currents. To ensure adequate expression and control for any influence of the γ2 subunit, we performed this study using cells transfected with α1β2-GKERγ2, α1β2-GKERD163Aγ2, α1R120Aβ2-GKERγ2, or α1R120Aβ2-GKERD163Aγ2, which shall be referred to from here on as wild type, D163A, R120A, or R120A/D163A, respectively.
The sensitivity of this region to mutations was additionally observed during our attempts to express charge reversal mutants of each residue (D163R and R120D), alone or in concert as a charge swap. Transfection with either charge reversal or the swap failed to express functional receptors when using the β2-GKER construct and the γ2 subunit.
Double-Mutant Cycle Analysis of D163A and R120A using EC50-GABA Yields a Weak Coupling Energy.
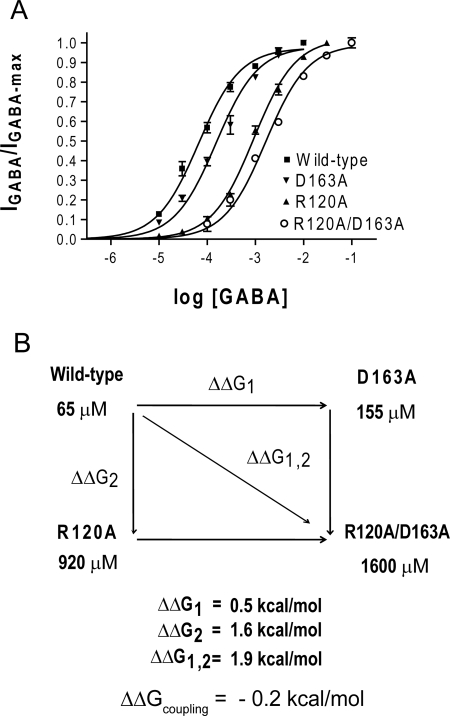
The primary goal of this study was to determine whether β2Asp163 and α1Arg120 participate in an intersubunit salt bridge. A tool that can be used to this end is the method of double-mutant cycle analysis, which quantifies the coupling energy between two mutated residues and clarifies the likelihood of two residues interacting (Horovitz, 1996). One parameter that has been commonly used for double-mutant cycle analysis in the study of LGICs is the apparent affinity for ligand, or EC50 (Kash et al., 2003; Price et al., 2007; Gleitsman et al., 2008). Therefore, the effects of the D163A and R120A mutations were initially characterized by determining the peak current EC50-GABA value through concentration-response experiments. The D163A mutation caused a 2.4-fold increase in EC50-GABA (155 μM) compared with the wild type (65 μM), whereas the R120A mutation had a much larger effect on EC50-GABA, causing a 14-fold shift to 900 μM. Receptors containing both mutations displayed a 25-fold increase in EC50-GABA (1600 μM) (Fig. 2A).
Fig. 2.
Mutant cycle analysis of EC50-GABA indicates α1Arg120 and β2Asp163 are weakly coupled. A, GABA concentration-response curves for single- and double-alanine mutations at α1Arg120 and β2Asp163 when measuring peak GABA responses. B, the mutant cycle for these alanine mutations. Equations for calculating the change in free energy associated with each mutation and the overall coupling energy are listed under Materials and Methods.
EC50 values were input into a mutant cycle to obtain a coupling energy of 0.2 kcal/mol (Fig. 2B and Table 2). If β2Asp163 and α1Arg120 are functionally independent with respect to EC50-GABA, we would expect the coupling energy to be 0 kcal/mol. Any value that deviates from zero may indicate coupling, but a more stringent criterion (i.e., a coupling energy of at least 0.5 kcal/mol) is typically used to identify direct interactions between two side chains.
TABLE 2.
Summary of macroscopic and microscopic parameters used for double-mutant cycle analysis
| Wild type | D163A | R120A | R120A/D163A | ΔΔGcoupling | |
|---|---|---|---|---|---|
| kcal/mol | |||||
| EC50-GABA | 65 μM | 155 μM | 920 μM | 1600 μM | 0.2 |
| Deactivation τw | 37.4 ms | 15.6 ms* | 3.5 ms* | 3.5 ms* | 0.5 |
| kon | 7.4 × 106 M−1 · s−1 | 4.3 × 106 M−1 · s−1* | 8.3 × 105 M−1 · s−1* | 5.0 × 105 M−1 · s−1* | 0.03 |
| koff1 | 8 s−1* | 21 s−1* | 118 s−1* | 108 s−1* | 0.63 |
| koff2 | 271 s−1* | 485 s−1* | 1072 s−1* | 1079 s−1* | 0.34 |
P < 0.05, significant differences between control and mutant parameters were calculated using ANOVA with a Dunnett's post test.
