Abstract
Phosphorylation of the μ opioid receptor (MOPr), mediated by several protein kinases, is a critical process in the regulation of MOPr signaling. Although G protein-coupled receptor kinases are known to play an essential role in the agonist-induced phosphorylation and desensitization of MOPr, evidence suggests that other protein kinases, especially protein kinase C (PKC), also participate in the regulation of MOPr signaling. In this study, we investigated the biochemical nature and downstream effects of PKC-mediated MOPr phosphorylation. We observed in vitro phosphorylation of the MOPr C terminus by purified PKC. Protein mass spectrometry and site-directed mutagenesis implicated Ser363 of MOPr as the primary substrate for PKC, and this was confirmed in Chinese hamster ovary cells stably expressing full-length MOPr using an antibody that specifically recognizes phosphorylated Ser363. Alanine mutation of Ser363 did not affect the affinity of MOPr-ligand binding and the efficiency of receptor G-protein coupling. However, the S363A mutation attenuated the desensitization of receptor G-protein coupling induced by phorbol 12-myristate. Our research thus has identified a specific PKC phosphorylation site in MOPr and demonstrated that PKC-mediated phosphorylation of MOPr induces receptor desensitization at the G protein coupling level.
Introduction
Opioid receptors belong to the G protein-coupled receptor (GPCR) superfamily and consist of three structurally distinct subtypes, μ, δ, and κ. Among them, the μ opioid receptor (MOPr) is the primary target through which most opioid drugs execute their biological effects (Pasternak, 2004). Similar to other GPCRs, MOPr is subject to a variety of regulatory processes, including agonist-induced adaptive changes at the receptor level, such as phosphorylation, desensitization, internalization, and down-regulation (Law and Loh, 1999). Adaptive changes of MOPr are believed to contribute to the development of opioid tolerance and dependence (Harrison et al., 1998).
Research on the adaptive changes in MOPr has shown that receptor phosphorylation plays a critical step in the initiation and regulation other adaptive changes (Yu et al., 1997; Burd et al., 1998; El Kouhen et al., 2001). Site-directed mutagenesis has revealed multiple sites of phosphorylation of MOPr on intracellular domains of the receptor, with most of them located near the C terminus (Pak et al., 1997; Burd et al., 1998; El Kouhen et al., 2001; Wang et al., 2002; Schulz et al., 2004). MOPr phosphorylation can be mediated by two types of serine/threonine protein kinases, G protein-coupled receptor kinases (GRKs) (Zhang et al., 1998; Celver et al., 2001), and second messenger-activated kinases, including protein kinase A (Bernstein and Welch, 1998), protein kinase C (PKC) (Zhang et al., 1996), and Ca2+/calmodulin-dependent protein kinase II (Koch et al., 1997). GRKs phosphorylate the agonist-occupied receptor and trigger binding of arrestin to the receptor. The binding of arrestin causes receptor desensitization by uncoupling of the receptor from the G protein and promotes receptor internalization by targeting the receptor to clathrin-coated pits (Ferguson et al., 1998). Much less is known about phosphorylation of MOPr mediated by the second messenger-activated kinases. However, these kinases, particularly PKC, have been shown to play important roles in regulating MOPr function.
The PKC family contains at least 10 isoforms. Based on requirements for second messengers in the activation of kinases, PKCs are divided into three subfamilies: conventional PKCs, including α, β, βII, and γ isoforms; novel PKCs, including δ, ε, η, and θ isoforms; and atypical PKCs, including ζ and λ isoforms (Mellor and Parker, 1998). The expression of each isoform in the central nervous system has been reported (Battaini, 2001). The first evidence for PKC regulation of MOPr was the finding that activation of PKC by PMA increased phosphorylation of MOPr 1(Zhang et al., 1996). However, it was unknown which residue was phosphorylated by PKC. Activation of PKC was also reported to attenuate the responses of MOPr to agonist stimulation (Wang et al., 1996; King et al., 1999). More recent work suggested that PKC was involved in the agonist-selective desensitization of MOPr (Bailey et al., 2009). Effects on the development of opioid tolerance of modifying PKC activity (Granados-Soto et al., 2000; Ueda et al., 2001) further support the importance of PKC in the regulation of MOPr signaling. Phosphorylation by PKC potentially resembles that by GRKs for regulating MOPr functions. However, the exact mechanisms are unclear, because PKC can phosphorylate and regulate multiple components in the MOPr signaling pathway, including G proteins (Chakrabarti and Gintzler, 2003), ion channels (King et al., 1999), and adenylate cyclase (AC) (Mandyam et al., 2002). Our limited knowledge about the details and effects of PKC-mediated phosphorylation of MOPr further obscures mechanisms underlying the regulation of MOPr by PKC.
In this study, using site-directed mutagenesis, protein mass spectrometry (MS), and a phosphospecific antibody, we investigated the phosphorylation of MOPr by PKC. We demonstrate that Ser363 near the C terminus of MOPr is specifically phosphorylated by PKC and that this phosphorylation desensitizes MOPr at the receptor G protein coupling level.
Materials and Methods
pGEX-5X-1 vector and glutathione-Sepharose 4B were from GE Healthcare (Chalfont St. Giles, Buckinghamshire, UK). pEGFP-N1 vector was from Clontech (Mountain View, CA), QuikChange mutagenesis kit was obtained from Stratagene (La Jolla, CA). [γ-32P]ATP (3000 Ci/mmol) was from PerkinElmer Life and Analytical Sciences (Waltham, MA). Rat brain PKC and trypsin were purchased from Promega (Madison, WI). PMA, proteinase inhibitor cocktail, phosphatase inhibitor cocktail, wheat germ agglutinin-agarose beads, and recombinant PKCα and ζ were from Sigma-Aldrich (St. Louis, MO). Recombinant PKC βII was from Calbiochem (San Diego, CA). Recombinant PKC γ and ε were obtained from Invitrogen (Carlsbad, CA). Anti-MOPr-C20 (rabbit) antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-pMOPrSer363 (mouse) was prepared by Precision Antibody (Columbia, MD). Anti-mouse and anti-rabbit horseradish peroxidase-linked antibodies were purchased from Kirkegaard and Perry Laboratories (Gaithersburg, MD).
Plasmid Constructions.
