Abstract
Multiple genetic alterations occur in melanoma, a lethal skin malignancy of increasing incidence1,2. These include mutations that activate Ras and two of its effector cascades, Raf and phosphoinositide 3-kinase (PI3K). Ras and Raf induction can occur via active N-Ras and B-Raf mutants as well as by gene amplification3–5. Activation of PI3K pathway components occurs by PTEN loss and by AKT amplification6–8. Melanomas also commonly display impairment of p16INK4A-CDK4-Rb and ARF-HDM2-p53 tumor suppressor pathways. CDKN2A mutations can produce p16INK4A and ARF protein loss5,9–11. Rb bypass can also occur through activating CDK4 mutations as well as by CDK4 amplification5,12. In addition to ARF deletion, p53 pathway disruption can result from dominant-negative TP53 mutations5,13. Other findings in melanoma include hTERT amplification5. The ability of any of these mutations to induce human melanocytic neoplasia, however, is unknown. The present work characterizes pathways sufficient to generate human melanocytic neoplasia and demonstrates that genetically altered human tissue facilitates functional analysis of mutations observed in human tumors.
Keywords: Skin, Cancer, Melanoma, Human
Determining whether mutations identified in specimens from melanomas and other human cancers have true oncogenic potential or represent only secondary associations has been difficult. Purely murine models may not entirely reflect human oncogene action14, as shown by species differences in pathways engaged by oncogenic Ras15. Moreover, murine tissue architecture can differ dramatically from human. For example, murine melanocytes reside within the hair follicle16 whereas the majority of human melanocytes and melanomas are located outside hair structures. Studies in mice suggest that oncogenic Ras in the context of p16INK4A and ARF loss increases melanoma frequency17,18. Oncogene activation during development, however, can produce very different effects from activation in the adult setting19, tempering extrapolation of these findings to the somatic cell carcinogenesis that occurs in adult humans. Moreover, melanomas observed in murine models arise stochastically in only a subset of animals, indicating that additional uncharacterized changes are required. Models testing oncogenic sufficiency would therefore benefit from the introduction of defined genetic alterations in a developmentally mature human tissue context. This may be possible in skin, which can be regenerated from primary human cells on immunodeficient mice to accurately recapitulate the three-dimensional architecture, matrix elements and gene expression patterns of adult human tissue20,21.
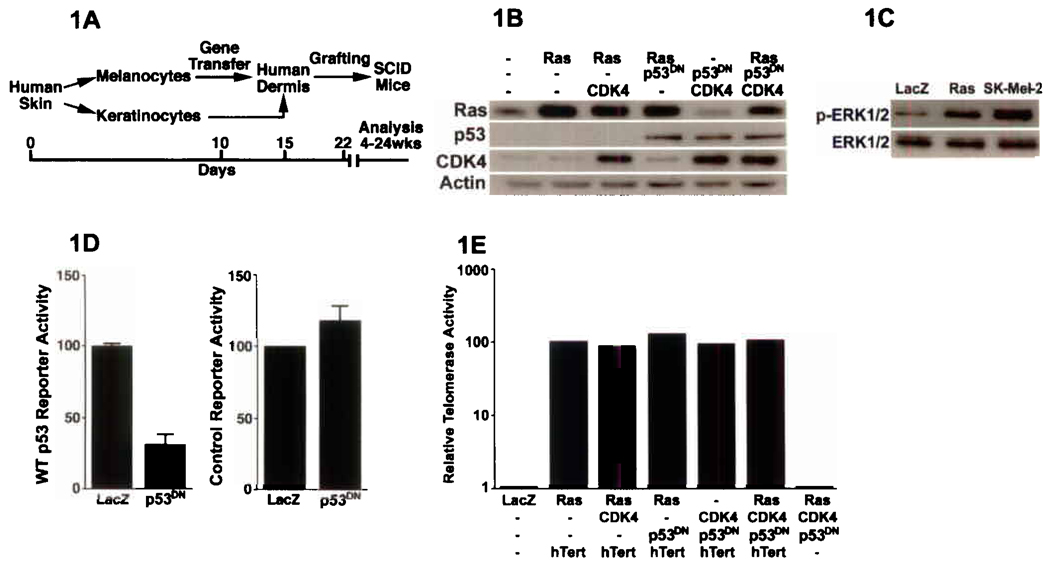
