Abstract
Nuclear factor (NF)-YB, a subunit of the transcription factor nuclear factor Y (NF-Y) complex, binds and activates CCAAT-containing promoters. Our previous work suggested that NF-YB may be a mediator of topoisomerase IIα (Top2α), working through the Top2α promoter. DNA topoisomerase II (Top2) is an essential nuclear enzyme and the primary target for several clinically important anticancer drugs. Our teniposide-resistant human lymphoblastic leukemia CEM cells (CEM/VM-1-5) express reduced Top2α protein compared with parental CEM cells. To study the regulation of Top2α during the development of drug resistance, we found that NF-YB protein expression is increased in CEM/VM-1-5 cells compared with parental CEM cells. This further suggests that increased NF-YB may be a negative regulator of Top2α in CEM/VM-1-5 cells. We asked what causes the up-regulation of NF-YB in CEM/VM-1-5 cells. We found by microRNA profiling that hsa-miR-485-3p is lower in CEM/VM-1-5 cells compared with CEM cells. MicroRNA target prediction programs revealed that the 3′-untranslated region (3′-UTR) of NF-YB harbors a putative hsa-miR-485-3p binding site. We thus hypothesized that hsa-miR-485-3p mediates drug responsiveness by decreasing NF-YB expression, which in turn negatively regulates Top2α expression. To test this, we overexpressed miR-485-3p in CEM/VM-1-5 cells and found that this led to reduced expression of NF-YB, a corresponding up-regulation of Top2α, and increased sensitivity to the Top2 inhibitors. Results in CEM cells were replicated in drug-sensitive and -resistant human rhabdomyosarcoma Rh30 cells, suggesting that our findings represent a general phenomenon. Ours is the first study to show that miR-485-3p mediates Top2α down-regulation in part by altered regulation of NF-YB.
Introduction
Nuclear factor-Y (NF-Y) is a conserved transcription factor that consists of three subunits—NF-YA, NF-YB, and NF-YC—and binds specifically to the CCAAT elements in promoters (Ronchi et al., 1995). Our previous work suggested that one of these subunits, NF-YB, may mediate Top2α expression, working through inverted CCAAT element 3 (ICE3) in the Top2α promoter (Morgan and Beck, 2001). DNA topoisomerase II (Top2) is an essential nuclear enzyme involved in many cellular processes (Champoux, 1990; Wang, 2002; Nitiss, 2009a) and mammalian cells have two isoforms of the type II enzymes: Top2α (170 kDa) (Tsai-Pflugfelder et al., 1988), and Top2β (180 kDa) (Chung et al., 1989). Top2-DNA covalent complexes serve as the cytotoxic target for many anticancer drugs such as doxorubicin, etoposide, teniposide, and amsacrine (Liu, 1989; Beck, 1996; Pommier et al., 2010). However, tumors frequently become refractory to treatments with Top2 inhibitors because of the emergence of drug resistance (Nitiss and Beck, 1996; Nitiss, 2009b). Our previous work suggested that the transcription factor NF-YB is a mediator of Top2α, working through the Top2α promoter (Morgan et al., 2000). Our current experiments revealed an inverse relationship between the expression of Top2α protein and NF-YB protein in drug-sensitive CEM cells compared with the teniposide-resistant CEM/VM-1-5 cells (Danks et al., 1988) and in drug-sensitive human rhabdomyosarcoma Rh30 cells and the etoposide-resistant Rh30/v1 cells (Bhat et al., 1999). We asked in the present study what causes the up-regulation of NF-YB in the CEM/VM-1-5 and Rh30/v1 cells that express decreased levels of Top2α.
There are reports that microRNAs, a group of nonprotein coding, single-stranded RNAs of 20 to 22 nucleotides often aberrantly expressed or mutated in cancer (Calin and Croce, 2006), may play roles as either oncogenes or tumor suppressors (Esquela-Kerscher and Slack, 2006), and may mediate drug-responsiveness (Mishra et al., 2007; To et al., 2008). Accordingly, we asked whether microRNAs might be involved in the regulation of NF-YB, which further mediates Top2α expression and drug-responsiveness in CEM and CEM/VM-1-5 cells and in Rh30 and Rh30/v1 cells. MicroRNAs function through perfect or near-perfect base pairing with protein coding mRNA 3′-untranslated regions (3′-UTRs) for mRNA degradation or translational repression (Bartel, 2004). We have found by microRNA profiling that a particular microRNA, hsa-miR-485-3p, may target the 3′-UTR of NF-YB to affect the expression of Top2α.
Materials and Methods
Cell Lines and Culture Conditions.
