Abstract
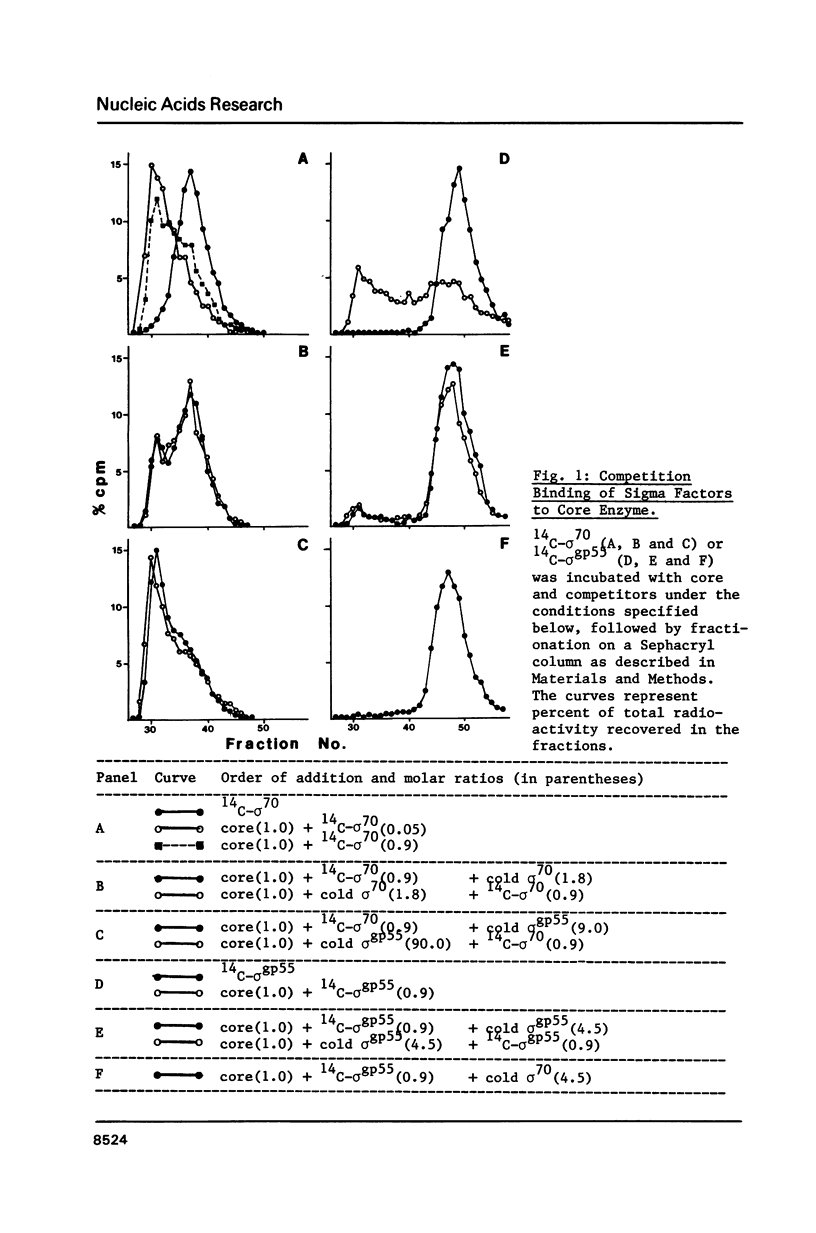
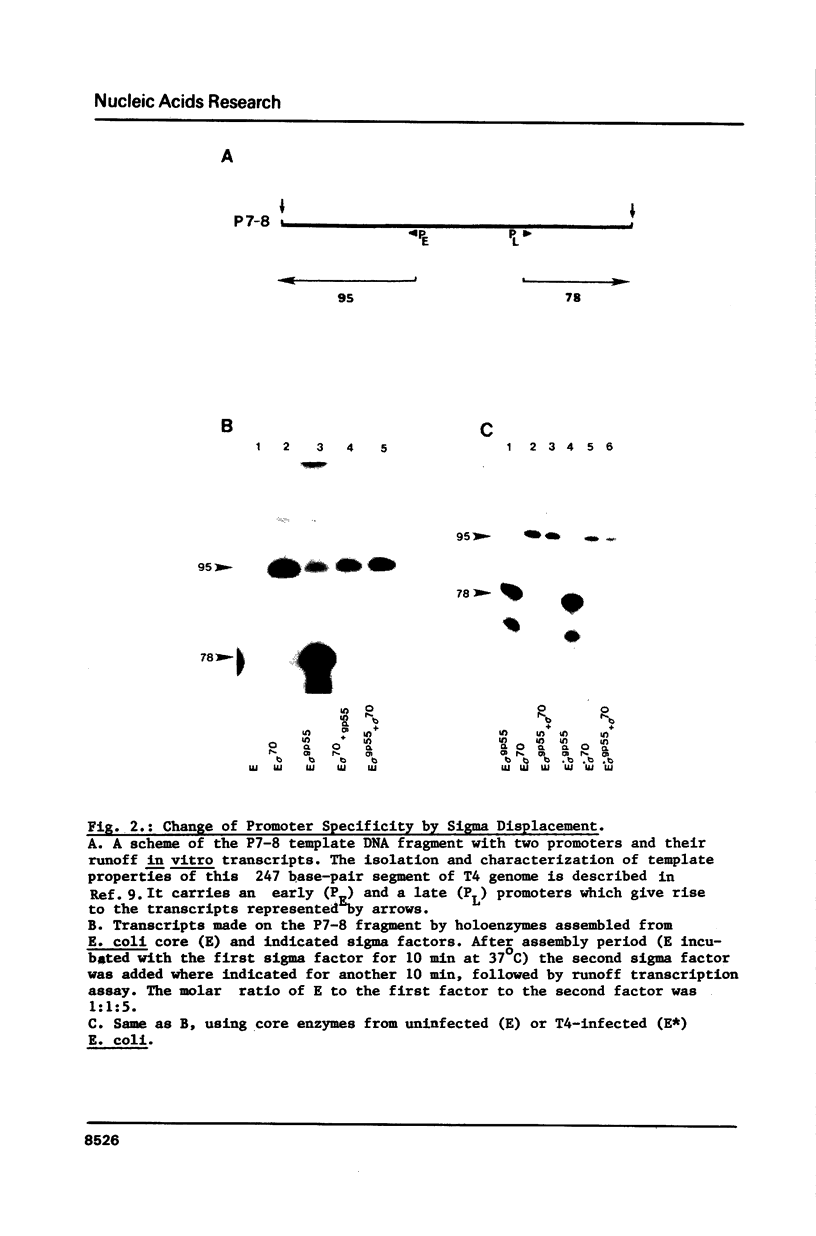
The switch of RNA polymerase specificity from early to late promoters of bacteriophage T4 is achieved by substitution of host sigma factor, sigma 70, with the T4 induced factor, sigma gp55. However, overproduction of sigma gp55 from an expression vector is not detrimental to Escherichia coli growth. Direct competition binding assays demonstrate that sigma 70 readily displaces sigma gp55 from RNA polymerase and thereby reverses the promoter specificity of the enzyme. The displacement also occurs with the core enzyme modified by bacteriophage T4 infection. We postulate that an antagonist of sigma 70 should be formed in T4-infected cells to aid sigma gp55 in the early/late switch.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker T. A., Grossman A. D., Gross C. A. A gene regulating the heat shock response in Escherichia coli also affects proteolysis. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6779–6783. doi: 10.1073/pnas.81.21.6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat J. F., Gilman M. Z., Chamberlin M. J. Bacillus subtilis sigma 28 and Escherichia coli sigma 32 (htpR) are minor sigma factors that display an overlapping promoter specificity. J Biol Chem. 1985 Feb 25;260(4):2038–2041. [PubMed] [Google Scholar]
- Doi R. H., Wang L. F. Multiple procaryotic ribonucleic acid polymerase sigma factors. Microbiol Rev. 1986 Sep;50(3):227–243. doi: 10.1128/mr.50.3.227-243.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Nomura T., Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase. Purification and properties of holoenzyme containing the heat-shock sigma subunit. J Biol Chem. 1987 Feb 5;262(4):1855–1859. [PubMed] [Google Scholar]
- Goff C. G., Setzer J. ADP ribosylation of Escherichia coli RNA polymerase is nonessential for bacteriophage T4 development. J Virol. 1980 Jan;33(1):547–549. doi: 10.1128/jvi.33.1.547-549.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb A., Broida J., Abelson J. Transcription in vitro of an isolated fragment of bacteriophage T4 genome. J Mol Biol. 1982 Oct 5;160(4):579–591. doi: 10.1016/0022-2836(82)90316-3. