Abstract
Palmitoylethanolamide (PEA) is an endogenous lipid amide that modulates pain and inflammation by engaging peroxisome proliferator-activated receptor type-α. Here, we show that the proinflammatory bacterial endotoxin lipopolysaccharide (LPS) decreases PEA biosynthesis in RAW264.7 macrophages by suppressing the transcription of N-acylphosphatidylethanolamine-specific phospholipase D (NAPE-PLD), which catalyzes the production of PEA and other lipid amides. Using a luciferase reporter construct and chromatin immunoprecipitation, we further show that LPS treatment reduces acetylation of histone proteins bound to the NAPE-PLD promoter, an effect that is blocked by the histone deacetylase inhibitor trichostatin A. The transcription factor Sp1 is involved in regulating baseline NAPE-PLD expression but not in the transcriptional suppression induced by LPS. The ability of LPS to down-regulate PEA biosynthesis is impaired in peritoneal macrophages from mutant NAPE-PLD-deficient mice, in which PEA is produced through a compensatory mechanism distinct from NAPE-PLD. Moreover, NAPE-PLD-deficient mice fail to mount a normal inflammatory reaction in response to carrageenan administration in vivo. Our findings suggest that proinflammatory stimuli suppress NAPE-PLD expression and PEA biosynthesis in macrophages and that this effect might contribute to the inflammatory response.
Introduction
PEA is a naturally occurring lipid amide that activates the nuclear receptor peroxisome proliferator-activated receptor-α (PPAR-α) (LoVerme et al., 2005). This interaction is probably responsible for the profound anti-inflammatory effects of PEA (Mazzari et al., 1996; Calignano et al., 1998; O'Sullivan, 2007), which are abrogated by genetic deletion of PPAR-α and closely mimicked by synthetic PPAR-α agonists (LoVerme et al., 2005, 2006). Although best known for its roles in the transcriptional regulation of lipid metabolism (Schoonjans et al., 1996), PPAR-α has been also implicated in the control of the inflammatory process, which it may influence by modulating the activities of nuclear factor-κB, activating protein-1, and inhibitor of κB kinase complex (Glass and Ogawa, 2006).
Endogenous PEA levels undergo striking changes during inflammation. Stimulation with proinflammatory agents such as LPS or carrageenan decreases PEA content in various cells and tissues of rodents, including intestine (Capasso et al., 2001), skin (LoVerme et al., 2005), and leukocytes (Endocannabinoid Research Group et al., 2010; Solorzano et al., 2009). Adding clinical relevance to these findings, it was shown that synovial fluid from patients with rheumatoid arthritis and osteoarthritis contains lower amounts of PEA compared with control subjects (Richardson et al., 2008). The possibility, suggested by these results, that endogenous PEA might participate in the inflammatory process is supported by experiments showing that pharmacological inhibition of PEA hydrolysis prevents the decrease in leukocyte PEA levels induced by inflammatory triggers and concomitantly blocks inflammation through a PPAR-α-dependent mechanism (Solorzano et al., 2009). A plausible interpretation of these results is that endogenous PEA acting at PPAR-α provides a stop signal that hinders the development of inflammation. Such a role is consistent with the phenotype of PPAR-α-deficient mice, which display a heightened sensitivity to proinflammatory agents (Devchand et al., 1996), and differentiates PEA from lipid mediators that either enhance the inflammatory process (e.g., prostaglandins) (Flower, 2006) or terminate it by promoting resolution and tissue healing (e.g., lipoxins and resolvins) (Serhan et al., 2008).
In the present study, we investigated the molecular mechanism through which the proinflammatory bacterial endotoxin LPS influences endogenous PEA levels in RAW264.7 cells and mouse peritoneal macrophages. We found that LPS suppresses the expression of the PEA-producing enzyme NAPE-PLD by influencing the acetylation state of histone proteins associated with the promoter region of the Nape-pld gene. In contrast, LPS does not alter the expression of two lipid amidases involved in PEA degradation: N-acylethanolamine-hydrolyzing acid amidase (NAAA) (Tsuboi et al., 2007), and fatty-acid amide hydrolase (FAAH) (McKinney and Cravatt, 2005). Our results further suggest that the down-regulation of NAPE-PLD expression may be functionally important because mutant NAPE-PLD-null mice, in which this regulatory process is defective, are unable to mount a normal inflammatory reaction in response to carrageenan.
Materials and Methods
Animals and Cells.
RAW264.7 cells were purchased from the American Type Culture Collection (Manassas, VA), and cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with fetal bovine serum (FBS, 10%; Invitrogen). C57BL/6J NAPE-PLD(−/−) mice were generated as described previously (Leung et al., 2006) and backcrossed 10 times to C57BL/6J wild-type mice (The Jackson Laboratory, Bar Harbor, ME). All procedures met the National Institutes of Health guidelines for the care and use of laboratory animals and were approved by the University of California, Irvine, Institutional Animal Care and Use Committee.
Plasmids.
We amplified a 2-kilobase sequence of the mouse Nape-pld promoter from mouse brain genomic DNA by polymerase chain reaction (PCR) using High-Fidelity PCR Master (Roche Diagnostics, Indianapolis, IN). We designed two primers using sequences obtained from the National Center for Biotechnology Information database: 5′-promoter (5′-CGTTCTGAGTCTTCCACTTCCTAAGAGAGAC-3′) and 3′-promoter (5′-AAACCTACTCGAAACCCGCTGCACTT-3′). The PCR product was subcloned into a pCR2.1-TOPO vector (Invitrogen) for screening and sequence, followed by digestion with BglI/NheI (Roche Diagnostics). The insert was subsequently cloned into the pGL3-Basic Vector (Promega, Madison, WI). Plasmids were transfected into RAW264.7 cells using Fugene HD 6 (Roche Diagnostics) following the manufacturer's instructions.
mRNA Extraction and Real-Time PCR.