Although studies of LGICs commonly use EC50 values to derive thermodynamic energies, these results can be confounded by the complex nature of the EC50 value, which depends on multiple microscopic processes that underlie both ligand affinity and channel gating (Colquhoun, 1998; Gleitsman et al., 2008). Therefore, coupling energies calculated from EC50 values may be skewed, particularly when exploring interactions that influence multiple parameters or exist only in certain receptor states. The significance of the weak coupling energy found here is unclear, and we cannot confirm or exclude an interaction between β2Asp163 and α1Arg120 with this evidence.
D163A and R120A Independently Reduce the GABA Binding Rate.
Because analysis of EC50-GABA was indeterminate, we directly measured the microscopic binding rates for GABA for each construct using race experiments, as described previously by Jones et al. (2001). In brief, the first step in this process is to directly measure the binding rate for a competitive antagonist, in this case SR-95531. Once the binding rate for SR-95531 (kon-SR) is determined, the binding rate of the agonist, GABA, can be measured by performing an experiment in which GABA and SR-95531 are coapplied. The resulting current is compared with the current evoked by an application of GABA alone. The extent to which the peak current is reduced by the presence of antagonist depends on the relative binding rates of the two compounds. Because the binding rate of SR-95531 has been determined, the binding rate of GABA (kon-GABA) can be calculated.
We characterized the binding kinetics of the competitive antagonist SR-95531 for each receptor type. Antagonist unbinding experiments were used as described previously (Jones et al., 2001), and we measured the dissociation constant (Kd-SR) and microscopic unbinding rate (koff-SR) for each receptor type (Supplemental Fig. 1). Each mutant construct caused a small, albeit significant (≈ 30%), reduction in koff-SR, but there were no measurable changes in Kd-SR (Table 1). kon-SR, which was determined using the equation kon-SR = koff-SR/Kd-SR, was also not significantly affected by any of the mutations.
TABLE 1.
Summary of microscopic binding and unbinding rates and macroscopic affinity for the competitive antagonist SR-95531
| KD-SR | koff-SR | kon-SR | |
|---|---|---|---|
| nM | s−1 | M−1 · s−1 | |
| Wild type | 140 | 15.9 ± 0.8 | 1.2 ± 0.1 × 108 |
| D163A | 110 | 10.3 ± 0.7* | 9.7 ± 0.6 × 107 |
| R120A | 100 | 11.5 ± 0.4* | 1.2 ± 0.1 × 108 |
| R120A/D163A | 100 | 11.1 ± 0.9* | 1.2 ± 0.1 × 108 |
P < 0.05, significant differences between control and mutant parameters were calculated using ANOVA with a Dunnett's post test.
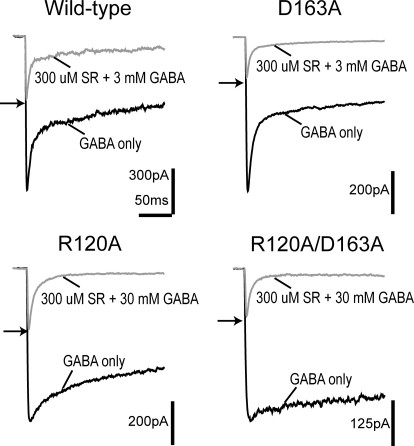
Figure 3A depicts the results of the race experiment. The ratio of the peak response of coapplication to the peak response to a control application of GABA alone is the result of the relative binding rates and concentrations of GABA and SR-95531. Using this ratio, termed Irace, the GABA binding rate can be computed as kon-GABA = [SR-95531] kon-SR/([GABA](1/Irace − 1)) (Jones et al., 1998). Application of 3 mM GABA and 300 μM SR-95531 gave an Irace of 0.41 ± 0.04 for the wild-type receptor, whereas for the same concentrations Irace was reduced to 0.27 ± 0.01 for D163A, indicating a slower binding rate for GABA. R120A and R120A/D163A receptors required a 10-fold increase in the concentration of GABA (30 mM) coapplied with 300 μM SR-95531 to obtain Irace values of 0.37 ± 0.03 and 0.31 ± 0.01, respectively, indicating even greater reductions in the GABA binding rate than seen for D163A receptors. Indeed, when these Irace values are used to calculate kon-GABA all of the mutants display significantly reduced GABA binding rates (wild type, 7.4 ± 0.4 × 106 M−1 · s−1; D163A, 4.3 ± 0.5 × 106 M−1 · s−1; R120A, 8.3 ± 0.8 × 105 M−1 · s−1; R120A/D163A, 5.0 ± 0.3 × 105 M−1 · s−1).
Fig. 3.