To construct the plasmid expressing GST-fused MOPr-CT proteins (pGEX-5X-1-MOPr-CT), the DNA coding MOPr-CT (residues 340–398) was amplified by PCR from the plasmid containing the full-length mouse MOPr cDNA. The PCR product was then inserted into pGEX-5X-1 vector, in which a GST-tag was fused to the N terminus of the recombinant protein. To construct the plasmid pEGFP-N1-MOPr, the DNA coding whole-mouse MOPr without stop codon was amplified by PCR from a plasmid containing the full-length mouse MOPr cDNA. The PCR product was then inserted into pEGFP-N1 vector, in which an EGFP-tag was fused to the C terminus of MOPr. To construct mutations, the QuikChange site-directed mutagenesis kit was used to mutate the putative phosphorylation residues to alanine using pGEX-5X-1-MOPr-CT plasmid or the pEGFP-N1-MOPr plasmid as templates. All constructions were confirmed by DNA sequencing.
Expression and Purification of Recombinant GST-Fusion Proteins.
pGEX-5X-1 plasmids containing either wild-type (WT) or mutated MOPr-CT were transformed into BL21 Escherichia coli. The bacteria were grown in Luria broth medium containing 100 mg/l ampicillin. Isopropyl-1-thio-β-galactopyranoside with a final concentration of 0.1 mM was used to induce the expression of recombinant proteins. Recombinant proteins were purified by affinity chromatography using a glutathione-Sepharose 4B column according to the manufacturer's protocol. All GST-fusion proteins were confirmed to have the expected molecular weight by SDS-polyacrylamide gel electrophoresis (PAGE). For MS, purified protein was digested by Factor Xa to release MOPr-CT from the fusion protein.
In Vitro Phosphorylation Assay.
Phosphorylation assays were performed according to the manufacturer's protocols. For all of the phosphorylation assays, 10 μg of purified protein was incubated with 0.5 μl of each protein kinase and 5 μCi 100 μM [γ-32P]ATP in the corresponding reaction buffer at 30°C for 30 min. Reaction was terminated by adding one-sixth volume of 6× SDS loading buffer and then heating at 95°C for 5 min. Protein samples were resolved by 12% SDS-PAGE. Total protein was quantified by densitometric scanning of the protein band stained with Coomassie Blue. Phosphorylated protein was visualized by autoradiography, and the corresponding radioactivity was measured by scintillation counting.
In-Gel Trypsin Digestion and Matrix-Assisted Laser Desorption Ionization-Time of Flight-Mass Spectrum Analyses.
MOPr-CT was incubated with PKC for 30 min under the conditions given above, except that [γ-32P]ATP was omitted, and then isolated by 12% SDS-PAGE. Protein bands of MOPr-CT were cut from the gel, reduced by incubation with 20 mM dithiothreitol for 1 h at 60°C, and then alkylated by incubation with 50 mM iodoacetamide for 45 min at room temperature. The gel was dried and trypsin-digested following the manufacturer's protocol. Digested protein was extracted with 50% acetonitrile and 0.1% trifluoroacetic acid, desalted with ZipTipc18, and then mixed at a ratio of 1:1 with 20 mg/ml dihydroxybenzoic acid and 1% phosphoric acid. The processed sample was analyzed by a bench-top matrix-assisted laser desorption ionization-time of flight-mass spectrum (MALDI-TOF-MS) (Omniflex; Bruker Daltonics, Billerica, MA) using a laser at 337 nm as the desorption/ionization source. Ions were accelerated at 19 kV and detected in reflective positive mode. Mass axis calibrations were accomplished by using peaks from trypsin autohydrolysis and human angiotensin II. The fragment ions observed in the mass spectra were analyzed by FindMod (http://ca.expasy.org/tools/findmod).
Cell Culture.
Chinese hamster ovary (CHO) cells were grown at 37°C in 5% CO2 in Ham's F-12 medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. To establish stable cell lines, the cells were transfected with either the wild-type EGFP-N1-MOPr or the S363A mutation plasmid using Lipofectamine 2000 reagent. After a limited dilution and 14 days of selection with 1.4 mg/ml G418 (Geneticin), colonies stably expressing wild type EGFP-MOPr or the S363A mutation were identified by radioligand-binding assay. Stable cell lines with similar receptor expression levels were selected and maintained under 0.6 mg/ml G418 selection pressure for the following research.
Western Blotting.
The CHO cells growing in a six-well plate were treated with or without 1 μM PMA for 20 min, washed with ice-scold PBS three times, and then lysed by adding 0.8 ml of lysis buffer (50 mM Tris-HCl pH 7.4, 100 mM NaCl, 5 mg/ml N-octyl-d-thioglucopyranoside, 1% NP-40, proteinase inhibitor cocktail, and phosphatase inhibitor cocktail) and incubated on ice for 30 min. The lysate was centrifuged at 14,000g in 4°C for 15 min. The protein concentration of the supernatant was measured using the Bradford assay. Samples containing equal amounts of protein (approximately 250 μg) were incubated with 30 μl of WGA-agarose beads at 4°C for 1 h with gentle shacking. Beads were washed three times with 1 ml of lysis buffer. The absorbed protein was eluted by adding 40 μl of 2× SDS loading buffer and boiled at 85°C for 10 min. Eluate (30 μl) was resolved by 8% SDS-PAGE and electrotransferred to a nitrocellulose membrane. The membrane was blocked with 5% milk in Tris-buffered saline containing 0.1% Tween 20 (TBS-T) for 1 h and then incubated with primary antibody diluted in TBS-T containing 5% milk overnight at 4°C. The membrane was washed three times for 10 min with TBS-T, followed by 1 h of incubation at room temperature with horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibodies at 1:5000 diluted in TBS-T containing 5% milk. Immunoreactive proteins were detected with a chemiluminescent regent.
[35S]GTPγS Binding.