We extracted total RNA using TRIzol (Invitrogen). cDNA was synthesized with 0.2 μg of total RNA and oligo(dT)12–18 primer using Superscript II RNase H-reverse transcriptase (Invitrogen). Quantitative real-time PCR was performed with an Mx3000P Real-Time PCR System (Stratagene, La Jolla, CA). We designed primer/probe sets using the Primer Express software based on gene sequences available from the GenBank database. Primers and fluorogenic probes were synthesized at TIB MolBiol (Adelphia, NJ). The primer/probe sequences for mouse genes were as follows: Nape-pld, forward, 5′-AACGAGCGGTTCGGCA-3′, reverse, 5′-ATCCAGTCAAGAAGGCCCAA-3′, probe, 5′-CGAGCTGCGGTGGTTTGTGCC-3′; Naaa, forward, 5′-CGGTGGCGCAGGTCA-3′, reverse, 5′-AATTTCTCCGACCATCCCG-3′, probe, 5′-TGGCGACAGGGTTCCCCAGTG-3′; Faah, forward, 5′-CCTTATGCCCTGGAGGTCCT-3′, reverse, 5′-GGAGAAAAGAGCAGCCACC A-3′, probe, 5′-TCGGCAGGTGGGCTGTTCAGTGT-3′; and glyceraldehyde 3-phosphate dehydrogenase, forward, 5′-TCACTGGCATGGCCTTCC-3′, reverse, 5′-GGCGGCACGTCAGATCC-3′, probe, 5′-TTCCTACCCCCAATGTGTCCGTCG-3′. RNA levels were normalized using glyceraldehyde 3-phosphate dehydrogenase as an internal standard.
Protein Analyses.
Protein concentrations were measured using the bicinchoninic acid assay (Pierce Chemical, Rockford, IL). Proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK). Overnight incubation in the presence of a previously characterized anti-NAPE-PLD antibody (1:2000) (Fu et al., 2007) at 4°C was followed by incubation with horseradish peroxidase conjugated anti-rabbit IgG antibody (1:3000; Sigma-Aldrich, St. Louis, MO) for 1 h at room temperature. The anti-NAPE-PLD antibody was kindly provided by Dr. Ken Mackie (Indiana University, Bloomington, IN) and was raised in rabbits as described previously (Fu et al., 2007). Protein bands were visualized using the ECL Plus kit (Amersham Biosciences).
Enzyme Assays.
We measured NAPE-PLD activity at 37°C for 30 min in Tris-HCl buffer (50 mM), pH 7.4, containing 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich), protein (0.1 mg), and 1,2-dipalmitoyl-sn-glycero-3-phospho-ethanolamine-N-heptadecenoyl (0.1 mM) as substrate, which was prepared as described previously (Fu et al., 2007). Reactions were stopped by adding chloroform/methanol [2:1 (v/v)] containing [2H4]oleoylethanolamide as internal standard. [2H4]Oleoylethanolamide was prepared as described previously (Fu et al., 2007). After centrifugation at 1500g at 4°C for 5 min, the organic layers were collected and dried under N2. The residues were suspended in 50 μl of chloroform/methanol [1:3 (v/v)] and analyzed by liquid chromatography/mass spectrometry (LC/MS), monitoring the [M+Na]+ ions of m/z = 334 for N-heptadecenoylethanolamide and m/z = 352 for [2H4]oleoylethanolamide. To measure FAAH activity, cell homogenates were incubated at 37°C for 30 min in Tris buffer (50 mM), pH 8.0, containing fatty acid-free bovine serum albumin (0.05%), and [ethanolamine-3H]anandamide (10,000 dpm; specific activity, 20 Ci/mmol). After stopping the reaction with a mixture of chloroform/methanol [1:1 (v/v)], we measured radioactivity in the aqueous layers by liquid scintillation counting. To measure NAAA activity, cell homogenates were incubated at 37°C for 30 min in 0.2 ml of sodium hydrogen phosphate buffer (50 mM), pH 4.5, containing 0.1% Triton X-100, 3 mM dithiothreitol, and 50 μM heptadecenoylethanolamide as substrate. The reaction was terminated by the addition of 0.2 ml of ice-cold methanol containing 1 nmol of heptadecanoic acid (NuChek Prep, Elysian, MN). Samples were analyzed by LC/MS. Heptadecenoic and heptadecanoic acids were eluted on an Eclipse XDB-C18 column (Agilent Technologies, Santa Clara, CA) isocratically at 2.2 ml/min for 1 min with a solvent mixture of 95% methanol and 5% water, both containing 0.25% acetic acid and 5 mM ammonium acetate. The column temperature was 50°C. Electrospray ionization was in the negative mode, capillary voltage was 4 kV, and fragmentor voltage was 100 V. N2 was used as drying gas at a flow rate of 13 L/min and a temperature of 350°C. Nebulizer pressure was set at 60 psi. We monitored [M − H]− in the selected-ion monitoring mode using heptadecanoic acid as internal standard. Calibration curves were generated using commercial heptadecanoic acid (m/z = 267; Nu-Chek Prep, Elysian, MN).
Lipid Analyses.
Lipids were extracted using a chloroform/methanol mixture [2:1 (v/v), 3 ml] containing appropriate internal standards. The organic phases were collected, dried under N2, and dissolved in methanol/chloroform [3:1 (v/v)] for LC/MS analyses.