GABA binds more slowly to R120A, D163A, and R120A/D163A. Race experiments of either single-alanine mutation or the double-alanine mutation of α1Arg120 and β2Asp163 are shown. For wild-type and D163A receptors, currents evoked by simultaneous application of 3 mM GABA and 300 μM SR-95531 (gray traces) were compared with the current evoked by 3 mM GABA alone (black traces). For R120A and R120A/D163A receptors, currents evoked by simultaneous application of 30 mM GABA and 300 μM SR-95531 (gray traces) were compared with the current evoked by 30 mM GABA alone (black traces). The two separate applications are overlaid for each receptor type, and the ratio of the peak currents is indicated by an arrow.
When these numbers are subjected to double-mutant cycle analysis the resulting coupling energy is effectively nil (−0.03 kcal/mol) (Table 2). Therefore, the mutations R120A and D163A demonstrate additive effects on the GABA binding rate, strongly indicative of an independent relationship. During the binding process, a salt bridge between these residues is either irrelevant or nonexistent.
D163A and R120A Accelerate the Deactivation Phase and Display Significant Coupling.
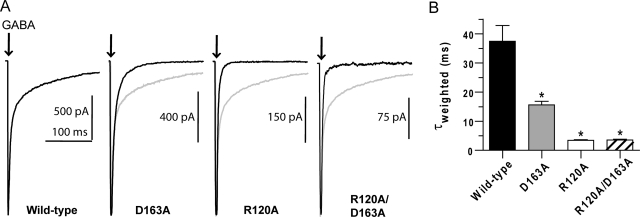
Each mutation causes significant changes in EC50 and the GABA binding rate; therefore, it is evident that these residues are important for normal receptor function. Examination of the macroscopic kinetics associated with GABA-evoked currents allowed us to further uncover the impact each mutation has on receptor function. We characterized the receptor kinetics of macroscopic deactivation after a brief (3 ms) pulse of saturating GABA, similar to that occurring during synaptic transmission (Fig. 4). When the deactivation time constants were extracted by fitting biexponential functions it was observed that both single mutations significantly accelerated deactivation (wild type, τw = 37.4 ± 5.5 ms; D163A, τw = 15.6 ± 1.2 ms; R120A, τw = 3.5 ± 0.2 ms). In addition, the double mutant has the same deactivation time constant as R120A (R120A/D163A: τw = 3.5 ± 0.3 ms).
Fig. 4.
Deactivation is faster for R120A, D163A, and R120A/D163A. A, macroscopic current responses to a 3-ms pulse of saturating GABA (indicated by arrow). Each mutant response is overlaid with the normalized wild-type response (light gray lines). B, summary of weighted time constants (τw) for deactivation, generated from biexponential fits of the macroscopic currents. τw is computed as Σai · τi/Σai, where ai and τi are the amplitude and time constant of component i, respectively. *, significant differences (P < 0.05) between control and mutant values were calculated using ANOVA with a Dunnett's post test.
Double-mutant cycle analysis of the deactivation time constant revealed a significant coupling energy of 0.5 kcal/mol (Table 2). This coupling energy suggests that the contributions of β2Asp163 and α1Arg120 to the function of deactivation are not independent and that they interact during this phase. Deactivation has a very complex nature and is comprised of numerous microscopic parameters, any of which could be functionally coupled for β2Asp163 and α1Arg120. Changes in the deactivation phase are often associated with changes in the microscopic unbinding rate, but other transitions such as desensitization and channel closing can influence deactivation (Jones and Westbrook, 1995). To investigate which of these microscopic transitions are responsible for the coupling found in the deactivation time constants of β2Asp163 and α1Arg120, we performed kinetic modeling as presented under Microscopic Unbinding Rates Are Strongly Coupled for D163A and R120A.
R120A Suppresses Desensitization.