The human lymphoblastic leukemia CEM cell line and its teniposide-resistant subline, CEM/VM-1-5, developed in our laboratory (Danks et al., 1988), were cultured in suspension in RPMI 1640 medium. The human rhabdomyosarcoma Rh30 cell line and its etoposide-resistant subline, Rh30/v1 (Bhat et al., 1999), were grown as monolayers and also cultured in RPMI 1640. HEK293T cells were cultured in DMEM. Both RPMI 1640 and DMEM (Lonza Walkersville, Inc., Walkersville, MD) were supplemented with 10% fetal bovine serum (Gemini, Irvine, CA), and 2 mM l-glutamine (Lonza Walkersville, Inc.). All cell lines were subcultured twice per week in either RPMI 1640 or DMEM and were incubated at 37°C in a humidified chamber containing 5% CO2/95% air.
Western Blot Analysis.
Nuclear protein extracts were prepared from logarithmically growing cells as described previously (Morgan and Beck, 2001). Protein concentration was measured by Bradford assay (Bio-Rad Laboratories, Hercules, CA). Equal amounts of protein samples (∼15–30 μg) were separated on a 4 to 12% Bis-Tris gradient gel (Invitrogen, Carlsbad, CA), electrophoretically transferred onto nitrocellulose, and incubated with either purified mouse anti-Top2α monoclonal antibody (BD Biosciences, San Jose, CA), rabbit anti-NF-YB polyclonal antibody, or mouse anti-NF-YA monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Bound antibody was detected using the enhanced chemiluminescence detection method (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) according to the manufacturer's instructions. Chemiluminescence signals were quantified by densitometric scanning using an Imaging Densitometer (Scion Corporation, Frederick, MD). Equal loading of nuclear protein was determined by blotting the membrane with anti-proliferating cell nuclear antigen (PCNA) antibody (Santa Cruz Biotechnology).
Isolation of Total RNA and Reverse Transcriptase-PCR.
Total RNAs were extracted from CEM and CEM/VM-1-5 and Rh30 and Rh30/v1 cells with TRIzol reagent (Invitrogen), according to the manufacturer's instructions. Single-stranded cDNA was synthesized by using the ThermoScript reverse transcriptase (RT)-PCR System (Invitrogen). The PCRs were carried out for each transcript under the following conditions: initial denaturation at 94°C for 5 min, and then 30 to 40 cycles at 94°C for 30 s, 50 to 60°C for 60 s, and 72°C for 90 s, and 1 cycle of 72°C for 10 min. The 3′-UTR sequences of NF-YB containing the putative mir-485-3p binding site were amplified using the following primers: NF-YB 3′-UTR: sense, 5′-TCT AGA AAG CAA GTG AAA GGT GCC AT-3′; and antisense, 5′-TCT AGA ATC ATG AAT TAA CCC AGC CG-3′. To delete the putative mir-485-3p binding site, we used the following primers: sense, 5′-TCT AGA AAG CAA GTG AAA GGT GCC AT-3′; and antisense, 5′-TCT AGA CCT GAT GCT TGA CTA ATT GAG G-3′, and the sequences were defined as NF-YB 3′-UTR-d.
Construction of Expression Vectors.
The miR-Crtl (pCDH-empty vector) and miR-485-3p expression vectors (pCDH-miR-485-3p) were packaged into lentiviruses by cotransfection of HEK293T cells with three plasmids: a lentiviral expression vector plus pMD2.G and psPAX2. (The latter two plasmids were generous gifts of Dr. Didier Trono, Department of Genetics and Microbiology, University of Geneva, Geneva, Switzerland.) CEM/VM-1-5 cells transduced with miR-Crtl or miR-485-3p expression vectors are defined as CEM/VM-1-5 miR-Crtl or CEM/VM-1-5 miR-485-3p, respectively (Same as Rh30/v1 cells). The NF-YB 3′-UTR containing the putative mir-485-3p binding site and NF-YB 3′-UTR-d without the putative mir-485-3p binding site were amplified by PCR and then cloned into the pGL3-thymidine kinase vector (generous gift of Dr. Hyun-Young Jeong, Department of Biopharmaceutical Sciences, University of Illinois at Chicago, Chicago, IL). The luciferase reporters containing the NF-YB 3′-UTR with or without the putative mir-485-3p binding site are defined as pGL3-NF-YB-3′-UTR or the pGL3-NF-YB-3′-UTR-d, respectively.
Cytotoxicity Assay.