Stable cell lines of CHO expressing either EGFP-MOPr or EGFP-MOPr-S363A were pretreated with or without 1 μM drug (DAMGO or PMA) for 20 min then washed with ice-cold PBS three times. Cells were harvested in buffer (50 mM Tris-HCl, pH 7.4, 10 mM EGTA, and 5 mM MgCl2) and homogenized with a hand-held homogenizer. The homogenate was centrifuged at 40,000g in 4°C for 20 min, and the pellet was resuspended in membrane buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, and 0.1% bovine serum albumin). Cell membranes (30 μg of protein) were first incubated with 100 μM GDP at 30°C for 10 min, and then incubated with appropriate concentrations of DAMGO (range, 10−7–10−10 M) in a final volume of 500 μl of assay buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 10 mM MgCl2, 1 mM EGTA, and 80 pM [35S]GTPγS) for 1 h. Basal binding was measured in the absence of DAMGO. Nonspecific binding was measured by adding 20 μM GTPγS to the assay. The reaction was terminated by the addition of 3 ml of ice-cold 20 mM Tris-HCl, pH 7.4, and the samples were rapidly filtered through glass filter and then washed with 3 ml of ice-cold 20 mM Tris-HCl pH 7.4 three times using a Brandel cell harvester. The amount of [35S]GTPγS bound to membrane on individual filter disks was then determined by Beckman LS 6500 scintillation counter (Beckman Coulter Inc., Fullerton, CA).
Statistical Analysis.
Values are the mean ± S.E.M. of at least three independent experiments. Statistical analysis was carried out using the one-way ANOVA with Tukey post test. In the figures, ** and *** refer to P values of <0.01 and <0.001, respectively, compared with wild type.
Results
Ser363 Is the Major PKC Phosphorylation Site at MOPr-CT In Vitro.
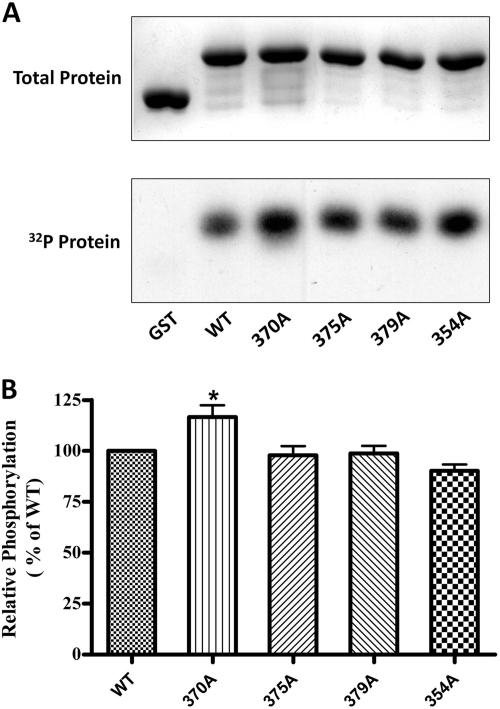
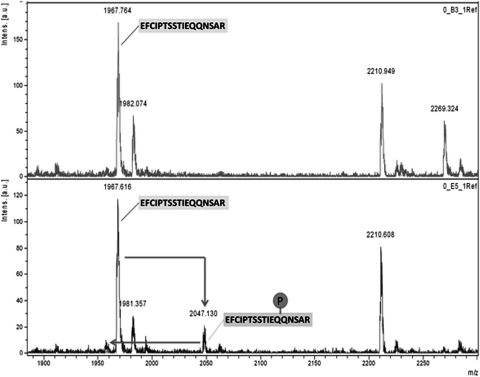
To determine whether PKC is capable of phosphorylating MOPr-CT in vitro, both GST and GST-fused MOPr-CT proteins were applied to the in vitro phosphorylation assay. Based on the kinase consensus sequences (NetPhos 2.0 program; http://www.cbs.dtu.dk/services/NetPhos/), 4 putative phosphorylation sites (Thr354, Thr370, Ser375, and Thr379) on MOPr C terminus were chosen and individually mutated to a nonphosphorylatable amino acid, alanine, as indicated in Fig. 1. When incubated with purified PKC, phosphate incorporation was observed in GST-fused MOPr-CT but not GST, indicating that PKC can directly phosphorylate MOPr-CT in vitro. However, none of the mutations significantly decreased PKC-mediated phosphorylation of the MOPr C terminus (Fig. 2), which suggested that PKC-mediated phosphorylation occurred at residue(s) other than the four predicted residues. To find these residues, a protein mass spectrometer approach was used. The phosphorylated MOPr C terminus was trypsin-digested and then analyzed by MALDI-TOF-MS (Fig. 3). The peak at 1967 m/z was identified as the peptide E349FCIPTSSTIEQQNSAR365, and the peak at 2047 m/z was identified as the monophosphate modification of the 1967 peak (1967 + 80). In addition, a small peak around 1950 m/z was identified as a neutral loss of H3PO4 (2047 − 98) from peak 2047. All of these data suggest that PKC phosphorylation occurs in the peptide of 1967 Da, which corresponds to in the region from Glu349 to Arg365 of MOPr. This region contains five serine and threonine residues. To identify which of them is phosphorylated, another two mutants, 354A4 and 363A (Fig. 1), were constructed and tested by in vitro phosphorylation assay. Mutation of Ser363 significantly decreased the phosphate incorporation by 75% compared with the wild type, whereas mutation of Thr354–SST357 had no significant effect on phosphate incorporation (Fig. 4). These results demonstrated that Ser363 is the major PKC phosphorylation site at the MOPr C terminus.
Fig. 1.
Schematic representation of MOPr-CT sequence and the mutants. The C terminus residues 340 to 398 of mouse wild-type MOPr are shown in single-letter amino acid codes. The potential phosphorylated residues predicted by software are italicized, whereas those identified by mass spectrometry are underlined. Mutants are named by the position and numbers of mutated residues. The letter A represents the corresponding serine/threonine residue being mutated to alanine, and dashes indicate unchanged residues.
Fig. 2.
In vitro phosphorylation of GST-MOPr-CT by PKC. PKC-phosphorylated proteins (5 μg), including GST and GST-fused MOPr C terminus, were resolved by 12% SDS-PAGE. A, top, protein bands stained with Coomassie Blue; bottom, autoradiograph of the same gel. B, the phosphorylation of each protein was measured by scintillation counting and normalized to total protein. Results are expressed relative to phosphorylated wild type protein (100%). Data are presented as means ± S.E.M. (n = 6). *, P < 0.05 compared with WT (one-way ANOVA with Tukey post test).
Fig. 3.
MALDI-TOF-MS of tryptic peptides of PKC phosphorylated MOPr-CT protein. Recombinant purified MOPr-CT (5 μg) was phosphorylated by PKC and then subjected to 12% SDS-PAGE. The protein band was excised from the gel and subjected to proteolytic digestion using trypsin. Top, MS result of the nonphosphorylated MOPr-CT; bottom, MS result of the PKC phosphorylated MOPr-CT. MALDI-TOF-MS found a phosphorylated peptide with m/z = 2047, which corresponds to amino acids 349 to 365 of MOPr.