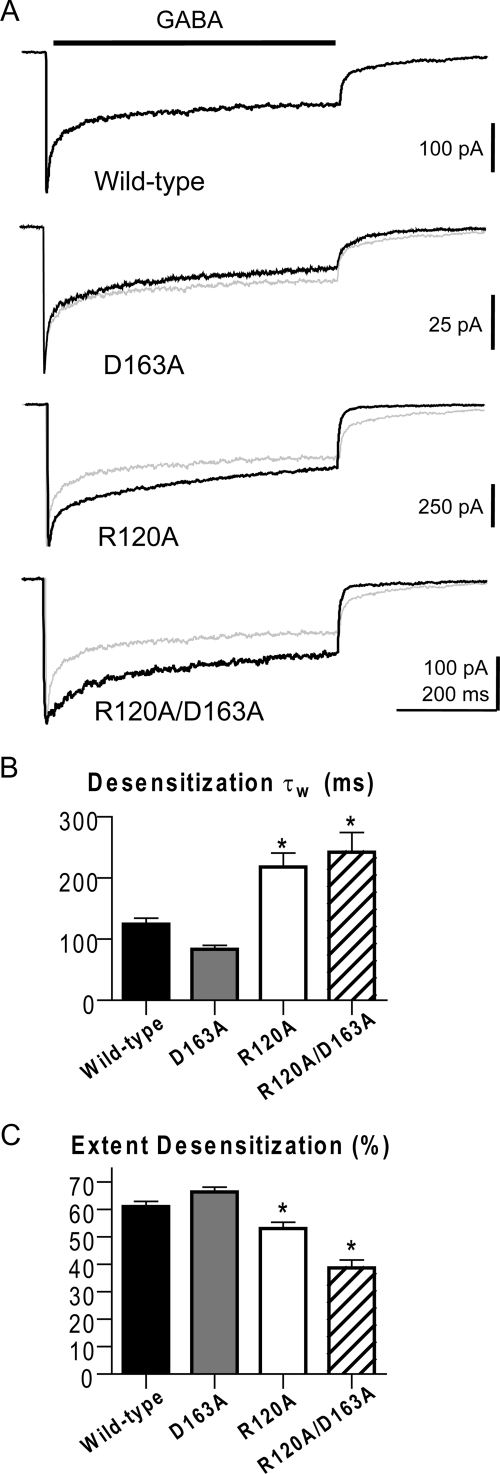
Further assessment of macroscopic data was conducted to explore the functional effect of D163A and R120A and provide additional constraint for kinetic modeling. Macroscopic desensitization was characterized during a long (500 ms) pulse of saturating GABA, and the resulting desensitization phase was fit with a biexponential function from which a weighted time constant was calculated (Fig. 5). Unlike D163A, which had little effect on desensitization, R120A and R120A/D163A showed a visible reduction of desensitization in raw traces (Fig. 5A). Comparison of the weighted τ values revealed R120A and R120A/D163A had significantly slower desensitization (wild type, τw = 124 ± 10 ms; D163A, τw = 85 ± 6 ms; R120A, τw = 218 ± 23 ms; R120A/D163A, τw = 242 ± 33 ms) (Fig. 5B). The extent of desensitization displayed by R120A and R120A/D163A was similarly reduced, unlike that of D163A, which remained normal (wild type, 61 ± 2%; D163A, 66 ± 2%; R120A, 53 ± 2%; R120A/D163A, 39 ± 3%) (Fig. 5C).
Fig. 5.
Macroscopic desensitization during a long GABA pulse. A, macroscopic current responses to a 500-ms pulse of saturating GABA (indicated by black bar above traces). Each mutant response is overlaid with the normalized wild-type response (light gray lines). B and C, macroscopic currents for all receptor types were fit with a biexponential equation, although several responses for R120A and R120A/D163A required only a monoexponential equation during fits. B, summary of weighted time constants for desensitization. C, summary of the extent of desensitization after 500 ms. *, significant differences (P < 0.05) between control and mutant values were calculated using ANOVA with a Dunnett's post test.
Microscopic Unbinding Rates Are Strongly Coupled for D163A and R120A.
To determine the microscopic basis for the coupling of deactivation effects caused by D163A and R120A we used a previously established seven-state kinetic model (Fig. 6A) (Jones and Westbrook, 1995; Wagner et al., 2004; Barberis et al., 2007; Goldschen-Ohm et al., 2010). This is a simplified model that recapitulates the dominant features present in our macroscopic αβγ receptor data. It incorporates several known features of GABAA receptor physiology, including two binding steps (Bormann and Clapham, 1985), multiple open states (Macdonald et al., 1989), and desensitized states that can occur before opening (Burkat et al., 2001). We also used an unlinked unbinding step, as was required by Goldschen-Ohm et al. (2010), which was necessary to consistently simulate our experimental data.
Before model optimization we performed nonstationary variance analysis (Supplemental Fig. 2) (Sigworth, 1980). This assessment provided a measure of both the single-channel conductance and maximal open probability (Po-max) of each receptor type, which can be used to further constrain our modeling. None of the mutations altered conductance (γ) at −60 mV or the Po-max (wild type, γ = 32 ± 3 pS, Po-max = 0.59 ± 0.05; D163A, γ = 33 ± 3 pS, Po-max = 0.55 ± 0.03; R120A, γ = 29 ± 3 pS, Po-max = 0.58 ± 0.06; R120A/D163A, γ = 31 ± 3 pS, Po-max = 0.57 ± 0.04).
We simultaneously fit current responses to 3- and 500-ms pulses of saturating GABA for each receptor type. Initially kon and Po-max were fixed to their experimentally determined rates and the remaining parameters were set at the values previously reported by Goldschen-Ohm et al. (2010). The model was then optimized under relatively tight constraints until values for the opening and closing rates (α1, β1, α2, β2), as well as for entry into D1, were obtained that gave consistently good fits to our data. These values were identical for all constructs except for the opening rate of the dominant open state (β2), which varied for each mutant to account for the slower rise times observed for the mutant constructs during responses to saturating concentrations of GABA (Supplemental Fig. 3). With these values constrained, a second optimization run was performed in which only the unbinding rates (koff) and the rates for entering and leaving the doubly bound desensitized state (D2) were allowed to vary.