Drug-induced cytotoxicity was measured by the MTT assay (Mosmann, 1983). Exponentially growing cells were plated at 4000 to 5000 cells/well in 96-well microtiter plates (100 μl/well) in triplicate. Etoposide, doxorubicin, or vinblastine (all from Sigma-Aldrich, St. Louis, MO) was added to the cells at various concentrations in a final volume of 200 μl/well, and the cells were incubated at 37°C for 72 h. After drug exposure, 20 μl of MTT compound (5 mg/ml in phosphate-buffered saline; Sigma-Aldrich) was added to each well, and the cells were incubated at 37°C for 3 h. The plates were centrifuged in a swinging bucket rotor (3000 rpm, 20 min, 4°C), and the medium was removed. After 200 μl of dimethyl sulfoxide) was added to each well, and the plates were incubated for 10 min at 37°C, the metabolic activity of the cells was measured by quantifying the conversion of the yellow MTT to a purple metabolite, MTT-formazan. Absorbance was read at 560 nm using a microplate reader. Experimental samples were measured for each drug concentration, and the experiments were replicated at least three times. The IC50 value (the concentration of drug that killed 50% of the cells) was calculated by Prism software (GraphPad Software Inc., San Diego, CA).
Luciferase Assay.
Luciferase assays were carried out in HEK293T cells. Cells were seeded in 24-well plates in triplicate and transfected with appropriate plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. After 24 h of transfection, cells were lysed and treated with Dual-Light assay system (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. Relative luciferase activities were measured and normalized against β-galactosidase activity. All of the experiments were replicated at least three times.
Results
Inverse Expression of Top2α and NF-YB Proteins in CEM and CEM/VM-1-5 Cells and Rh30 and Rh30/v1 Cells.
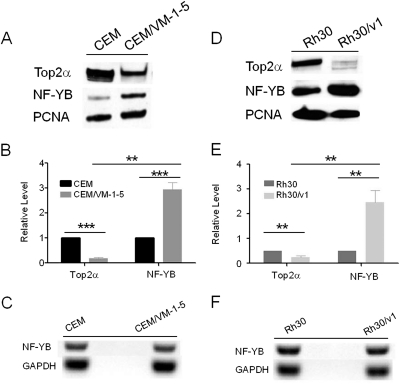
Previous work from our laboratory suggested that the transcription factor NF-YB is a mediator of Top2α, working through the ICE3 in the Top2α promoter (Morgan and Beck, 2001). We found that NF-YB protein levels were 3-fold higher in CEM/VM-1-5 cells compared with CEM cells (Fig. 1, A and B), and there is an inverse relation between the protein levels of Top2α and NF-YB in CEM and CEM/VM-1-5 cells by Western blots (Fig. 1A). To determine whether this is a cell line-specific phenomenon, we examined the human rhabdomyosarcoma Rh30 cell line and compared it with its etoposide-resistant subline, Rh30/v1 (Bhat et al., 1999). We observed a similar result: Top2α is down-regulated in the etoposide-resistant Rh30/v1 cell line, and there is an inverse relation between the protein levels of Top2α and NF-YB (Fig. 1, D and E). In addition, the expression of NF-YA protein is similar between drug-sensitive and -resistant cells (Supplemental Fig. S1). These data suggest that increased NF-YB may be either related to or the cause of reduced Top2α in CEM/VM-1-5 and Rh30/v1 cells. Furthermore, we found that NF-YB mRNA levels were similar in CEM, CEM/VM-1-5 and Rh30, Rh30/v1 cells (Fig. 1, C and F), indicating that the observed differences in NF-YB protein levels are probably a consequence of regulation at the posttranscriptional level. Post-transcriptional repression is a major mechanism by which microRNAs regulate gene expression (Carrington and Ambros, 2003; Bartel, 2004). Accordingly, we then asked whether microRNAs are involved in the regulation of NF-YB protein during the development of drug resistance.
Fig. 1.
Inverse relationship between the protein levels of Top2α and NF-YB in CEM and CEM/VM-1-5 cells and Rh30 and Rh30/v1 cells. Western blots of nuclear Top2α and NF-YB expression in CEM and CEM/VM-1-5 cells (A) and Rh30 and Rh30/v1 cells (D). PCNA served as loading control for nuclear proteins. Average of Top2α and NF-YB levels from three independent experiments ± S.D. are shown, determined by densitometric scanning on Western blots and normalized to PCNA signal; either CEM (B) or Rh30 (E) was assigned a value of 1 for comparison. **, p < 0.005; ***, p < 0.0001. C and F, NF-YB mRNA expression was analyzed by semiquantitative RT-PCR in CEM and CEM/VM-1-5 cells (C) and Rh30 and Rh30/v1 cells (F).
Expression of Micro-RNAs in Human Lymphoblastic Leukemia CEM and CEM/VM-1-5 Cells.