PEA.
We used an Agilent 1100-LC system coupled to a 1946A-MS detector equipped with an electrospray ionization interface (Agilent Technologies). PEA was separated on a Eclipse XDB-C18 column (50 × 4.6 mm internal diameter, 1.8 μm; Zorbax, Agilent Technologies) with a gradient of methanol in water (from 85 to 90% methanol in 2.0 min and 90 to 100% in 3.0 min) at a flow rate of 1.5 ml/min. Column temperature was kept at 40°C. MS detection was in the positive ionization mode, capillary voltage was 3 kV, and fragmentor voltage was 120 V. N2 was used as drying gas at a flow rate of 13 L/min and a temperature of 350°C. Nebulizer pressure was set at 60 psi. Detection and analysis was by Agilent Chemstation.
N-Palmitoyl-phosphatidylethanolamine.
We used an 1100-LC system equipped with an Ion Trap XCT (both from Agilent Technologies). N-Palmitoyl-phosphatidylethanolamine was separated on a 300SB C-18 column (75 × 2.1 mm internal diameter, 5 μm; Poroshell, Agilent Technologies) with a gradient methanol in water containing 5 mM ammonium acetate and 0.25% acetic acid (from 85 to 100% of water in 4 min) at a flow rate of 1.0 ml/min. Column temperature was kept at 50°C. Detection was set in the negative mode, capillary voltage was 4.5 kV, and fragmentor voltage was 120 V. N2 was used as drying gas at a flow rate of 13 L/min and a temperature of 350°C. Nebulizer pressure was set at 80 psi. Helium was used as the collision gas. Cell-derived NAPEs were identified by comparison of their LC retention times and MSn fragmentation patterns with those of authentic standards. Extracted ion chromatograms were used to quantify each NAPE precursor ion by monitoring characteristic product ions as follows: 1-stearoyl-2-arachidonoyl-sn-glycero-phosphoethanolamine-N-palmitoyl (m/z 1004.8 > 718.8) and 1-O-hexadecyl-2-palmitoyl-sn-glycero-3-phospho-N-palmitoyl-ethanolamide (m/z 914.8 > 676.8), which was used as internal standard. Detection and analysis were controlled by Agilent/Bruker Daltonics (Billerica, MA) software version 5.2, and three-dimensional maps were generated using MS Processor from Advanced Chemistry Development, Inc. (Toronto, ON, Canada).
Chromatin Immunoprecipitation.
Chromatin immunoprecipitation (ChIP) assays were conducted using a histone H3 ChIP assay kit following the manufacturer's protocol (Millipore Corporation, Billerica, MA). In brief, ∼3 × 107 RAW264.7 cells were cross-linked with 1% formaldehyde at 37°C for 10 min, followed by incubation with 125 mM glycine for 10 min. Cells were then rinsed twice with ice-cold phosphate-buffered saline and harvested by brief centrifugation. Pellets were suspended in SDS lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS, 0.5 mM PMSF, and protease inhibitor cocktail; Roche Diagnostics). After 20-min incubation on ice, sonication was performed on ice to achieve chromatin fragments ranging between 200 and 1000 bp in size, followed by centrifugation at 15,000g for 15 min at 4°C. Supernatants were collected and diluted 10-fold in ChIP dilution buffer (50 mM Tris-HCl, pH 8.0, 167 mM NaCl, 1.1% Triton X-100, 0.11% sodium deoxycholate, 0.5 mM PMSF, and protease inhibitor cocktail). Samples were subjected to preimmunoprecipitation clearing with 60 μl of a mixture of salmon sperm DNA/protein A/protein G at 4°C with rotation for 4 to 6 h. A 0.2-ml aliquot was removed to serve as an input sample. Immunoprecipitation was carried out with 5 μg of anti-Sp1 (Millipore Corporation, Billerica, MA) or anti-acetylhistone H3 (Millipore Corporation) antibodies at 4°C overnight with rotation. After immunoprecipitation, 20 μl of a mixture of salmon sperm DNA/protein A/protein G was added and incubated at 4°C with rotation for 3 h and followed by brief centrifugation. The precipitates were washed once for 5 min at 4°C with low-salt buffer, once with high-salt buffer, and once with LiCl buffer. The precipitates were washed again with Tris-EDTA buffer twice for 5 min each at 4°C, and immune complexes were extracted twice with 0.2 ml of elution buffer (10 mM Tris-HCl, pH 8.0, 0.3 M NaCl, 5 mM EDTA, pH 8.0, and 0.5% SDS). Eluates and input were heated at 65°C overnight to reverse the cross-link. After RNase A and proteinase K treatment, DNA was extracted with a phenol/chloroform/isoamyl alcohol solution and precipitated with ethanol. The recovered DNA was resuspended in 20 μl of distilled water, and 2 μl was used for PCR. The PCR products were analyzed on a 2% agarose gel. Primer sequence was the following: 5′-TCG AGG CCT CTG CGC CTC TGT CC-3′ and 5′-GGG AGG CTC ACC CGC AGG CG-3′. Anti-FLAG antibody (Sigma-Aldrich) served as negative control.
Reporter Assays.
The NAPE-PLD-luciferase and LacZ constructs (0.5 μg) were transfected into RAW264.7 cells using FuGENE HD 6 following the manufacturer's instructions. Six hours after transfection, the cells were incubated with vehicle or drugs (LPS, 100 ng/ml; mithramycin A, 0.1, 0.3, and 1.0 μM; trichostatin A, 1 μM; nicotinamide, 1 mM; all from Sigma-Aldrich) for 12 h in culture medium supplemented with 10% FBS. Cells were harvested and tested for luciferase activity using a dual-luciferase reporter assay system (Promega) on a MIX Microtiter plate luminometer (Dynex Technologies, Chantilly, VA).