Fig. 4.
In vitro phosphorylation of GST-MOPr-CT by PKC. PKC phosphorylated proteins (5 μg), including GST and GST-fused MOPr C terminus, were resolved by 12% SDS-PAGE. A, top, represents protein bands stained with Coomassie Blue; bottom, autoradiograph of the same gel. B, the phosphorylation of each protein was measured by scintillation counting and normalized to total protein. Results are expressed relative to phosphorylated wild type protein (100%). Data are presented as means ± S.E.M. (n = 6). ***, P < 0.001 compared with WT (one-way ANOVA with Tukey post test).
Substrate Selectivity of PKC Isoforms on the Phosphorylation of MOPr-CT.
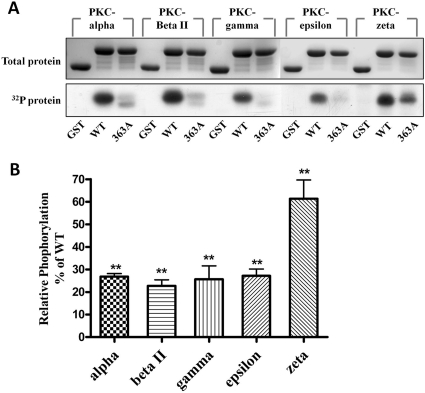
PKC is a family of protein kinases containing at least 10 isozymes. PKC purified from the rat brain used in the above assay is mainly a mixture of PKC α, β, and γ isozymes. To identify the substrate selectivity of PKC isozymes on MOPr C-terminal phosphorylation, five PKC isozymes, including α, βII, γ, ξ, and ε, were chosen for the in vitro phosphorylation assay. Phosphate incorporation was observed with all five isozymes, which indicates that MOPr-CT serves as the substrate for all five PKC isozymes (Fig. 5A). However, when the S363A mutated protein was incubated with the PKC isozymes, the extents of phosphate incorporation varied among the different PKC isozymes. PKC α, βII, γ, and ε-mediated phosphorylation decreased by 75%, whereas PKC ξ-mediated phosphorylation only decreased by approximately 25% compared with the wild type (Fig. 5B). This result suggested that different PKC isozymes may have phosphorylation target preference, and Ser363 serves as the major phosphorylation site for PKC α, βII, γ, and ε, but not for PKC ξ.
Fig. 5.
In vitro phosphorylation of GST-MOPr-CT by PKC isozymes. PKC phosphorylated proteins (5 μg), including GST and GST-fused MOPr C terminus, were resolved by 12% SDS-PAGE. A, top, protein bands stained with Coomassie Blue; bottom, autoradiograph of the same gel. B, the phosphorylation of each protein was measured by scintillation counting and normalized to total protein. Results are expressed relative to phosphorylated wild-type protein (100%). Data are presented as means ± S.E.M. (n = 6). ***, P < 0.001 compared with WT (one-way ANOVA with Tukey post test).
Characterization of the Phosphospecific Antibody against pMOPr (Ser363).
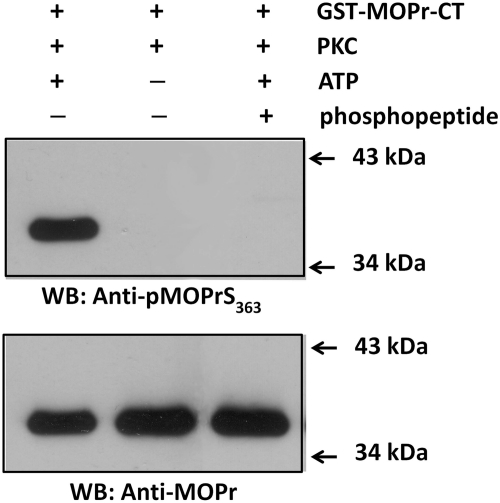
Based on the in vitro phosphorylation result, rabbit anti-serum against the phosphopeptide that corresponds to the sequence surrounding the phospho-Ser363 of MOPr was raised, and the specificity of the antibody (anti-pMOPrS363 hereafter) was assessed. The recombinant GST-MOPr-CT was incubated with PKC in the presence or absence of ATP. As shown at the top of Fig. 6, the anti-pMOPrS363 only recognized GST-MOPr-CT incubated with PKC in the presence of ATP. This result suggested that the antibody is capable of recognizing the PKC phosphorylated GST-MOPr-CT. The antibody signal was completely blocked by adding the antigen phosphopeptide to the blotting system (lane 3), which demonstrated that the anti-pMOPrS363 does specifically recognize the PKC-mediated phosphorylation of MOPr-CT at Ser363. The presence of equal amounts of GST-MOPr-CT protein in the samples was proved by probing the Western blots with an antibody against MOPr (Fig. 6, bottom).
Fig. 6.
Phosphospecific antibody against MOPr (Ser363). Recombinant GST-MOPr-CT protein was incubated at 30°C for 30 min with PKC in the presence (lanes 1 and 3) or absence (lane 2) of ATP. Proteins (20 ng) were separated by 12% SDS-PAGE. Top, Western blot with the anti-pMOPrS363 in the absence (lanes 1 and 2) or the presence (lane 3) of phosphopeptide block. Bottom, the same membrane was stripped and reblotted with anti-MOPr antibody.
PKC-Mediated Phosphorylation of MOPr in CHO Cells.