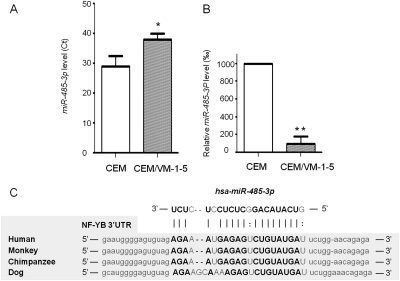
We used microRNA profiling by the Applied Biosystems real-time PCR array (TaqMan Human MicroRNA Array v1.0) to analyze microRNA expression in CEM and CEM/VM-1-5 cells. The array carries unique hairpin-loop RT primer sets that allow for detection of 365 mature human microRNAs in the cells. Compared with CEM cells, microRNA profiling revealed that CEM/VM-1-5 cells with acquired resistance to teniposide demonstrated substantial changes in microRNA expression. Compared with parental controls, 32 microRNAs were consistently either up- or down-regulated in CEM/VM-1-5 cells, as determined by Ct (cycle threshold) values from three separate microRNA profiling experiments (T.-T. Ho, A. D. Arslan, Y.-Y. Mo, X. He, and W. T. Beck, unpublished observations). Because higher Ct values indicate lower expression levels, one of the microRNAs, hsa-miR-485-3p, was found to be consistently substantially lower in CEM/VM-1-5 cells compared with CEM cells after conversion (Wu et al., 2009) to expression level (Fig. 2, A and B). It is noteworthy that the decreased expression of miR-485-3p in CEM/VM-1-5 cells is inversely related to the overexpression of NF-YB protein (Fig. 1, A and B).
Fig. 2.
NF-YB is a putative target for hsa-miR-485-3p. Expression of hsa-miR-485-3p in CEM and CEM/VM-1-5 cells was determined by Applied Biosystems real-time PCR array (TaqMan Human MicroRNA Array version 1.0) according to the manufacturer's instructions. Ct values (A) or relative expression levels of miR-485-3p (B) are means of three pairs of microRNA profile results from CEM and CEM/VM-1-5 cells ± S.D. *, p < 0.05; **, p < 0.01. C, sequence alignment of putative has-miR-485-3p binding sites in 3′-UTR of NF-YB gene of four species. The base pairing nucleotides are in boldface type.
Using miRanda (http://www.microrna.org/microrna/home.do), Target Scan (http://www.targetscan.org/), and MicroCosm (http://microrna.sanger.ac.uk/cgi-bin/targets/v5/search.pl) prediction programs, we found that the 3′-UTR of NF-YB harbors a putative hsa-miR-485-3p binding site (Fig. 2C), which is conserved in human, Rhesus monkeys, chimpanzees, and dogs. Moreover, we found by sequencing that there is no mutation in the region of NF-YB 3′-UTR, to which miR-485-3p binds in either CEM or CEM/VM-1-5 cells (data not shown), suggesting that the binding of microRNAs to the NF-YB 3′-UTRs is unlikely to be altered.
NF-YB Is a Direct Target for miR-485-3p.
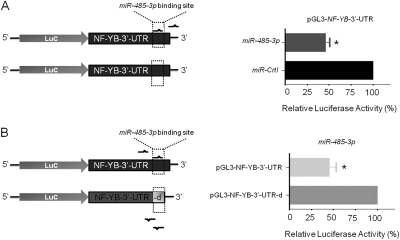
To validate that NF-YB is a direct target of miR-485-3p, we constructed a luciferase reporter (pGL3-thymidine kinase) carrying the NF-YB 3′-UTR with the putative miR-485-3p binding site. We cotransfected miR-485-3p expression vector and luciferase reporter into HEK293T cells. Relative luciferase activity of the NF-YB 3′-UTR with miR-485-3p binding site, which was cotransfected with the miR-485-3p expression vector (pCDH-miR-485-3p), was significantly lower (∼46%) than that of the miR-Crtl (pCDH-empty vector) cotransfected cells (Fig. 3A). Moreover, the relative luciferase activity of the pGL3-NFYB-3′-UTR with the miR-485-3p binding site was also lower (∼45%) than that of the pGL3-NF-YB-3′-UTR-d (pGL3-NF-YB 3′-UTR without miR-485-3p binding site), which was carried out in HEK293T cells cotransfected with the miR-485-3p expression vector (Fig. 3B).
Fig. 3.
MiR-485-3p targets NF-YB 3′-UTR. A, luciferase reporter containing a putative miR-485-3p binding site, NF-YB-3′-UTR (pGL3-NF-YB-3′-UTR), was cotransfected with either miR-Crtl (pCDH-empty vector) or the miR-485-3p expression vector (pCDH-miR-485-3p) with schematic diagram on the left. B, MiR-485-3p expression vector (pCDH-miR-485-3p) was cotransfected with either luciferase reporter containing NF-YB-3′-UTR with or without putative miR-485-3p binding site, which was defined as pGL3-NF-YB-3′-UTR or pGL3-NF-YB-3′-UTR-d, respectively, with schematic diagram (left). Relative luciferase activities were measured and normalized against β-galactosidase activity. Values are average of three separate experiments done in triplicate ± S.E.; *, p < 0.05.
MiR-485-3p Modulates the Sensitivity of CEM/VM-1-5 Cells to Top2 Inhibitors.