Carrageenan-Induced Inflammation.
Sterile polyethylene sponges (1 cm3) were implanted under the dorsal skin of C57BL/6J wild-type and C57BL/6J NAPE-PLD(−/−) mice. Carrageenan (0.1–0.3% in 100 μl sterile water/sponge) was instilled into the sponges, wounds were sutured, and the animals were allowed to recover. After 6 to 72 h, mice were sacrificed, and sponges were collected. Exudate volume was measured and cells were counted using a hemocytometer.
Peritoneal Macrophage Cultures.
Resident peritoneal macrophages were harvested from C57BL/6J wild-type and C57BL/6J NAPE-PLD(−/−) mice by peritoneal lavage using sterile phosphate-buffered saline at 4°C. The macrophages were plated in 6-cm dishes at a density of 5 × 106 cells/dish in Dulbecco's modified Eagle's medium containing 10% FBS and 1% penicillin and streptomycin.
Statistics.
Results are expressed as the mean ± S.E.M. of n observations. They were analyzed by one-way analysis of variance followed by Bonferroni post hoc test for multiple comparisons. P values <0.05 were considered to be significant.
Results
Inflammation Suppresses NAPE-PLD Expression in RAW264.7 Macrophages.
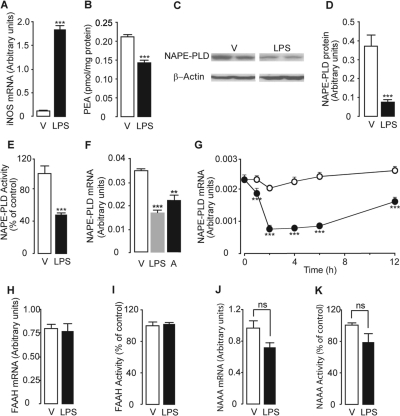
Stimulation of RAW264.7 cells with LPS (100 ng/ml, 6 h) increased inducible nitric-oxide synthase mRNA levels (Fig. 1A), an index of macrophage activation (Ohmori and Hamilton, 2001), and decreased cellular PEA content (Solorzano et al., 2009) (Fig. 1B) without altering the levels of the PEA precursor, N-palmitoyl-phosphatidylethanolamine (in picomoles per milligram of protein: vehicle, 6.15 ± 0.86; LPS, 7.79 ± 0.204; P = 0.5358, n = 3). These effects were accompanied by a reduction in NAPE-PLD expression, as documented by immunoblot, enzymatic, and reverse transcriptase-PCR analyses (Fig. 1, C–F). The time course of NAPE-PLD mRNA down-regulation, illustrated in Fig. 1G, shows that the response was maximal within 2 h of LPS application. By contrast, LPS did not significantly affect NAAA and FAAH expression (Fig. 1, H–K). Prior work has shown that the proinflammatory effects of LPS are mediated by activation of Toll-like receptor-4 (TLR4) (Poltorak et al., 1998). Accordingly, stimulation of RAW264.7 cells with the selective TLR4 agonist 3-deoxy-d-mannose-octulosonic acid-lipid A (100 ng/ml, 6 h) (Raetz et al., 2006) reduced NAPE-PLD mRNA levels (Fig. 1F). These findings indicate that LPS lowers PEA levels in RAW264.7 macrophages by suppressing NAPE-PLD-dependent PEA biosynthesis.
Fig. 1.
LPS suppresses PEA production in RAW264.7 macrophages through down-regulation of NAPE-PLD expression. Effects of vehicle (V) or LPS (100 ng/ml, 6 h) on inducible nitric-oxide synthase mRNA levels (A) and PEA levels (B). C to E, effects of vehicle or LPS on NAPE-PLD protein levels (C and D) and NAPE-PLD activity levels(E). F, effects of vehicle, LPS (100 ng/ml, 6 h), or the TLR4 agonist 3-deoxy-d-mannose-octulosonic acid-lipid A (A, 100 ng/ml, 6 h) on NAPE-PLD mRNA levels. G, time course of the effects of vehicle (○) or LPS (●) on NAPE-PLD mRNA levels. H to K, effects of vehicle or LPS (100 ng/ml, 6 h) on FAAH mRNA levels (H), FAAH activity (I), NAAA mRNA levels (J); and NAAA activity (K). **, P < 0.01 versus vehicle; ***, P < 0.001 versus vehicle (n = 4–6).
LPS Regulates NAPE-PLD Transcription.
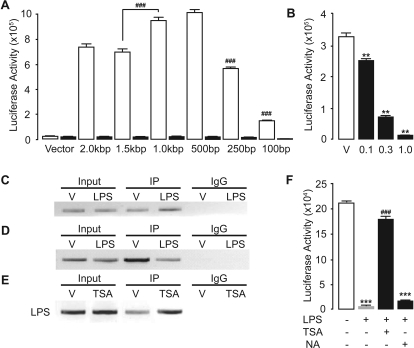
To identify mechanisms through which LPS regulates the expression of NAPE-PLD, we cloned a 2-kilobase sequence of the Nape-pld promoter and ligated it upstream of the luciferase reporter gene. Reporter assays in RAW264.7 cells using serially truncated promoter constructs revealed the presence of a repressive element in the −1500- to −1000-bp region upstream of the putative transcription start site (Fig. 2A). Transcriptional activity by the promoter construct decreased significantly when the sequence was shortened to less than 500 bp, suggesting that this region contains consensus sequences for the binding of transcription factors (Fig. 2A). Computational prediction of the 250-bp 5′-untranslated region suggested the existence of binding sites for Sp1, which is known to interact with GC-rich sequences (Kadonaga et al., 1988). Consistent with this possibility, the Sp1 inhibitor mithramycin A (Chatterjee et al., 2001) impaired, in a concentration-dependent manner, transcriptional activity by the NAPE-PLD promoter construct (Fig. 2B). Confirming that Sp1 regulates baseline NAPE-PLD expression, chromatin immunoprecipitation experiments showed that Sp1 binds to the proximal segment of the NAPE-PLD promoter (Fig. 2C). It is noteworthy, however, that all NAPE-PLD promoter constructs were fully responsive to the transcriptional suppression induced by LPS (Fig. 2A), and LPS did not decrease the binding of Sp1 to the NAPE-PLD promoter (Fig. 2C). These results suggest that Sp1 is involved in the basal control of NAPE-PLD transcription but not in the regulation of NAPE-PLD transcription by LPS.