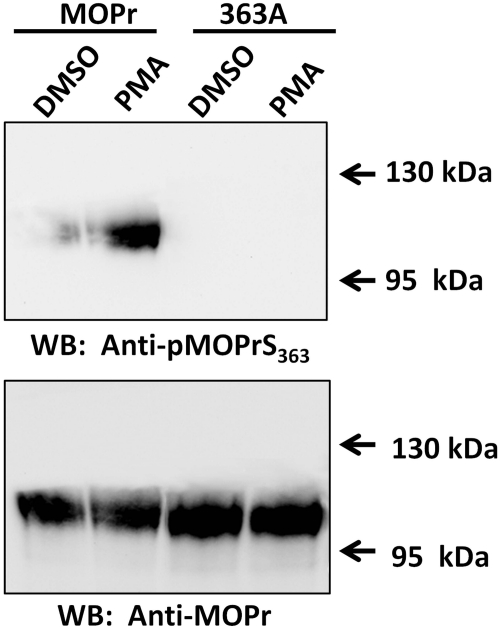
These results are all based on an in vitro phosphorylation assay. We also examined the PKC-meditated phosphorylation of MOPr using a phosphospecific antibody in CHO cells stably expressing EGFP-tagged MOPr, either wild-type or 363A mutant. The cells were treated with 1 μM PMA (PKC activator) or DMSO (control) for 20 min. The cell lysate was partially purified with WGA-agarose beads and then separated by 7% SDS-PAGE. Western blotting with the anti-pMOPrS363 (Fig. 7 upper panel) showed that the PMA treatment significantly increased the signal of the anti-MOPrpSer363 compared with the DMSO control. When the Ser363 was mutated to alanine, the signal of was completely abolished. Thus, in agreement with the in vitro phosphorylation, activation of PKC in the CHO cells also increases the MOPr phosphorylation at Ser363, as specifically recognized by the anti-MOPrpSer363. The presence of equal amount of MOPr proteins, both wild-type and mutated, was confirmed by probing the same membrane with the antibody against total MOPr (Fig. 7, bottom).
Fig. 7.
PKC-mediated phosphorylation of MOPr in CHO cells. Stable cell lines expressing either wild-type EGFP-MOPr (abbreviated as MOPr) or EGFP-MOPr-S363A (abbreviated as 363A) were treated with 1 μM PMA for 20 min, with DMSO as treatment control. Cell lysate was partially purified by WGA-agarose beads then separated by 8% SDS-PAGE. Top, Western blot with anti-pMOPrS363. Bottom, same membrane was stripped and reblotted with anti-MOPr.
Effect of PKC-Mediated Phosphorylation of MOPr on Receptor Desensitization.
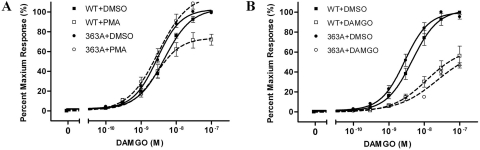
To investigate how PKC-mediated receptor phosphorylation regulates MOPr desensitization, receptor coupling to G protein, which is the initiating event of the MOPr signaling cascade, was measured by a [35S]GTPγS binding assay. Stable cell lines expressing wild-type MOPr or 363A mutant were used for the assay. Radioligand binding showed that the receptor expression levels of the two cell lines were similar and the mutation did not change the ligand binding affinity (Bmax = 2.57 ± 0.16 pmol/mg protein, Kd = 1.20 ± 0.04 nM for wild type versus Bmax = 2.65 ± 0.06 pmol/mg protein, Kd = 1.23 ± 0.12 nM for 363A, P > 0.05). [35S]GTPγS binding demonstrated that, compared with the wild-type MOPr, the 363A mutant had no significant difference in G-protein coupling of MOPr (EC50 = 4.57 ± 0.8 nM for wild type versus EC50 = 2.82 ± 0.9 nM for 363A mutant). Moreover, the wild-type and 363A mutant showed similar responses to the DAMGO-induced MOPr desensitization (percentage of maximum response = 64.5 ± 6.9%, EC50 = 55.0 ± 5.4 nM for wild type, versus 62.6 ±7.3%, EC50 = 22.9 ± 5.1 nM for 363A; P > 0.05), which suggested that the mutation of the PKC phosphorylation site on the C terminus had no effect on the agonist-induced desensitization of MOPr (Fig. 8B and Table 1). When the cells were pretreated with PMA, an activator of PKC, the G-protein coupling of the receptor was decreased (percentage of maximum response = 103.3 ±3.1% for DMSO-pretreated versus 73.8 ± 4.4% for PMA-pretreated, P < 0.001), which suggested that PKC-mediated receptor phosphorylation decreases the capacity of MOPr coupling to G protein. This finding was confirmed by results with the 363A mutant: mutating the PKC phosphorylation site completely abolished the effect of PMA pretreatment (percentage of maximum response = 102.9 ± 1.7% for DMSO-pretreated versus 117.3 ± 5.2% for PMA-pretreated; P > 0.05) (Fig. 8A and Table 1).
Fig. 8.
[35S]GTPγS binding of MOPr and S363A mutation. Stable cell lines expressing either wild-type EGFP-MOPr (abbreviated as MOPr) or EGFP-MOPr-S363A (abbreviated as 363A) were pretreated with 1 μM PMA (A) or 1 μM DAMGO (B) (□, MOPr; ○, 363A) for 20 min using DMSO as the untreated control (■, MOPr; ●, 363A) followed by three washes with ice-cold PBS. Cell membranes were prepared and subjected to [35S]GTPγS binding assay as described under Materials and Methods. Data are shown as a percentage of the response to highest concentration of DAMGO and fitted by nonlinear regression using the Prism program (GraphPad Software Inc., San Diego, CA). Results are means ± S.E.M. of at least three experiments performed in duplicate.
TABLE 1.
Effect of DAMGO and PMA pretreatment on DAMGO-stimulated [35S]GTPγS binding in cells expressing MOPr or 363A
Data are presented as mean value ± S.E.M. (n ≥ 3).
| Pretreatment | Maximum Response | EC50 | Pretreatment | Maximum Response | EC50 | |
|---|---|---|---|---|---|---|
| % | nM | % | nM | |||
| MOPr | DMSO | 102.6 ± 4.4 | 4.57 ± 0.8 | DMSO | 103.3 ± 3.1 | 4.57 ± 0.9 |
| DAMGO | 64.5 ± 6.9** | 55.0 ± 5.4*** | PMA | 73.8 ± 4.4*** | 2.63 ± 0.8 | |
| 363A | DMSO | 100.2 ± 3.2 | 2.75 ± 0.9 | DMSO | 102.9 ± 1.7 | 2.82 ± 0.9 |
| DAMGO | 62.6 ± 7.3** | 22.9 ± 5.1*** | PMA | 117.3 ± 5.2††† | 3.02 ± 0.9 |
P < 0.01 compared with same cell line DMSO pretreatment.
P < 0.001 compared with same cell line DMSO pretreatment.
P < 0.001 compared with cell line expressing wild-type MOPr (one-way ANOVA with Tukey post test).