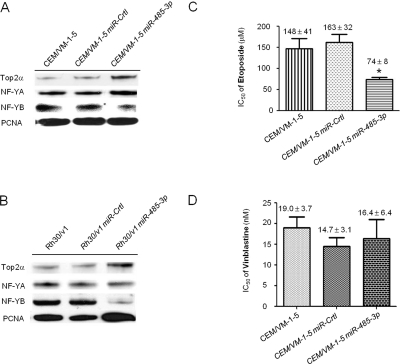
To ascertain whether miR-485-3p regulates NF-YB, we examined the effects of overexpressing miR-485-3p on NF-YB expression in CEM/VM-1-5 cells, which have reduced endogenous miR-485-3p expression (Fig. 2A). Thus, we transduced CEM/VM-1-5 cells with either the pCDH-miR-485-3p or the miR-Crtl (pCDH-empty vector) virus particles. We validated miR-485-3p expression in CEM/VM-1-5 miR-485-3p cells by Taqman real-time PCR (data not shown). Western blot analysis revealed decreased NF-YB protein levels in pCDH-miR-485-3p virus stably transduced CEM/VM-1-5 cells, compared with the miR-Crtl transduced CEM/VM-1-5 cells and no difference in NF-YA protein levels was seen in response to miR-485-3p overexpression cells (Fig. 4A).
Fig. 4.
MiR-485-3p inhibition on NF-YB and mediation on drug responsiveness. Western blots of nuclear Top2α, NF-YB, and NF-YA expression in CEM/VM-1-5 (A) and Rh30/v1 cells (B). CEM/VM-1-5 and Rh30/v1 cells were transduced with either miR-485-3p expression virus or control virus (miR-Crtl). PCNA served as loading control for nuclear protein. IC50 values of cells exposed to etoposide (C) or vinblastine (D) at various concentrations were calculated from the percentage of viable cells after exposure to treatment obtained from MTT assay. Values are average of three independent experiments done in triplicate ± S.E. *, p < 0.05.
To determine whether this was a cell line-specific phenomenon, we examined the etoposide-resistant human rhabdomyosarcoma Rh30/v1 cells. Introduction of miR-485-3p into these cells also decreased their NF-YB protein levels in Rh30/v1 cells (Fig. 4B), indicating that the miR-485-3p-mediated regulation of NF-YB is not cell line-specific.
Because miR-485-3p targets NF-YB and NF-YB may negatively regulate the expression of Top2α, the above findings suggest an interesting relationship between NF-YB, miR-485-3p, and Top2α. Accordingly, we analyzed the levels of Top2α in CEM/VM-1-5 miR-485-3p cells (Fig. 4A). It is noteworthy that we found that the level of Top2α protein is up-regulated, whereas the level of NF-YB is down-regulated compared with CEM/VM-1-5 miR-Crtl cells, suggesting that NF-YB regulates Top2α expression via miR-485-3p. A similar effect was observed in the etoposide-resistant Rh30 subline Rh30/v1 (Fig. 4B). We thus hypothesized that hsa-miR-485-3p may regulate drug-responsiveness by increasing NF-YB expression, which in turn negatively regulates Top2α expression. To test this, we performed the following experiments.
We examined the effects of miR-485-3p on the drug sensitivity of CEM/VM-1-5 cells. MTT assays revealed that CEM/VM-1-5 miR-485-3p cells exhibited enhanced sensitivity, 2-fold to etoposide, compared with CEM/VM-1-5 miR-Crtl cells, as indicated by decreased IC50 values (Fig. 4C). Similar results were seen in doxorubicin-treated cells (data not shown). However, no significant difference in responsiveness to vinblastine, a microtubule inhibitor, was found in miR-485-3p overexpression CEM/VM-1-5 cells (Fig. 4D). Taken together, miR-485-3p-mediated down-regulation of NF-YB in CEM/VM-1-5 cells was accompanied by increased sensitivity of CEM/VM-1-5 cells to Top2 inhibitors.
Discussion
In this study, we confirmed our previous observation that Top2α was down-regulated in drug-resistant CEM/VM-1-5 and Rh30/v1 cells. Moreover, we found that Top2α down-regulation was accompanied by up-regulation of transcription factor NF-YB in resistant cells compared with parental drug-sensitive cells. Recent studies indicate that microRNAs are involved in mediating drug sensitivity and resistance. Therefore, based on microRNA profiling results and computer- based microRNA target prediction programs, we studied the role of miR-485-3p that is differentially expressed in drug-sensitive and -resistant cells and has a putative target to the 3′-UTR of NF-YB. We first examined the repression effect of miR-485-3p to its target, NF-YB, by overexpressing miR-485-3p in the cells. We also confirmed, by luciferase assay, the binding of miR-485-3p to NF-YB 3′-UTR. Overexpressing miR-485-3p in CEM/VM-1-5 cells led to reduced expression of NF-YB, a corresponding up-regulation of Top2α, and increased sensitivity to the Top2 inhibitors.