Fig. 2.
Transcriptional regulation of NAPE-PLD expression in RAW264.7 macrophages. A, effects of vehicle (□) or LPS (■) on luciferase activity levels of serially truncated NAPE-PLD promoter constructs. B, effects of vehicle (V) or the Sp1 inhibitor mithramycin A (0.1–1.0 μM) on NAPE-PLD promoter activity. C, chromatin immunoprecipitation using anti-Sp1 antibody on cells treated with vehicle or LPS. D and E, histone acetylation levels at NAPE-PLD promoter region assessed by chromatin immunoprecipitation using anti-acetyl histone H3 antibody following either treatment with LPS (D) or HDAC inhibitor trichostatin A (TSA) (E). F, effect of TSA (1 μM) or nicotinamide (NA, 1 mM) on the effects induced by LPS on the transcriptional activity of the NAPE-PLD promoter. **, P < 0.01 versus vehicle; ***, P < 0.001 versus vehicle/vehicle; ###, P < 0.001 versus LPS/vehicle or the 1.5-kilobase pair construct (n = 3–5).
Treatment with LPS increases the expression of histone deacetylase (HDAC) enzymes in macrophages, resulting in the modulation of inflammation-responsive genes (Aung et al., 2006). Furthermore, Sp1 and HDACs physically interact and mediate the silencing of various genes through histone deacetylation (Won et al., 2002; Enya et al., 2008; Liao et al., 2008). These data prompted us to ask whether histone deacetylation might be involved in the transcriptional repression of NAPE-PLD caused by LPS. Chromatin immunoprecipitation analysis of the NAPE-PLD promoter with an anti-acetyl histone H3 antibody showed that LPS causes a reduction in the amount of acetylated histone bound to the NAPE-PLD promoter (Fig. 2D). Moreover, LPS-induced NAPE-PLD down-regulation was prevented by the broad-spectrum HDAC inhibitor, trichostatin A (Yoshida et al., 1990) (Fig. 2F), which also increased histone H3 acetylation (Fig. 2E). By contrast, nicotinamide, a competitive sirtuin inhibitor (Avalos et al., 2005), did not affect the transcriptional repression of NAPE-PLD by LPS (Fig. 2F), implying that NAD+-dependent class III HDACs are not involved. The results suggest that LPS suppresses NAPE-PLD expression by changing the acetylation state of histone proteins bound to the NAPE-PLD promoter.
Inflammation Does Not Suppress PEA Production in NAPE-PLD(−/−) Mice.
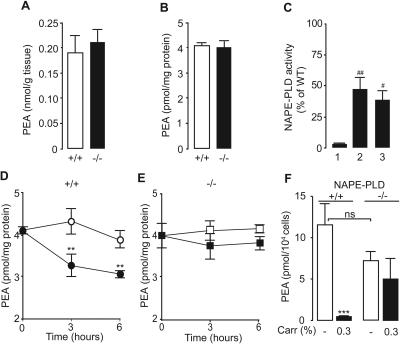
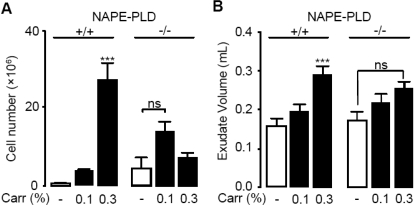
Prior work has shown that mutant mice in which the Nape-pld gene is disrupted by homologous recombination produce PEA through an alternative enzymatic pathway, which remains unidentified (Leung et al., 2006; Simon and Cravatt, 2006). Consistent with those results, we found that primary cultures of peritoneal macrophages from NAPE-PLD-deficient [NAPE-PLD(−/−)] mice contain normal amounts of PEA and significant levels of NAPE-PLD-like activity (Fig. 3A–C). It is noteworthy that when NAPE-PLD(−/−) macrophages were exposed to LPS (100 ng/ml), they failed to exhibit the down-regulation of PEA production observed in wild-type macrophages (Fig. 3, D and E). This suggests that macrophages from NAPE-PLD(−/−) mice cannot suppress PEA production in response to proinflammatory challenges, likely because the as-yet-unidentified enzyme(s) that produces PEA in NAPE-PLD(−/−) mice lacks the regulatory elements responsible for the inflammation-dependent suppression of NAPE-PLD expression. In vivo experiments provided evidence supporting this interpretation. We induced a localized inflammatory reaction in mice by implanting subcutaneously polyethylene sponges instilled with the proinflammatory polysaccharide carrageenan and collected infiltrating neutrophils and macrophages 3 days after surgery (Solorzano et al., 2009). LC/MS analyses of the lipid extracts showed that carrageenan decreases PEA levels in infiltrating leukocytes of wild-type mice but not NAPE-PLD(−/−) mutants (Fig. 3F). Furthermore, in vivo administration of carrageenan elicited a dose-dependent increase in leukocyte infiltration and edema in wild-type mice, whereas this effect was greatly attenuated in NAPE-PLD(−/−) mice, in which a dose of carrageenan that caused maximal leukocyte infiltration in wild-type mice was virtually ineffective (Fig. 4). We interpret these findings to indicate that mutant NAPE-PLD(−/−) mice cannot suppress PEA production in response to LPS or carrageenan.