Our model quantitatively reproduced both wild-type and mutant data (Fig. 6, B and C). The models for all three of our mutant constructs display slower binding and faster unbinding of GABA relative to wild type. The models for R120A and R120A/D163A also have slower entry and a more rapid exit from the doubly bound desensitized state (D2). When double-mutant cycle analysis is applied to the modeled unbinding rates, coupling energies of 0.63 and 0.34 kcal/mol were calculated for koff1 and koff2, respectively (Table 2). This indicates that coupled effects on unbinding are the basis for the coupling seen in deactivation.
Discussion
To build on our understanding of the structure of the GABA binding site and the functional significance of specific residues, we experimentally tested for the existence of an interaction across the β/α interface between β2Asp163 and α1Arg120. It has been demonstrated that mutating either residue alters the GABA concentration-response curve, but whether either of these residues is involved in binding, gating, or desensitization has been speculative. Here, the residues were mutated to alanine, and double-mutant cycle analysis was applied to a variety of macroscopic and microscopic parameters. Intriguingly, the residues seemed to be completely independent when considering the binding of GABA, but they were coupled when looking at the unbinding of GABA. These results suggest that β2Asp163 and α1Arg120 do not interact in the unbound state but form an interaction upon binding of GABA.
Evidence for a Salt Bridge between β2Asp163 and α1Arg120.
The effects on unbinding (koff1) for β2D163A and α1R120A were coupled with an energy of 0.63 kcal/mol. This energy is slightly lower than, but consistent with, energies reported for confirmed surface salt bridges in other proteins (0.86 and 0.95 kcal/mol) (Horovitz et al., 1990; Makhatadze et al., 2003). Low coupling energies were expected because salt bridges at solvent-exposed surfaces of a protein have significantly weaker interactions compared with those buried in the hydrophobic interior. In the case of β2Asp163 and α1Arg120, both are exposed to the aqueous environment, as demonstrated by SCAM studies (Newell et al., 2004; Kloda and Czajkowski, 2007). In addition, the strength of a salt bridge is influenced by the distance between the two residues. In homology models of the GABAA receptor, the nearest charged groups of β2Asp163 and α1Arg120 are 3.6 Å apart (O'Mara et al., 2005), just within range for a salt bridge. Because the coupling energy between pairs of residues decreases with distance, the weaker coupling energy measured in our experiments may be the consequence of this distance. Therefore, we believe the coupling energy observed with β2D163A and α1R120A represents the loss of a salt bridge.
It is also important to acknowledge additional interpretations to a significant coupling energy. A predicted interaction may not exclusively be direct, but could result from secondary interactions through a third side chain or could be the result of indirect coupling caused by broader structural rearrangements or conformational changes. Although we cannot rule out such possibilities, the presumed proximity of these residues suggests a direct interaction.
A State-Dependent Interaction.
Transient salt bridges may provide a mechanism for governing conformational changes and stabilization of specific receptor states. In this study, double-mutant cycle analysis revealed that an interaction between β2Asp163 and α1Arg120 occurred during unbinding, but not binding steps, indicating that an interaction between these two residues stabilizes the bound-closed states (B1 and B2 on our model). A state-dependent interaction is not difficult to envision, where these residues are coupled during the GABA-bound state, but not during the unbound state. In the unbound receptor no interaction is present; when GABA binds, conformational changes occur that move the residues into position to interact. Conformational rearrangements in response to GABA binding are expected and have been repeatedly observed in studies of the binding pocket (Wagner and Czajkowski, 2001; Newell et al., 2004; Muroi et al., 2006). In fact, β2Asp163 itself, along with other residues on loop B of the β2 subunit, has been shown to undergo rearrangements in response to receptor activation (Newell et al., 2004). This movement may underlie the state dependence of the interaction between β2Asp163 and α1Arg120. State-dependent electrostatic interactions for residues coupled to the open state of nicotinic acetylcholine receptors have also been proposed (Kash et al., 2003; Gleitsman et al., 2008). In addition, there is growing evidence in the field of protein structure supporting the occurrence of salt-bridge switching, especially where networks of charged residues are involved (Law and Lightstone, 2009).