To characterize the functional impact of genetic alterations associated with human melanoma, we generated human skin tissue containing melanocytes selectively engineered to express specific mutations found in human melanoma. Because melanoma arises embedded in stratified epithelial tissue, we combined freshly isolated primary human keratinocytes with melanocytes on human dermis and regenerated human skin in vivo on immunodeficient mice (Fig. 1A). First, multiple genes were introduced into melanocytes via rapid serial retroviral transductions21. Mutants found in human melanoma, which produce activated Ras (N-RasG12V), as well as disruptions of the Rb (active CDK4R24C) and p53 pathways (dominant-negative p53R248W [p53DN]), were expressed in human melanocytes (Fig. 1B) along with the wild-type catalytic subunit of human telomerase (hTERT); none of these mutants, singly or in combination, significantly altered proliferation or cell viability (Suppl. Fig. S1 and data not shown). Expression of Ras mutants increased active ERK MAP kinases to levels seen in SK-Mel-2 melanoma cells that contain an activating mutation in an endogenous NRAS allele, confirming Ras pathway action comparable to that occurring in at least some human melanoma cells (Fig. 1C). p53DN function was confirmed using p53 reporter activity (Fig. 1D). Activity of delivered hTERT was confirmed by TRAP assay (Fig. 1E). These data confirm introduction of functionally active genetic elements found in human melanoma to primary human melanocytes.
Figure 1.
Expression of mutant proteins in human melanocytes. (A) Experimental schematic. (B) Confirmation of protein expression; Ras=active N-RasG12V, CDK4=active CDK4R24C, p53DN=dominant-negative p53R248W. (C) Levels of active phosphorylated and total ERK1/2 in primary human melanocytes engineered to express active N-Ras compared to SK-Mel-2 melanoma cells harboring a known activating mutation at endogenous NRAS. (D) Reporter activity from wild-type (WT, left panel) and non-functional mutant control p53 binding sites (right panel) as a function of expression of p53DN or LacZ control; relative reporter activity is shown, with LacZ control assigned a value of 100%. (E) Telomerase activity with combinatorial expression of hTERT.
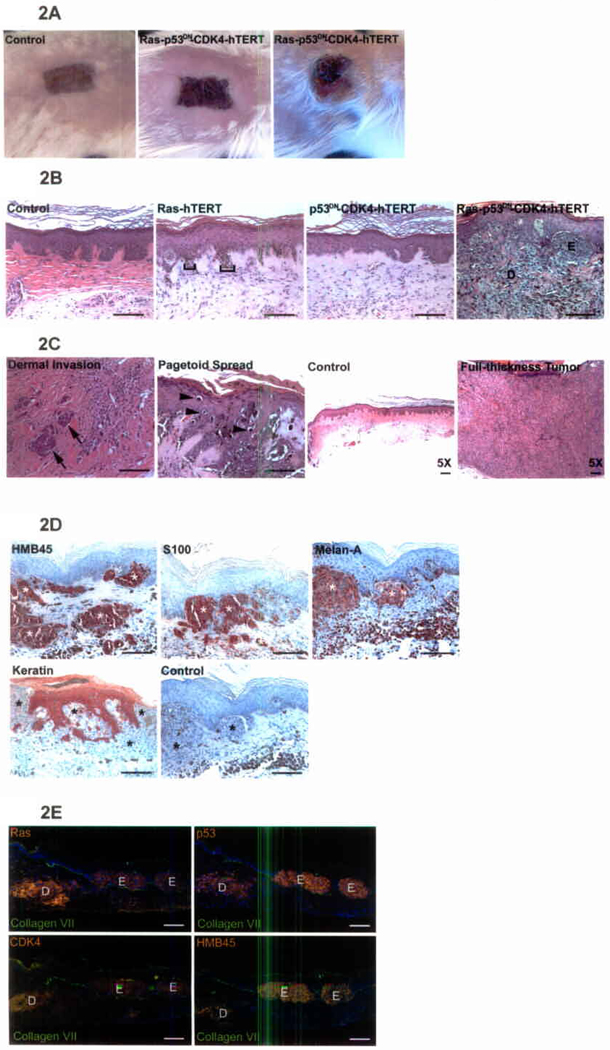
Human skin tissue regenerated with melanocytes expressing combinations of the genes noted was analyzed starting 4 weeks post-grafting (n≥5 mice/combination; Suppl. Table S1). While Rb and p53 pathway inhibition in concert with hTERT produced no effects, addition of active Ras to this combination produced clinical and histologic features of invasive human melanoma. Clinically, this manifested as darkly pigmented skin that progressed to ulcerated tumor nodules (Fig. 2A). Histologic features were characteristic of melanoma, including asymmetric junctional melanocytic hyperplasia, pagetoid spread upwards into the epidermis, deep vertical invasion into the dermis and tumor vascularization (Fig. 2B, C; Suppl. Fig. S2). Later timepoints displayed ulceration and full-thickness replacement of skin tissue with melanocytic tumor (Fig. 2C, far right