Fig. 3.
Proinflammatory stimuli fail to suppress PEA biosynthesis in NAPE-PLD(−/−) mice. A and B, PEA levels in spleen (A) and peritoneal macrophages (B) from wild-type C57BL/6J (+/+) and NAPE-PLD(−/−) (−/−) mice. C, NAPE-PLD activity in brain (1), spleen (2), and peritoneal macrophage (3) homogenates of NAPE-PLD(−/−) mice, expressed as a percentage of activity in same tissues from wild-type mice. D and E, effects of vehicle (open symbols) or LPS (100 ng/ml, closed symbols) on PEA levels in macrophages from wild-type (D) and NAPE-PLD(−/−) mice (E). F, dose-dependent effects of carrageenan (Carr, 0.1–0.3%) on PEA levels in infiltrating leukocytes from wild-type (+/+) and NAPE-PLD(−/−) (−/−) mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus vehicle; #, P < 0.05; ##, P < 0.01 versus brain (n = 4–8); ns, not significant.
Fig. 4.
Defective responses to pro-inflammatory stimuli in NAPE-PLD(−/−) mice. A and B, dose-dependent effects of carrageenan (Carr, 0.1–0.3%) on leukocyte infiltration (A) and exudate volume (B) in wild-type (+/+) and NAPE-PLD(−/−) (−/−) mice. ***, P < 0.001 versus vehicle; ns, not significant.
Discussion
The main finding of the present study is that the bacterial endotoxin LPS, a powerful proinflammatory stimulus, interrupts PEA biosynthesis in macrophages by suppressing NAPE-PLD expression. Promoter reporter and chromatin immunoprecipitation experiments suggest that basal NAPE-PLD expression is controlled by the transcription factor Sp1, whereas LPS-induced suppression of NAPE-PLD expression requires decreased acetylation of histone proteins associated with the promoter region of the Nape-pld gene. This regulatory mechanism is strikingly absent in NAPE-PLD(−/−) mice, which fail to reduce PEA production in response to LPS and carrageenan. The results suggest that proinflammatory stimuli down-regulate NAPE-PLD expression and consequently reduce the production of the anti-inflammatory lipid amide PEA.
PEA is structurally similar to the endocannabinoid anandamide but does not interact with cannabinoid receptors (Devane et al., 1992; Lo Verme et al., 2005). Rather, PEA produces a well characterized series of analgesic and anti-inflammatory effects in animals and humans, which are mediated by activation of PPAR-α receptors (Masek et al., 1974; Kahlich et al., 1979; Mazzari et al., 1996; Costa et al., 2002; Lo Verme et al., 2005; Kemeny et al., 2007; Genovese et al., 2008; Endocannabinoid Research Group et al., 2010). Nevertheless, the intrinsic roles of PEA in the response to proinflammatory triggers remain unclear. Evidence that this lipid amide serves as an endogenous anti-inflammatory factor was provided by experiments that showed that stimulation with either LPS or carrageenan lowers PEA levels in mouse leukocytes (Solorzano et al., 2009). The same study also reported that NAAA inhibition normalizes PEA levels in activated leukocytes and blunts inflammatory responses to LPS in vitro and carrageenan in vivo (Solorzano et al., 2009). The present results provide additional support for a role of PEA as an endogenous anti-inflammatory mediator. In particular, our experiments with mutant NAPE-PLD(−/−) mice indicate that the ability of activated macrophages to down-regulate NAPE-PLD expression and reduce PEA production is critical for the normal development of inflammation.
Our results also show that NAPE-PLD expression in leukocytes is controlled by two distinct transcriptional mechanisms. Under unstimulated conditions, NAPE-PLD expression is maintained through a steady-state level of histone acetylation, which renders the Nape-pld gene accessible to transcriptional regulators such as Sp1. When stimulated with LPS, macrophages initiate a molecular cascade that results in the transcriptional suppression of NAPE-PLD, which is associated with decreased acetylation of histone proteins bound to the NAPE-PLD promoter. This model is supported by the effects of the HDAC inhibitor trichostatin A, which blocks the transcriptional control of NAPE-PLD expression by LPS and concomitantly increases histone acetylation. Further experiments are needed to identify the effectors responsible for mediating histone deacetylation at the NAPE-PLD promoter, as well as the histones involved in this response. Additional experiments will also need to investigate the relative roles of NAPE-PLD and NAAA in PEA turnover. Our studies show indeed that the incomplete suppression in NAPE-PLD expression caused by LPS (approximately 50% of control) is paralleled by a similar reduction in PEA levels (approximately 30% of control), which is suggestive that NAPE-PLD represents a rate-limiting step in the tonic production of PEA under nonstimulated conditions.
In conclusion, our results provide new information on a signaling system, mediated by PEA, which negatively regulates macrophage activation and facilitates the maintenance of a homeostatic “anti-inflammatory” state in tissues (Nathan, 2002). Future investigations on the role of PEA in inflammation might help to define novel therapeutic strategies for the treatment of chronic inflammatory conditions.