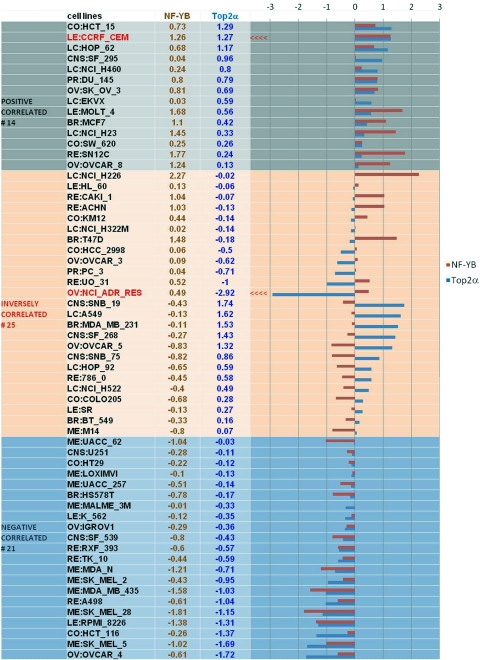
Cell cycle-dependent regulation of Top2α expression is mediated in part by the transcription factor NF-Y (Magan et al., 2003), a conserved transcription factor that consists of three different subunits, NF-YA, NF-YB, and NF-YC, and binds specifically to the CCAAT elements in the promoter (Ronchi et al., 1995). NF-Y has been reported to regulate the expression of several key cell cycle regulators, such as cyclin B1/B2 and cdc25 C (Manni et al., 2001). Some studies show that knockout of NF-YB causes early mouse embryo lethality (Bhattacharya et al., 2003) and overexpressing a dominant-negative mutant of NF-YA, which contains amino acid substitutions within the DNA-binding domain, also inhibits cell proliferation and growth (Hu and Maity, 2000). Because we showed earlier (Morgan and Beck, 2001) that NF-YB may be a mediator of Top2α at ICE3 in the Top2α promoter, and other studies also suggested the regulatory role for NF-Y in the Top2α promoter (Magan et al., 2003), we further examined the possibility that NF-YB causes down-regulation of Top2α in the drug-resistant cells. We have now shown that the NF-YB protein is up-regulated in the CEM/VM-1-5 cells that have decreased levels of Top2α compared with the drug-sensitive CEM cells. We observed a similar result in the human rhabdomyosarcoma Rh30 cell line and its etoposide-resistant subline, Rh30/v1, which also has down-regulated Top2α protein compared with the drug-sensitive parents. This strongly suggests that NF-YB is a negative regulator of Top2α. However, examination of Top2α and NF-YB gene expression levels in the National Cancer Institute (NCI)-60 panel of human tumor cancer cell lines (http://discover.nci.nih.gov/cellminer/) (Shankavaram et al., 2009) suggests that there is no significant overall correlation, negative or positive, between Top2α and NF-YB at the mRNA level (Fig. 5). Some cell lines show a positive relationship between the two genes, others show a negative relationship, and yet others show an inverse relationship, as we have demonstrated here. The CEM cell line in this figure shows that both genes are overexpressed. One explanation for this apparent contradiction with our present results is that CEM cell line in the NCI-60 panel is diploid (Roschke et al., 2003) instead of near tetraploid as are our CEM and CEM/VM-1-5 cell lines (Kusumoto et al., 1996). By contrast, some of the cell lines showing an inverse relationship between Top2α and NF-YB are derived from solid tumors, and, of possible relevance to our results, one is in the drug-resistant NCI/ADR-RES cell line. Moreover, examination of the genes of cell lines in which both genes are inversely expressed (Fig. 5, middle group), the ratio of Top2α to NF-YB, and the differential (opposite) expression of both genes tends to be higher than in the rest of the cells (i.e., the top and bottom groups in Fig. 5, in which genes are expressed in the same direction). We can speculate that the inverse relationship that we have observed herein and in those NCI-60 cells with inverse expression of these genes may reflect aspects of the biology of drug resistance. Clearly, examination of the expression of these genes in more pairs of drug-sensitive and drug-resistant cell lines is warranted.
Fig. 5.
NF-YB and Top2α mRNA expression levels in the NCI-60 panel of tumor cell lines. Shown is a graphical summary of NF-YB and Top2α mRNA levels in the form of a mean graph of the NCI-60 cells. Expression above the mean levels is drawn to the right of the centerline, and below the mean is drawn to the left.