Discussion
Previous studies have suggested that phosphorylation of MOPr, especially phosphorylation of its C terminus, plays an important role in the regulation of MOPr functions. Several residues on the receptor can be phosphorylated, including Ser354, Thr357, Ser363, Thr370, Ser375, and Thr394 (Pak et al., 1997; Burd et al., 1998; Wolf et al., 1999; Deng et al., 2000; El Kouhen et al., 2001). Moreover, multiple protein kinases have been reported to be involved in MOPr phosphorylation, including GRK, PKC, protein kinase A, and Ca2+/calmodulin-dependent protein kinase II (Zhang et al., 1996, 1998; Koch et al., 1997; Kovoor et al., 1997; Bernstein and Welch, 1998). However, little information is available regarding the specific site(s) and the regulatory effects of PKC-mediated phosphorylation of MOPr. In this study, we provide direct evidence that the C terminus of MOPr is a substrate of PKC and that Ser363 is the major site for PKC-mediated phosphorylation. We also show that PKC-mediated phosphorylation of Ser363 interrupts the desensitization of MOPr to G protein coupling.
Ser363 was identified as the primary PKC phosphorylation site on the C terminus of MOPr using a combination of in vitro phosphorylation, site-direct mutagenesis, and protein mass spectrometry. A problem with mutagenesis is that a mutation-induced decrease in phosphorylation can be caused either by changing the consensus sequence recognized by PKC or by replacing the real phosphorylation residue. Because MS of the phosphorylated C terminus showed that phosphorylation occurs in the region of Glu349 to Arg365 of receptor, this concern is most likely unimportant in our study. Although there are still four serine and threonine residues in this region besides Ser363, which may potentially be phosphorylated by PKC, mutation of these four residues (354A4) did not affect the C-terminal phosphorylation. This result indicates that PKC phosphorylation of MOPr does not occur on residues other than Ser363, and the S363A-induced decrease of phosphorylation was caused by the replacement of a PKC phosphorylation residue on the C terminus.
Previous studies, cited above, showed that most phosphorylation sites are near the C terminus of the receptor and that phosphorylation of MOPr C-terminal is a critical process in the regulation of MOPr functions. It should be noted that other intracellular domains might also be phosphorylated by PKC. Moreover, several receptor splice variants have also been identified in which the last 12 amino acids at the C terminus of MOPr being examined in the current study are replaced by various lengths of sequences up to 82 amino acids (Pan et al., 1999; Pasternak, 2004). It is noteworthy that these splice variants differ in agonist-induced receptor phosphorylation, desensitization, and resensitization (Koch et al., 1998; Oldfield et al., 2008). Because some of the splice variants contain additional serine and threonine residues, which can potentially be phosphorylated by PKC, phosphorylation of these additional residues by PKC may also contribute to the observed differences among the MOPr splice variants. A strategy similar to that used in the current research could be adopted to investigate this possibility in the future.
Another concern about the in vitro phosphorylation study is that a fused protein containing only part of MOPr was used for the assay. The C terminus of MOPr might lack the proper conformation found in the whole receptor expressed in living cells. To address this concern, results of the in vitro PKC phosphorylation were confirmed using CHO cells expressing MOPr and a phosphospecific antibody. Western blotting indicated that activation of PKC in living cells also induced MOPr phosphorylation at Ser363. This demonstrated that Ser363 is a PKC phosphorylation site in the intact receptor.
The initial receptor phosphorylation assay was performed using a PKC mixture containing predominantly α, β, and γ isoforms. To identify whether different isoforms of PKC use the same phosphorylation site, based on previous reports (Kramer and Simon, 1999a,b; Granados-Soto et al., 2000), five PKC isoforms, including α, β, γ, ε, and ζ, were chosen for the in vitro phosphorylation test. The results revealed that all five isoforms are capable of phosphorylating the MOPr C terminus and that Ser363 serves as the major phosphorylation site for the α, β, γ, and ε isoforms but not for the ζ isoform. However, this experiment did not identify which isoforms of PKC have been activated and are involved in the phosphorylation of MOPr. Moreover, the specificity of the PKC isoforms observed in the in vitro phosphorylation assay may be different from that in intact living cells. A future study using selective activators and/or inhibitors of different PKC isoforms could clarify these issues.
The regulatory effects of PKC on MOPr signaling have been evaluated by monitoring changes in function of several downstream signaling components, including ion channels (Zhang et al., 1996; King et al., 1999) and AC (Mandyam et al., 2002). However, PKC-mediated phosphorylation can also regulate other downstream signaling components besides the receptor, such as G protein (Chakrabarti and Gintzler, 2003), AC (Lin et al., 2002), and ion channels (Breitwieser, 2005; Haller et al., 2008). Because of this complexity, it remains unclear whether PKC-mediated phosphorylation of the receptor directly contributes to functional changes in MOPr. In the current study, receptor-G protein coupling, the initial step of MOPr signaling, was measured to monitor the activation of MOPr. [35S]GTPγS binding showed that the activation of PKC reduced the capacity of MOPr-G protein coupling, and the decreased coupling was completely prevented by mutating the PKC phosphorylation site on MOPr. This finding supports the idea that PKC-mediated desensitization of MOPr is induced, at least in part, by directly interrupting MOPr-G protein coupling. At present, the exact mechanism of regulation of receptor-G protein coupling by PKC-mediated phosphorylation of MOPr remains unknown. It has been proposed that the binding of β-arrestin-2 to the phosphorylated GPCR mainly depends on an increase of bulk negative charge on the intracellular surface of the receptor rather than the precise location of the phosphorylation sites (Gurevich and Gurevich, 2006). Based on the fact that Ser363 is in close proximity to Ser375, which we have found to be a GRK2 phosphorylation site (B. Feng, unpublished data), a possible explanation is that phosphorylation of Ser363 may also induce the binding of β-arrestin-2, thus disrupting receptor-G protein coupling, just as in the GRK-mediated phosphorylation and desensitization of the receptor.
Like desensitization, internalization is an important adaptive change regulated by receptor phosphorylation (Wolf et al., 1999). Although the regulation of MOPr internalization by PKC was not explored in the current research, it has been reported previously that MOPr with the S363A mutation internalizes faster than the wild-type MOPr (El Kouhen et al., 2001). This observation, together with our current finding that Ser363 is the PKC phosphorylation site, suggests that PKC-mediated MOPr phosphorylation may inhibit MOPr internalization. This conclusion is supported by another study that found inhibition of PKC activity to facilitate MOPr internalization (Ueda et al., 2001). If this is true, it will be interesting to know how PKC affects receptor internalization in a manner opposite to GRK, even both kinases show a similar regulatory effect on MOPr-G protein coupling. One possible explanation is that arrestin adopts a distinct conformation when it binds to the PKC phosphorylated receptor, and that this conformation affects receptor internalization differently from that produced by GRK-mediated phosphorylation. The concept that arrestin adopts a variety of conformations depending on the type of phosphorylation site on the receptor has been supported by studies on two other GPCRs, rhodopsin (Vishnivetskiy et al., 2007) and N-formylpeptide receptors (Key et al., 2003).