Acknowledgments
We thank Dr. B. Cravatt for the gift of NAPE-PLD heterozygous mice [NAPE-PLD(+/−)]; Drs. J. Fu, K.M. Jung, and R. Edwards for experimental advice; and J. Lockney, C. Moazam, and Y. Zahedi for assistance with experiments.
This work was supported by the Sandler Asthma Foundation [Grant 02-0075]; and the National Institutes of Health National Institute on Drug Abuse [R01-DA012413].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/mol.110.070201.
ABBREVIATIONS:
- PPAR-α
- peroxisome proliferator-activated receptor-α
- PEA
- palmitoylethanolamide
- NAPE-PLD
- N-acylphosphatidylethanolamine-specific phospholipase D
- LPS
- lipopolysaccharide
- HDAC
- histone deacetylase
- NAAA
- N-acylethanolamine-hydrolyzing acid amidase
- FAAH
- fatty-acid amide hydrolase
- FBS
- fetal bovine serum
- PCR
- polymerase chain reaction
- PMSF
- phenylmethylsulfonyl fluoride
- MS
- mass spectrometry
- LC/MS
- liquid chromatography/mass spectrometry
- ChIP
- chromatin immunoprecipitation
- bp
- base pair
- TLR4
- Toll-like receptor-4.
Authorship Contributions
Participated in research design: Zhu, Solorzano, Sahar, Fung, Sassone-Corsi, and Piomelli.
Conducted experiments: Zhu, Solorzano, Realini, and Fung.
Contributed new reagents or analytic tools: Zhu, Sassone-Corsi, and Piomelli.
Performed data analysis: Zhu, Solorzano, Sahar, Realini, Fung, and Piomelli.
Wrote or contributed to the writing of the manuscript: Zhu, Solorzano, Fung, Sassone-Corsi, and Piomelli.
References
- Aung HT, Schroder K, Himes SR, Brion K, van Zuylen W, Trieu A, Suzuki H, Hayashizaki Y, Hume DA, Sweet MJ, et al. (2006) LPS regulates proinflammatory gene expression in macrophages by altering histone deacetylase expression. FASEB J 20:1315–1327 [DOI] [PubMed] [Google Scholar]
- Avalos JL, Bever KM, Wolberger C. (2005) Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol Cell 17:855–868 [DOI] [PubMed] [Google Scholar]
- Calignano A, La Rana G, Giuffrida A, Piomelli D. (1998) Control of pain initiation by endogenous cannabinoids. Nature 394:277–281 [DOI] [PubMed] [Google Scholar]
- Capasso R, Izzo AA, Fezza F, Pinto A, Capasso F, Mascolo N, Di Marzo V. (2001) Inhibitory effect of palmitoylethanolamide on gastrointestinal motility in mice. Br J Pharmacol 134:945–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Zaman K, Ryu H, Conforto A, Ratan RR. (2001) Sequence-selective DNA binding drugs mithramycin A and chromomycin A3 are potent inhibitors of neuronal apoptosis induced by oxidative stress and DNA damage in cortical neurons. Ann Neurol 49:345–354 [PubMed] [Google Scholar]
- Costa B, Conti S, Giagnoni G, Colleoni M. (2002) Therapeutic effect of the endogenous fatty acid amide, palmitoylethanolamide, in rat acute inflammation: inhibition of nitric oxide and cyclo-oxygenase systems. Br J Pharmacol 137:413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. (1992) Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science 258:1946–1949 [DOI] [PubMed] [Google Scholar]
- Devchand PR, Keller H, Peters JM, Vazquez M, Gonzalez FJ, Wahli W. (1996) The PPARalpha-leukotriene B4 pathway to inflammation control. Nature 384:39–43 [DOI] [PubMed] [Google Scholar]
- Endocannabinoid Research Group, De Filippis D, D'Amico A, Cipriano M, Petrosino S, Orlando P, Di Marzo V, Iuvone T. (2010) Levels of endocannabinoids and palmitoylethanolamide and their pharmacological manipulation in chronic granulomatous inflammation in rats. Pharmacol Res 61:321–328 [DOI] [PubMed] [Google Scholar]
- Enya K, Hayashi H, Takii T, Ohoka N, Kanata S, Okamoto T, Onozaki K. (2008) The interaction with Sp1 and reduction in the activity of histone deacetylase 1 are critical for the constitutive gene expression of IL-1 alpha in human melanoma cells. J Leukoc Biol 83:190–199 [DOI] [PubMed] [Google Scholar]
- Flower RJ. (2006) Prostaglandins, bioassay and inflammation. Br J Pharmacol 147:S182–S192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Astarita G, Gaetani S, Kim J, Cravatt BF, Mackie K, Piomelli D. (2007) Food intake regulates oleoylethanolamide formation and degradation in the proximal small intestine. J Biol Chem 282:1518–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese T, Esposito E, Mazzon E, Di Paola R, Meli R, Bramanti P, Piomelli D, Calignano A, Cuzzocrea S. (2008) Effects of palmitoylethanolamide on signaling pathways implicated in the development of spinal cord injury. J Pharmacol Exp Ther 326:12–23 [DOI] [PubMed] [Google Scholar]
- Glass CK, Ogawa S. (2006) Combinatorial roles of nuclear receptors in inflammation and immunity. Nat Rev Immunol 6:44–55 [DOI] [PubMed] [Google Scholar]
- Kadonaga JT, Courey AJ, Ladika J, Tjian R. (1988) Distinct regions of Sp1 modulate DNA binding and transcriptional activation. Science 242:1566–1570 [DOI] [PubMed] [Google Scholar]
- Kahlich R, Klíma J, Cihla F, Franková V, Masek K, Rosický M, Matousek F, Bruthans J. (1979) Studies on prophylactic efficacy of N-2-hydroxyethyl palmitamide (Impulsin) in acute respiratory infections. Serologically controlled field trials. J Hyg Epidemiol Microbiol Immunol 23:11–24 [PubMed] [Google Scholar]