The mechanism underlying the differential expression of miR-485-3p in drug-resistant and -sensitive cells is not clear. Some studies suggest that epigenetic alterations (Grady et al., 2008), deregulation of microRNA processing factors (Melo et al., 2009), and chromosomal abnormalities (Zhang et al., 2006) can contribute to down-regulation or up-regulation of microRNAs. MiR-485-3p resides on chromosome 14q32.31, a region that includes miR-127 (Saito et al., 2006), and miR-370 (Meng et al., 2008), which has been suggested to be epigenetically regulated. Furthermore, chromosome 14q32.31 is a region in which allelic deletions (Suzuki et al., 1989) and translocations (Avet-Loiseau et al., 2002) are frequently identified. Because microRNAs are expressed differently in drug-sensitive versus drug-resistant cells, and they have been shown to mediate drug response (Mishra et al., 2007; To et al., 2008; Bourguignon et al., 2009), studying the regulation of microRNAs during the emergence of drug resistance may reveal new therapeutic opportunities.
In conclusion, we believe that ours is the first study to demonstrate that miR-485-3p expression can mediate etoposide sensitivity indirectly by fine-tuning Top2α expression through the modification of NF-YB expression. Accordingly, miR-485-3p can be a putative therapeutic target to modulate etoposide resistance in tumor cells.
Supplementary Material
Acknowledgments
We thank Martina Vaskova for outstanding administrative support.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
This work was supported by the National Institutes of Health National Cancer Institute [Grant CA40570]; the University of Illinois at Chicago; and the National Institutes of Health National Center for Research Resources [Grant C06-RR15482].
Details of the microRNA profiling experiments have been provided previously: He X, Arslan AD, Ho T-T, and Beck WT (2009) MicroRNA profiling in drug-resistant leukemic cells, in Proceedings of the 100th Annual Meeting of the American Association for Cancer Research; 2009 Apr 18–22; Denver, CO. Abstract 5514. American Association for Cancer Research, Philadelphia.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/mol.110.069633.
ABBREVIATIONS:
- NF-Y
- nuclear factor Y
- 3′-UTR
- 3′-untranslated region
- hsa-miR-485-3p
- human microRNA-485-3p
- ICE
- inverted CCAAT element
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
- PCNA
- proliferating cell nuclear antigen
- PCR
- polymerase chain reaction
- RT
- reverse transcriptase
- Top2
- DNA topoisomerase II
- Top2α
- topoisomerase IIα
- HEK
- human embryonic kidney
- DMEM
- Dulbecco's modified Eagle's medium
- Ct
- cycle threshold.
Authorship Contributions
Participated in research design: Chen, He, and Beck.
Conducted experiments: Chen.
Contributed new reagents or analytic tools: Mo.
Performed data analysis: Chen, He, Arslan, Mo, Reinhold, Pommier, and Beck.
Wrote or contributed to the writing of the manuscript: Chen and Beck.
References
- Avet-Loiseau H, Facon T, Grosbois B, Magrangeas F, Rapp MJ, Harousseau JL, Minvielle S, Bataille R. Intergroupe Francophone du Myélome (2002) Oncogenesis of multiple myeloma: 14q32 and 13q chromosomal abnormalities are not randomly distributed, but correlate with natural history, immunological features, and clinical presentation. Blood 99:2185–2191 [DOI] [PubMed] [Google Scholar]
- Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- Beck WT. (1996) DNA topoisomerases and tumor cell resistance to their inhibitors, in Principles of Antineoplastic Drug Development and Pharmacology (Shilsky R, Milano G, Ratain M. eds) pp 487–502, M. Dekker, New York [Google Scholar]
- Bhat UG, Raychaudhuri P, Beck WT. (1999) Functional interaction between human topoisomerase IIalpha and retinoblastoma protein. Proc Natl Acad Sci USA 96:7859–7864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Deng JM, Zhang Z, Behringer R, de Crombrugghe B, Maity SN. (2003) The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res 63:8167–8172 [PubMed] [Google Scholar]
- Bourguignon LY, Spevak CC, Wong G, Xia W, Gilad E. (2009) Hyaluronan-CD44 interaction with protein kinase C(epsilon) promotes oncogenic signaling by the stem cell marker Nanog and the Production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J Biol Chem 284:26533–26546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6:857–866 [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. (2003) Role of microRNAs in plant and animal development. Science 301:336–338 [DOI] [PubMed] [Google Scholar]
- Champoux JJ. (1990) Mechanistic aspects of type-II topoisomerases, in DNA Topology and Its Biological Effects (Cozzarelli NZ, Wang JC. eds) pp 217–242, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Chung TD, Drake FH, Tan KB, Per SR, Crooke ST, Mirabelli CK. (1989) Characterization and immunological identification of cDNA clones encoding two human DNA topoisomerase II isozymes. Proc Natl Acad Sci USA 86:9431–9435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danks MK, Schmidt CA, Cirtain MC, Suttle DP, Beck WT. (1988) Altered catalytic activity of and DNA cleavage by DNA topoisomerase II from human leukemic cells selected for resistance to VM-26. Biochemistry 27:8861–8869 [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. (2006) Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 6:259–269 [DOI] [PubMed] [Google Scholar]