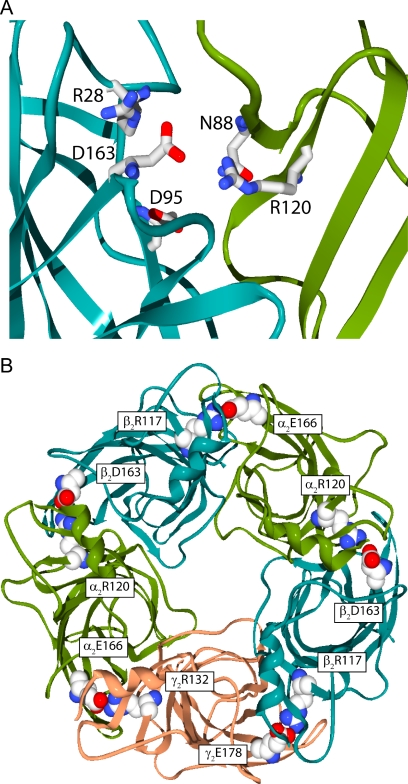
Aspartic acid and arginine are capable of forming multiple electrostatic interactions. Aspartic acid has two partial negative charges distributed between the two oxygen atoms of the carboxylic group, and arginine has three nitrogens in its guanidinium group. Therefore, it is likely that β2Asp163 and α1Arg120 participate in a larger ionic network. Although no specific additional interactions for β2Asp163 and α1Arg120 have been identified, a number of polar side chains are found in close proximity. Candidates for participation in this extended ionic network include, but are not limited to, α1Asn88, β2Arg28, β2Asp95, and nearby backbone amides and carbonyls (Fig. 7A). α1Asn88 is just one example of numerous polar residues located nearby that could also participate in an ionic network. The homology model based on the acetylcholine binding protein actually shows β2Arg28 participating in a ternary ionic network with β2Asp163 and α1Arg120 (Cromer et al., 2002).
Fig. 7.
Possible intersubunit ionic networks exist in homology models of the GABAA receptor. A, side view of the extracellular domain at a single β/α interface. Polar side chains in the vicinity of β2Asp163 and α1Arg120, which may contribute to the ionic network, are depicted. B, view from the extracellular side looking through the channel pore. β2Asp163 and α1Arg120 are located at the β/α intersubunit interfaces and are also conserved at the other interfaces.
β2Asp95, located on loop A and proximal to α1Arg120, is particularly intriguing. The equivalent residues in the glycine receptor (α1Asp97 and α1Arg119) participate in a state-dependent intersubunit electrostatic interaction (Todorovic et al., 2010). This interaction is proposed to exist when the glycine receptor is in the unbound state and is broken upon the binding of agonist. This result from the glycine receptor argues for a model in which α1Arg120 of the GABA receptor interacts with β2Asp95 (on loop A) in the unbound state and is handed off to β2Asp163 (on loop B) upon binding of agonist.
Discontinuity in Functional Effects.
β2Asp163 and α1Arg120 seem to interact in the ligand-bound state; however, the residues have asymmetric roles in desensitization. α1R120A displays greatly reduced desensitization, whereas β2D163A displays desensitization indistinguishable from wild type. We propose that these residues, as part of a dynamic region, can stabilize the bound-closed receptor without necessarily influencing the rate-limiting transitions of desensitization. The additional effect observed for α1R120A may be the byproduct of its involvement in a complex electrostatic network. It is highly probable that a simple binary interaction is not occurring between β2Asp163 and α1Arg120. Several examples of an arginine residue simultaneously making multiple interactions have been identified (Horovitz et al., 1990; Borders et al., 1994). The multifaceted functional group of α1Arg120 may be participating in an additional interaction that is separate from β2Asp163 and is critical to desensitization transitions. Under these circumstances, an interaction between β2Asp163 and α1Arg120 could exist in all bound receptor states and still yield asymmetric effects when either is mutated.
Functional Role of β2Asp163-α1Arg120 Interaction.
The interaction between β2Asp163 and α1Arg120 seems to be highly conserved, not just at the β/α interface where a negative and a positive residue are found at these positions on every isoform of β and α subunits, but also at other intersubunit interfaces (Fig. 7B). A corresponding interaction at the γ/β interface has been identified (Goldschen-Ohm et al., 2010). This interaction consisted of a triad of charge residues (β2Arg117, γ2Glu178, and γ2Arg43) forming a salt-bridge network. The additional arginine (γ2Arg43) is conserved at the β/α interface (β2Arg28), but was not investigated in our current study. This conserved motif may play an important role in establishing the architecture at each subunit interface.
Goldschen-Ohm et al. (2010) reported that mutation at any of the residues in this motif slowed deactivation after a GABA response. Not only is this significant because it demonstrates that a homologous interaction influences ligand binding at other subunit interfaces, but it is also significant because disruption of this interaction at the γ/β interface had the opposite effect of disruption of the corresponding interaction at the β/α interface. Mutation of either β2Asp163 or α1Arg120 significantly increases the rate of deactivation. We propose this intersubunit motif is involved in subunit positioning. In this scenario, breaking of the interaction at the β/α interface may increase the distance between the β and α subunits, speeding unbinding, whereas breaking the interaction at the γ/β interface may reduce the distance between the β and α subunits at the β/α interface, slowing unbinding.