panel). Melanocytic neoplasms displayed the wide range of findings seen in human melanoma, including variation in cell morphology and degree of pigmentation. As in spontaneous human melanomas, invasive tumor cells displayed S100 and Melan-A melanocyte markers as well as the HMB45 melanoma marker, while lacking epithelial keratins (Fig. 2D). Genetically induced human melanocytic neoplasia thus displays the cardinal clinical, histologic and immunophenotypic features of invasive melanoma.
Figure 2.
Invasive human melanocytic neoplasia. (A) Clinical appearance. Note ulceration with hemorrhagic crust in right panel. (B) Histology. Note mild junctional melanocytic hyperplasia [brackets] in tissue with melanocytes expressing active Ras and hTERT compared with invasive melanocytic neoplasia seen with Ras, hTERT, CDK4 and p53DN. Representative histology is shown at 6–12 weeks post-grafting (E=epidermis; D=dermis; scale bars=100µm). (C) Features of melanocytic neoplasia. Note deep dermal invasion (arrows), and pagetoid spread upwards into the epidermis (arrowheads). Note also full-thickness tumor and ulceration seen in low power view of 24 week timepoint (far right panel); scale bars=50µm. (D) Immunoperoxidase staining demonstrating that melanocytic neoplasms express HMB45, S100, Melan-A and lack epithelial keratins. Large melanocytic clusters are seen replacing keratin[+] epidermal cells; “*” denotes melanocytes; control=no primary antibody; scale bars=100µm. (E) Immunofluorescence staining demonstrating retention of introduced genes in melanocytic neoplasms. Ras, p53, CDK4 (orange), collagen VII epidermal basement membrane zone marker (green), Hoechst 33342 nuclei (blue), are shown in serial tissue sections. Note retention of introduced genes in both junctional/epidermal [E] and dermally invasive [D] HMB45[+] melanocytic regions (scale bars=100µm).
Tumor cells expressed all introduced genes (Fig. 2E and Suppl. Fig. S3). Five independent attempts failed to re-isolate and grow melanocytes from tumors in adequate numbers for analysis; however, Giemsa banding karyotypic analysis of cells expressing oncogenic combinations grown in parallel in vitro for 8 months displayed no evidence of genetic instability that might introduce additional genetic changes. Heterogeneous marker gene detection in cutaneous tumors generated from melanocytes marked at low efficiency indicated that tumors are polyclonal (Suppl. Fig. S4). In spite of aggressive local invasion, no spontaneous visceral metastases were observed. Although it also produced locally invasive tumors, the A375 human melanoma cell line, which forms pulmonary tumors when injected intravenously into nude mice22, also failed to metastasize when incorporated with keratinocytes into regenerated human skin grafts (Suppl. Fig. S5). Human dermal and epithelial elements may therefore comprise a barrier to metastasis, the basis for which requires further study.