- Kemeny L, Koreck A, Kis K, Kenderessy-Szabo A, Bodai L, Cimpean A, Paunescu V, Raica M, Ghyczy M. (2007) Endogenous phospholipid metabolite containing topical product inhibits ultraviolet light-induced inflammation and DNA damage in human skin. Skin Pharmacol Physiol 20:155–161 [DOI] [PubMed] [Google Scholar]
- Leung D, Saghatelian A, Simon GM, Cravatt BF. (2006) Inactivation of N-acyl phosphatidylethanolamine phospholipase D reveals multiple mechanisms for the biosynthesis of endocannabinoids. Biochemistry 45:4720–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Zhang Y, Dufau ML. (2008) Protein kinase Calpha-induced derepression of the human luteinizing hormone receptor gene transcription through ERK-mediated release of HDAC1/Sin3A repressor complex from Sp1 sites. Mol Endocrinol 22:1449–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Verme J, Fu J, Astarita G, La Rana G, Russo R, Calignano A, Piomelli D. (2005) The nuclear receptor peroxisome proliferator-activated receptor-alpha mediates the anti-inflammatory actions of palmitoylethanolamide. Mol Pharmacol 67:15–19 [DOI] [PubMed] [Google Scholar]
- LoVerme J, La Rana G, Russo R, Calignano A, Piomelli D. (2005) The search for the palmitoylethanolamide receptor. Life Sci 77:1685–1698 [DOI] [PubMed] [Google Scholar]
- LoVerme J, Russo R, La Rana G, Fu J, Farthing J, Mattace-Raso G, Meli R, Hohmann A, Calignano A, Piomelli D. (2006) Rapid broad-spectrum analgesia through activation of peroxisome proliferator-activated receptor-alpha. J Pharmacol Exp Ther 319:1051–1061 [DOI] [PubMed] [Google Scholar]
- Masek K, Perlík F, Klíma J, Kahlich R. (1974) Prophylactic efficacy of N-2-hydroxyethyl palmitamide (impulsin) in acute respiratory tract infections. Eur J Clin Pharmacol 7:415–419 [DOI] [PubMed] [Google Scholar]
- Mazzari S, Canella R, Petrelli L, Marcolongo G, Leon A. (1996) N-(2-hydroxyethyl)hexadecanamide is orally active in reducing edema formation and inflammatory hyperalgesia by down-modulating mast cell activation. Eur J Pharmacol 300:227–236 [DOI] [PubMed] [Google Scholar]
- McKinney MK, Cravatt BF. (2005) Structure and function of fatty acid amide hydrolase. Annu Rev Biochem 74:411–432 [DOI] [PubMed] [Google Scholar]
- Nathan C. (2002) Points of control in inflammation. Nature 420:846–852 [DOI] [PubMed] [Google Scholar]
- O'Sullivan SE. (2007) Cannabinoids go nuclear: evidence for activation of peroxisome proliferator-activated receptors. Br J Pharmacol 152:576–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori Y, Hamilton TA. (2001) Requirement for STAT1 in LPS-induced gene expression in macrophages. J Leukoc Biol 69:598–604 [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. (1998) Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085–2088 [DOI] [PubMed] [Google Scholar]
- Raetz CR, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC, Jr., Ribeiro AA, Murphy RC, Ulevitch RJ, Fearns C, Reichart D, Glass CK, Benner C, Subramaniam S, Harkewicz R, Bowers-Gentry RC, Buczynski MW, Cooper JA, Deems RA, Dennis EA. (2006) Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J Lipid Res 47:1097–1111 [DOI] [PubMed] [Google Scholar]
- Richardson D, Pearson RG, Kurian N, Latif ML, Garle MJ, Barrett DA, Kendall DA, Scammell BE, Reeve AJ, Chapman V. (2008) Characterisation of the cannabinoid receptor system in synovial tissue and fluid in patients with osteoarthritis and rheumatoid arthritis. Arthritis Res Ther 10:R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonjans K, Staels B, Auwerx J. (1996) The peroxisome proliferator activated receptors (PPARS) and their effects on lipid metabolism and adipocyte differentiation. Biochim Biophys Acta 1302:93–109 [DOI] [PubMed] [Google Scholar]
- Serhan CN, Chiang N, Van Dyke TE. (2008) Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol 8:349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon GM, Cravatt BF. (2006) Endocannabinoid biosynthesis proceeding through glycerophospho-N-acyl ethanolamine and a role for alpha/beta-hydrolase 4 in this pathway. J Biol Chem 281:26465–26472 [DOI] [PubMed] [Google Scholar]
- Solorzano C, Zhu C, Battista N, Astarita G, Lodola A, Rivara S, Mor M, Russo R, Maccarrone M, Antonietti F, et al. (2009) Selective N-acylethanolamine-hydrolyzing acid amidase inhibition reveals a key role for endogenous palmitoylethanolamide in inflammation. Proc Natl Acad Sci USA 106:20966–20971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi K, Takezaki N, Ueda N. (2007) The N-acylethanolamine-hydrolyzing acid amidase (NAAA). Chem Biodivers 4:1914–1925 [DOI] [PubMed] [Google Scholar]
- Won J, Yim J, Kim TK. (2002) Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells. J Biol Chem 277:38230–38238 [DOI] [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, Beppu T. (1990) Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem 265:17174–17179 [PubMed] [Google Scholar]