The GABA Binding Site Is Involved in Desensitization.
This study is the first to offer in-depth characterization of α1Arg120. We provide results that demonstrate α1Arg120 has important roles in GABA binding/unbinding and desensitization. It should not be lost in this discussion that α1Arg120 is the first residue at the GABA binding pocket that shows a dramatic influence in desensitization (particularly the early, fast phase). Most work regarding desensitization has focused on the transmembrane domains, and previous studies have demonstrated a significant role for the pre-M1 region in transducing desensitization to the pore surrounded by the transmembrane 2 domain (Bianchi and Macdonald, 2002). The identification of α1Arg120 may provide a unique focus for studying how desensitization is transduced from the GABA binding site.
Supplementary Material
Acknowledgments
We thank Marcel Goldschen-Ohm for assistance with data analysis.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
This research was supported by the National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant NS055793].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/mol.110.068619.
ABBREVIATIONS:
- LGIC
- ligand-gated ion channel
- SCAM
- substituted cysteine accessibility method
- HEK
- human embryonic kidney
- SR-95531
- 6-imino-3-(4-methoxyphenyl)-1(6H)-pyridazinebutanoic acid hydrobromide
- ANOVA
- analysis of variance
- 5-HT
- serotonin.
Authorship Contributions
Participated in research design: Laha and Wagner.
Conducted experiments: Laha.
Performed data analysis: Laha and Wagner.
Wrote or contributed to the writing of the manuscript: Laha and Wagner.
Other: Wagner acquired funding for the research.
References
- Barberis A, Mozrzymas JW, Ortinski PI, Vicini S. (2007) Desensitization and binding properties determine distinct α1β2γ2 and α3β2γ2 GABAA receptor-channel kinetic behavior. Eur J Neurosci 25:2726–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann SW, Baur R, Sigel E. (2002) Forced subunit assembly in α1β2γ2 GABAA receptors. Insight into the absolute arrangement. J Biol Chem 277:46020–46025 [DOI] [PubMed] [Google Scholar]
- Benke D, Fritschy JM, Trzeciak A, Bannwarth W, Mohler H. (1994) Distribution, prevalence, and drug binding profile of γ-aminobutyric acid type A receptor subtypes differing in the β-subunit variant. J Biol Chem 269:27100–27107 [PubMed] [Google Scholar]
- Bianchi MT, Macdonald RL. (2002) Slow phases of GABAA receptor desensitization: structural determinants and possible relevance for synaptic function. J Physiol 544:3–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollan K, King D, Robertson LA, Brown K, Taylor PM, Moss SJ, Connolly CN. (2003) GABAA receptor composition is determined by distinct assembly signals within α and β subunits. J Biol Chem 278:4747–4755 [DOI] [PubMed] [Google Scholar]
- Borders CL, Jr, Broadwater JA, Bekeny PA, Salmon JE, Lee AS, Eldridge AM, Pett VB. (1994) A structural role for arginine in proteins: multiple hydrogen bonds to backbone carbonyl oxygens. Protein Sci 3:541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormann J, Clapham DE. (1985) γ-Aminobutyric acid receptor channels in adrenal chromaffin cells: a patch-clamp study. Proc Natl Acad Sci USA 82:2168–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. (2001) Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411:269–276 [DOI] [PubMed] [Google Scholar]
- Burkat PM, Yang J, Gingrich KJ. (2001) Dominant gating governing transient GABAA receptor activity: a first latency and Po/o analysis. J Neurosci 21:7026–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D. (1998) Binding, gating, affinity and efficacy: the interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. Br J Pharmacol 125:924–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D, Hawkes AG. (1995a) The principles of the stochastic interpretation of ion-channel mechanisms, in Single-Channel Recording (Sakmann B, Neher E. eds) pp 397–482, Plenum, New York [Google Scholar]
- Colquhoun D, Hawkes AG. (1995b) A Q-matrix cookbook. How to write only one program to calculate the single-channel and macroscopic predictions for any kinetic mechanism, in Single-Channel Recording (Sakmann B, Neher E. eds) pp 589–633, Plenum, New York [Google Scholar]
- Cromer BA, Morton CJ, Parker MW. (2002) Anxiety over GABAA receptor structure relieved by AChBP. Trends Biochem Sci 27:280–287 [DOI] [PubMed] [Google Scholar]
- Gleitsman KR, Kedrowski SM, Lester HA, Dougherty DA. (2008) An intersubunit hydrogen bond in the nicotinic acetylcholine receptor that contributes to channel gating. J Biol Chem 283:35638–35643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschen-Ohm MP, Wagner DA, Petrou S, Jones MV. (2010) An epilepsy-related region in the GABAA receptor mediates long-distance effects on GABA and benzodiazepine binding sites. Mol Pharmacol 77:35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz A. (1996) Double-mutant cycles: a powerful tool for analyzing protein structure and function. Fold Des 1:R121–R126 [DOI] [PubMed] [Google Scholar]