- Grady WM, Parkin RK, Mitchell PS, Lee JH, Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Willson JK, Kaz AM, et al. (2008) Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene 27:3880–3888 [DOI] [PubMed] [Google Scholar]
- Hu Q, Maity SN. (2000) Stable expression of a dominant negative mutant of CCAAT binding factor/NF-Y in mouse fibroblast cells resulting in retardation of cell growth and inhibition of transcription of various cellular genes. J Biol Chem 275:4435–4444 [DOI] [PubMed] [Google Scholar]
- Kusumoto H, Rodgers QE, Boege F, Raimondi SC, Beck WT. (1996) Characterization of novel human leukemic cell lines selected for resistance to merbarone, a catalytic inhibitor of DNA topoisomerase II. Cancer Res 56:2573–2583 [PubMed] [Google Scholar]
- Liu LF. (1989) DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem 58:351–375 [DOI] [PubMed] [Google Scholar]
- Magan N, Szremska AP, Isaacs RJ, Stowell KM. (2003) Modulation of DNA topoisomerase II alpha promoter activity by members of the Sp (specificity protein) and NF-Y (nuclear factor Y) families of transcription factors. Biochem J 374:723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni I, Mazzaro G, Gurtner A, Mantovani R, Haugwitz U, Krause K, Engeland K, Sacchi A, Soddu S, Piaggio G. (2001) NF-Y mediates the transcriptional inhibition of the cyclin B1, cyclin B2, and cdc25C promoters upon induced G2 arrest. J Biol Chem 276:5570–5576 [DOI] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. (2009) A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet 41:365–370 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Meng F, Wehbe-Janek H, Henson R, Smith H, Patel T. (2008) Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene 27:378–386 [DOI] [PubMed] [Google Scholar]
- Mishra PJ, Humeniuk R, Mishra PJ, Longo-Sorbello GS, Banerjee D, Bertino JR. (2007) A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci USA 104:13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan SE, Beck WT. (2001) Role of an inverted CCAAT element in human topoisomerase IIalpha gene expression in ICRF-187-sensitive and -resistant CEM leukemic cells. Mol Pharmacol 59:203–211 [DOI] [PubMed] [Google Scholar]
- Morgan SE, Cadena RS, Raimondi SC, Beck WT. (2000) Selection of human leukemic CEM cells for resistance to the DNA topoisomerase II catalytic inhibitor ICRF-187 results in increased levels of topoisomerase IIalpha and altered G(2)/M checkpoint and apoptotic responses. Mol Pharmacol 57:296–307 [PubMed] [Google Scholar]
- Mosmann T. (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63 [DOI] [PubMed] [Google Scholar]
- Nitiss JL. (2009a) DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer 9:327–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss JL. (2009b) Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer 9:338–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss JL, Beck WT. (1996) Antitopoisomerase drug action and resistance. Eur J Cancer 32A:958–966 [DOI] [PubMed] [Google Scholar]
- Pommier Y, Leo E, Zhang H, Marchand C. (2010) DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol 17:421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchi A, Bellorini M, Mongelli N, Mantovani R. (1995) CCAAT-box binding protein NF-Y (CBF, CP1) recognizes the minor groove and distorts DNA. Nucleic Acids Res 23:4565–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschke AV, Tonon G, Gehlhaus KS, McTyre N, Bussey KJ, Lababidi S, Scudiero DA, Weinstein JN, Kirsch IR. (2003) Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res 63:8634–8647 [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. (2006) Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9:435–443 [DOI] [PubMed] [Google Scholar]
- Shankavaram UT, Varma S, Kane D, Sunshine M, Chary KK, Reinhold WC, Pommier Y, Weinstein JN. (2009) CellMiner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics 10:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Yokota J, Mugishima H, Okabe I, Ookuni M, Sugimura T, Terada M. (1989) Frequent loss of heterozygosity on chromosome 14q in neuroblastoma. Cancer Res 49:1095–1098 [PubMed] [Google Scholar]
- To KK, Zhan Z, Litman T, Bates SE. (2008) Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol Cell Biol 28:5147–5161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai-Pflugfelder M, Liu LF, Liu AA, Tewey KM, Whang-Peng J, Knutsen T, Huebner K, Croce CM, Wang JC. (1988) Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21–22. Proc Natl Acad Sci USA 85:7177–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC. (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3:430–440 [DOI] [PubMed] [Google Scholar]
- Wu H, Zhu S, Mo YY. (2009) Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res 19:439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, et al. (2006) microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA 103:9136–9141 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.