In summary, we have identified for the first time Ser363 as the major PKC phosphorylation site on the MOPr. In vitro assays revealed that this site is specific for PKC α, β, γ, and ε isoforms, but not the ζ isoform. We also provide evidence that PKC-mediated receptor phosphorylation regulates receptor desensitization by direct interruption of receptor-G protein coupling. These findings move us one step closer to understanding the detailed mechanisms underlying PKC regulation of MOPr functions and to overcoming the problems of opioid tolerance and addiction.
Acknowledgments
We are grateful for the technical assistant in mass spectrum analysis from Julie Ray.
This work was supported by the National Institutes of Health National Institutes of Drug Abuse [Grant DA11925].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/mol.110.069096.
ABBREVIATIONS:
- GPCR
- G protein-coupled receptor
- CHO
- Chinese hamster ovary
- CT
- carboxyl terminal
- DAMGO
- [d-Ala2-N-Me-Phe4,Gly5-ol]-enkephalin
- EGFP
- enhanced green fluorescence protein
- GRK
- G protein-coupled receptor kinase
- GST
- glutathione transferase
- MALDI-TOF-MS
- matrix-assisted laser desorption ionization-time of flight-mass spectrometry
- MOPr
- μ opioid receptor
- PMA
- phorbol 12-myristate 12-acetate
- PKC
- protein kinase C
- WGA
- wheat germ agglutinin
- WT
- wild type
- AC
- adenylate cyclase
- MS
- mass spectrometry
- PCR
- polymerase chain reaction
- PAGE
- polyacrylamide gel electrophoresis
- TBS-T
- Tris-buffered saline/Tween 20
- ANOVA
- analysis of variance
- DMSO
- dimethyl sulfoxide
- PBS
- phosphate-buffered saline
- [35S]GTPγS
- guanosine 5′-O-(3-[35S]thio)triphosphate.
Authorship Contributions
Participated in research design: Feng and Wang.
Conducted experiments: Feng and Li.
Performed data analysis: Feng.
Wrote or contributed to the writing of the manuscript: Feng and Wang.
References
- Bailey CP, Llorente J, Gabra BH, Smith FL, Dewey WL, Kelly E, Henderson G. (2009) Role of protein kinase C and mu-opioid receptor (MOPr) desensitization in tolerance to morphine in rat locus coeruleus neurons. Eur J Neurosci 29:307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaini F. (2001) Protein kinase C isoforms as therapeutic targets in nervous system disease states. Pharmacol Res 44:353–361 [DOI] [PubMed] [Google Scholar]
- Bernstein MA, Welch SP. (1998) mu-Opioid receptor down-regulation and cAMP-dependent protein kinase phosphorylation in a mouse model of chronic morphine tolerance. Brain Res Mol Brain Res 55:237–242 [DOI] [PubMed] [Google Scholar]
- Breitwieser GE. (2005) GIRK channels: hierarchy of control. Focus on “PKC-delta sensitizes Kir3.1/3.2 channels to changes in membrane phospholipid levels after M3 receptor activation in HEK-293 cells”. Am J Physiol Cell Physiol 289:C509–C511 [DOI] [PubMed] [Google Scholar]
- Burd AL, El-Kouhen R, Erickson LJ, Loh HH, Law PY. (1998) Identification of serine 356 and serine 363 as the amino acids involved in etorphine-induced down-regulation of the mu-opioid receptor. J Biol Chem 273:34488–34495 [DOI] [PubMed] [Google Scholar]
- Celver JP, Lowe J, Kovoor A, Gurevich VV, Chavkin C. (2001) Threonine 180 is required for G-protein-coupled receptor kinase 3- and beta-arrestin 2-mediated desensitization of the mu-opioid receptor in Xenopus oocytes. J Biol Chem 276:4894–4900 [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Gintzler AR. (2003) Phosphorylation of Gbeta is augmented by chronic morphine and enhances Gbetagamma stimulation of adenylyl cyclase activity. Brain Res Mol Brain Res 119:144–151 [DOI] [PubMed] [Google Scholar]
- Deng HB, Yu Y, Pak Y, O'Dowd BF, George SR, Surratt CK, Uhl GR, Wang JB. (2000) Role for the C-terminus in agonist-induced mu opioid receptor phosphorylation and desensitization. Biochemistry 39:5492–5499 [DOI] [PubMed] [Google Scholar]
- El Kouhen R, Burd AL, Erickson-Herbrandson LJ, Chang CY, Law PY, Loh HH. (2001) Phosphorylation of Ser363, Thr370, and Ser375 residues within the carboxyl tail differentially regulates mu-opioid receptor internalization. J Biol Chem 276:12774–12780 [DOI] [PubMed] [Google Scholar]
- Ferguson SS, Zhang J, Barak LS, Caron MG. (1998) Molecular mechanisms of G protein-coupled receptor desensitization and resensitization. Life Sci 62:1561–1565 [DOI] [PubMed] [Google Scholar]
- Granados-Soto V, Kalcheva I, Hua X, Newton A, Yaksh TL. (2000) Spinal PKC activity and expression: role in tolerance produced by continuous spinal morphine infusion. Pain 85:395–404 [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. (2006) The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol Ther 110:465–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller VL, Bernstein MA, Welch SP. (2008) Chronic morphine treatment decreases the Cav1.3 subunit of the L-type calcium channel. Eur J Pharmacol 578:101–107 [DOI] [PubMed] [Google Scholar]
- Harrison LM, Kastin AJ, Zadina JE. (1998) Opiate tolerance and dependence: receptors, G-proteins, and antiopiates. Peptides 19:1603–1630 [DOI] [PubMed] [Google Scholar]
- Key TA, Foutz TD, Gurevich VV, Sklar LA, Prossnitz ER. (2003) N-formyl peptide receptor phosphorylation domains differentially regulate arrestin and agonist affinity. J Biol Chem 278:4041–4047 [DOI] [PubMed] [Google Scholar]