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Overexpression and purification of the sigma subunit of Escherichia coli RNA polymerase. Gene. 1983 Dec;26(2-3):109–118. doi: 10.1016/0378-1119(83)90180-4. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986 Aug 26;14(16):6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Kruse U., Rüger W. The region of phage T4 genes 34, 33 and 59: primary structures and organization on the genome. Nucleic Acids Res. 1986 Dec 9;14(23):9311–9327. doi: 10.1093/nar/14.23.9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz H. R. Polypeptide bound to the host RNA polymerase is specified by T4 control gene 33. Nat New Biol. 1973 Aug 1;244(135):137–140. doi: 10.1038/newbio244137a0. [DOI] [PubMed] [Google Scholar]
- Hsu T., Wei R. X., Dawson M., Karam J. D. Identification of two new bacteriophage T4 genes that may have roles in transcription and DNA replication. J Virol. 1987 Feb;61(2):366–374. doi: 10.1128/jvi.61.2.366-374.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Geiduschek E. P. Defining a bacteriophage T4 late promoter: bacteriophage T4 gene 55 protein suffices for directing late promoter recognition. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5101–5105. doi: 10.1073/pnas.81.16.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R., Pero J. Cascades of Sigma factors. Cell. 1981 Sep;25(3):582–584. doi: 10.1016/0092-8674(81)90164-1. [DOI] [PubMed] [Google Scholar]
- Malik S., Dimitrov M., Goldfarb A. Initiation of transcription by bacteriophage T4-modified RNA polymerase independently of host sigma factor. J Mol Biol. 1985 Sep 5;185(1):83–91. doi: 10.1016/0022-2836(85)90184-6. [DOI] [PubMed] [Google Scholar]
- Malik S., Goldfarb A. The effect of a bacteriophage T4-induced polypeptide on host RNA polymerase interaction with promoters. J Biol Chem. 1984 Nov 10;259(21):13292–13297. [PubMed] [Google Scholar]
- Williams K. P., Kassavetis G. A., Esch F. S., Geiduschek E. P. Identification of the gene encoding an RNA polymerase-binding protein of bacteriophage T4. J Virol. 1987 Feb;61(2):597–599. doi: 10.1128/jvi.61.2.597-599.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkin M. D. The sigma-like product of sporulation gene spoIIAC of Bacillus subtilis is toxic to Escherichia coli. Mol Gen Genet. 1986 Jan;202(1):55–57. doi: 10.1007/BF00330516. [DOI] [PubMed] [Google Scholar]