- Horovitz A, Serrano L, Avron B, Bycroft M, Fersht AR. (1990) Strength and co-operativity of contributions of surface salt bridges to protein stability. J Mol Biol 216:1031–1044 [DOI] [PubMed] [Google Scholar]
- Jones MV, Jonas P, Sahara Y, Westbrook GL. (2001) Microscopic kinetics and energetics distinguish GABAA receptor agonists from antagonists. Biophys J 81:2660–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MV, Sahara Y, Dzubay JA, Westbrook GL. (1998) Defining affinity with the GABAA receptor. J Neurosci 18:8590–8604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MV, Westbrook GL. (1995) Desensitized states prolong GABAA channel responses to brief agonist pulses. Neuron 15:181–191 [DOI] [PubMed] [Google Scholar]
- Kash TL, Jenkins A, Kelley JC, Trudell JR, Harrison NL. (2003) Coupling of agonist binding to channel gating in the GABAA receptor. Nature 421:272–275 [DOI] [PubMed] [Google Scholar]
- Kash TL, Trudell JR, Harrison NL. (2004) Structural elements involved in activation of the γ-aminobutyric acid type A (GABAA) receptor. Biochem Soc Trans 32:540–546 [DOI] [PubMed] [Google Scholar]
- Kloda JH, Czajkowski C. (2007) Agonist-, antagonist-, and benzodiazepine-induced structural changes in the α1 Met113-Leu132 region of the GABAA receptor. Mol Pharmacol 71:483–493 [DOI] [PubMed] [Google Scholar]
- Law RJ, Lightstone FC. (2009) Modeling neuronal nicotinic and GABA receptors: important interface salt-links and protein dynamics. Biophys J 97:1586–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald RL, Rogers CJ, Twyman RE. (1989) Kinetic properties of the GABAA receptor main conductance state of mouse spinal neurones in culture. J Physiol 410:479–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhatadze GI, Loladze VV, Ermolenko DN, Chen X, Thomas ST. (2003) Contribution of surface salt bridges to protein stability: guidelines for protein engineering. J Mol Biol 327:1135–1148 [DOI] [PubMed] [Google Scholar]
- McKernan RM, Whiting PJ. (1996) Which GABAA-receptor subtypes really occur in the brain? Trends Neurosci 19:139–143 [DOI] [PubMed] [Google Scholar]
- Muroi Y, Czajkowski C, Jackson MB. (2006) Local and global ligand-induced changes in the structure of the GABAA receptor. Biochemistry 45:7013–7022 [DOI] [PubMed] [Google Scholar]
- Newell JG, McDevitt RA, Czajkowski C. (2004) Mutation of glutamate 155 of the GABAA receptor β2 subunit produces a spontaneously open channel: a trigger for channel activation. J Neurosci 24:11226–11235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mara M, Cromer B, Parker M, Chung SH. (2005) Homology model of the GABAA receptor examined using Brownian dynamics. Biophys J 88:3286–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett CL, Hanek AP, Lester HA, Dougherty DA, Lummis SC. (2007) Unnatural amino acid mutagenesis of the GABAA receptor binding site residues reveals a novel cation-π interaction between GABA and β2Tyr97. J Neurosci 27:886–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price KL, Millen KS, Lummis SC. (2007) Transducing agonist binding to channel gating involves different interactions in 5-HT3 and GABAC receptors. J Biol Chem 282:25623–25630 [DOI] [PubMed] [Google Scholar]
- Sigworth FJ. (1980) The variance of sodium current fluctuations at the node of Ranvier. J Physiol 307:97–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PM, Thomas P, Gorrie GH, Connolly CN, Smart TG, Moss SJ. (1999) Identification of amino acid residues within GABAA receptor β subunits that mediate both homomeric and heteromeric receptor expression. J Neurosci 19:6360–6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorovic J, Welsh BT, Bertaccini EJ, Trudell JR, Mihic SJ. (2010) Disruption of an intersubunit electrostatic bond is a critical step in glycine receptor activation. Proc Natl Acad Sci USA 107:7987–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DA, Czajkowski C. (2001) Structure and dynamics of the GABA binding pocket: a narrowing cleft that constricts during activation. J Neurosci 21:67–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DA, Czajkowski C, Jones MV. (2004) An arginine involved in GABA binding and unbinding but not gating of the GABAA receptor. J Neurosci 24:2733–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westh-Hansen SE, Witt MR, Dekermendjian K, Liljefors T, Rasmussen PB, Nielsen M. (1999) Arginine residue 120 of the human GABAA receptor α1 subunit is essential for GABA binding and chloride ion current gating. Neuroreport 10:2417–2421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.