- King AP, Hall KE, Macdonald RL. (1999) Kappa- and mu-opioid inhibition of N-type calcium currents is attenuated by 4beta-phorbol 12-myristate 13-acetate and protein kinase C in rat dorsal root ganglion neurons. J Pharmacol Exp Ther 289:312–320 [PubMed] [Google Scholar]
- Koch T, Kroslak T, Mayer P, Raulf E, Höllt V. (1997) Site mutation in the rat mu-opioid receptor demonstrates the involvement of calcium/calmodulin-dependent protein kinase II in agonist-mediated desensitization. J Neurochem 69:1767–1770 [DOI] [PubMed] [Google Scholar]
- Koch T, Schulz S, Schröder H, Wolf R, Raulf E, Höllt V. (1998) Carboxyl-terminal splicing of the rat mu opioid receptor modulates agonist-mediated internalization and receptor resensitization. J Biol Chem 273:13652–13657 [DOI] [PubMed] [Google Scholar]
- Kovoor A, Nappey V, Kieffer BL, Chavkin C. (1997) Mu and delta opioid receptors are differentially desensitized by the coexpression of beta-adrenergic receptor kinase 2 and beta-arrestin 2 in xenopus oocytes. J Biol Chem 272:27605–27611 [DOI] [PubMed] [Google Scholar]
- Kramer HK, Simon EJ. (1999a) Role of protein kinase C (PKC) in agonist-induced mu-opioid receptor down-regulation: I. PKC translocation to the membrane of SH-SY5Y neuroblastoma cells is induced by mu-opioid agonists. J Neurochem 72:585–593 [DOI] [PubMed] [Google Scholar]
- Kramer HK, Simon EJ. (1999b) Role of protein kinase C (PKC) in agonist-induced mu-opioid receptor down-regulation: II. Activation and involvement of the alpha, epsilon, and zeta isoforms of PKC. J Neurochem 72:594–604 [DOI] [PubMed] [Google Scholar]
- Law PY, Loh HH. (1999) Regulation of opioid receptor activities. J Pharmacol Exp Ther 289:607–624 [PubMed] [Google Scholar]
- Lin TH, Lai HL, Kao YY, Sun CN, Hwang MJ, Chern Y. (2002) Protein kinase C inhibits type VI adenylyl cyclase by phosphorylating the regulatory N domain and two catalytic C1 and C2 domains. J Biol Chem 277:15721–15728 [DOI] [PubMed] [Google Scholar]
- Mandyam CD, Thakker DR, Christensen JL, Standifer KM. (2002) Orphanin FQ/nociceptin-mediated desensitization of opioid receptor-like 1 receptor and mu opioid receptors involves protein kinase C: a molecular mechanism for heterologous cross-talk. J Pharmacol Exp Ther 302:502–509 [DOI] [PubMed] [Google Scholar]
- Mellor H, Parker PJ. (1998) The extended protein kinase C superfamily. Biochem J 332:281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield S, Braksator E, Rodriguez-Martin I, Bailey CP, Donaldson LF, Henderson G, Kelly E. (2008) C-terminal splice variants of the mu-opioid receptor: existence, distribution and functional characteristics. J Neurochem 104:937–945 [DOI] [PubMed] [Google Scholar]
- Pak Y, O'Dowd BF, George SR. (1997) Agonist-induced desensitization of the mu opioid receptor is determined by threonine 394 preceded by acidic amino acids in the COOH-terminal tail. J Biol Chem 272:24961–24965 [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Bolan E, Abbadie C, Chang A, Zuckerman A, Rossi G, Pasternak GW. (1999) Identification and characterization of three new alternatively spliced mu-opioid receptor isoforms. Mol Pharmacol 56:396–403 [DOI] [PubMed] [Google Scholar]
- Pasternak GW. (2004) Multiple opiate receptors: déjà vu all over again. Neuropharmacology 47:312–323 [DOI] [PubMed] [Google Scholar]
- Schulz S, Mayer D, Pfeiffer M, Stumm R, Koch T, Höllt V. (2004) Morphine induces terminal micro-opioid receptor desensitization by sustained phosphorylation of serine-375. EMBO J 23:3282–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Inoue M, Matsumoto T. (2001) Protein kinase C-mediated inhibition of mu-opioid receptor internalization and its involvement in the development of acute tolerance to peripheral mu-agonist analgesia. J Neurosci 21:2967–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Raman D, Wei J, Kennedy MJ, Hurley JB, Gurevich VV. (2007) Regulation of arrestin binding by rhodopsin phosphorylation level. J Biol Chem 282:32075–32083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HL, Chang WT, Hsu CY, Huang PC, Chow YW, Li AH. (2002) Identification of two C-terminal amino acids, Ser(355) and Thr(357), required for short-term homologous desensitization of mu-opioid receptors. Biochem Pharmacol 64:257–266 [DOI] [PubMed] [Google Scholar]
- Wang L, Medina VM, Rivera M, Gintzler AR. (1996) Relevance of phosphorylation state to opioid responsiveness in opiate naive and tolerant/dependent tissue. Brain Res 723:61–69 [DOI] [PubMed] [Google Scholar]
- Wolf R, Koch T, Schulz S, Klutzny M, Schröder H, Raulf E, Bühling F, Höllt V. (1999) Replacement of threonine 394 by alanine facilitates internalization and resensitization of the rat mu opioid receptor. Mol Pharmacol 55:263–268 [DOI] [PubMed] [Google Scholar]
- Yu Y, Zhang L, Yin X, Sun H, Uhl GR, Wang JB. (1997) Mu opioid receptor phosphorylation, desensitization, and ligand efficacy. J Biol Chem 272:28869–28874 [DOI] [PubMed] [Google Scholar]
- Zhang J, Ferguson SS, Barak LS, Bodduluri SR, Laporte SA, Law PY, Caron MG. (1998) Role for G protein-coupled receptor kinase in agonist-specific regulation of mu-opioid receptor responsiveness. Proc Natl Acad Sci USA 95:7157–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu Y, Mackin S, Weight FF, Uhl GR, Wang JB. (1996) Differential mu opiate receptor phosphorylation and desensitization induced by agonists and phorbol esters. J Biol Chem 271:11449–11454 [DOI] [PubMed] [Google Scholar]