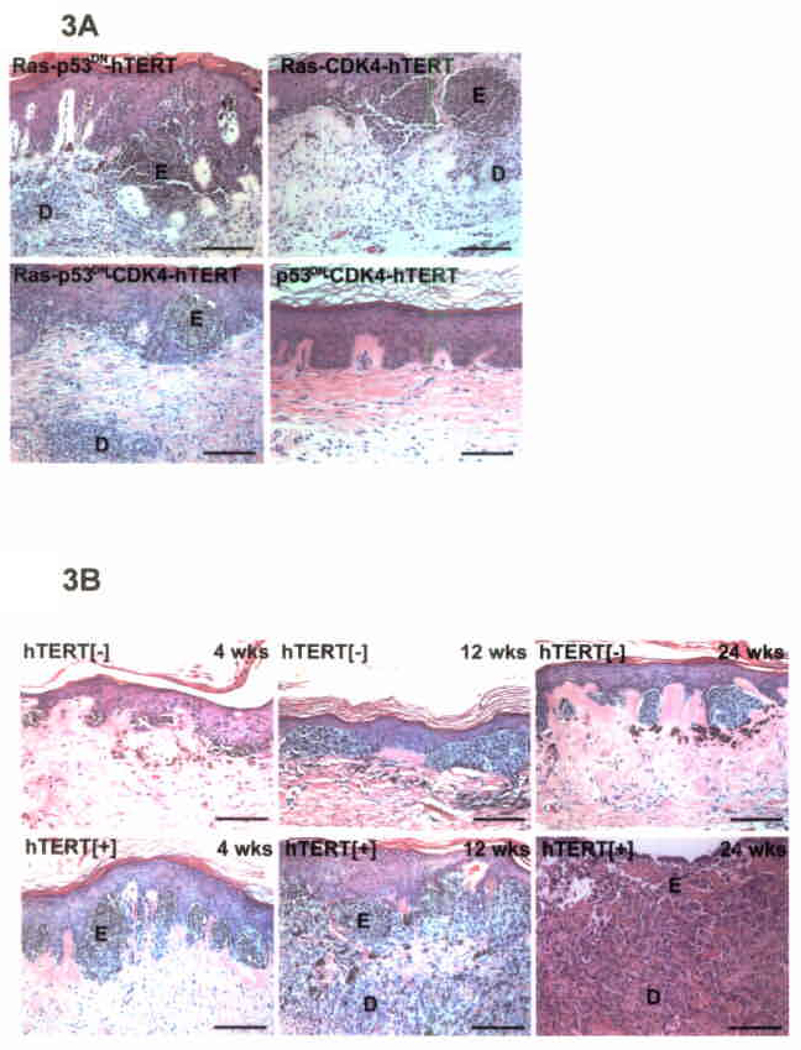
CDKN2A deletions in melanoma may alter the function of both p16INK4A and ARF simultaneously, with resultant inhibition of Rb and p53 pathways. We next determined if interfering with either pathway alone is sufficient to trigger human melanocytic neoplasia. Co-expression of either CDK4 or p53DN with oncogenic Ras and hTERT induced invasive melanocytic neoplasia in human tissue (Fig. 3A), indicating that disruption of either Rb or p53 pathway function alone can cooperate with Ras and hTERT in this process. Rb hyperphosphorylation occurred in melanocytes when Ras was co-expressed with either CDK4 or p53DN (Suppl. Fig. S6), suggesting bypass of the Rb block occurs by both gene sets. We next examined the role of elevated telomerase activity. hTERT supported progressively invasive melanocytic neoplasia whereas only junctional melanocytic hyperplasia with minimal invasion developed without hTERT addition (Fig 3B). Melanocytes in tissue that did not receive hTERT maintained mitotic activity over the course of 6 months in vivo (Suppl. Fig. S7), suggesting hTERT action in this context was not merely due to prevention of replicative exhaustion.
Figure 3.
Effect of disrupting function of p53 and Rb pathways and the impact of hTERT. (A) Invasive melanocytic neoplasia occurs with Ras and hTERT with CDK4 as well as p53DN. Representative histology is shown at 6 weeks; scale bars=100µm. (B) Tumor progression at 4, 12 and 24 weeks in the presence or absence of delivered hTERT. Note progression from junctional hyperplasia to deep dermal invasion and full-thickness ulceration of skin tissue that occurs with hTERT co-expression with Ras, CDK4 and p53DN compared to the milder, predominantly junctional hyperplasia that occurs without hTERT co-expression; scale bars=100µm.
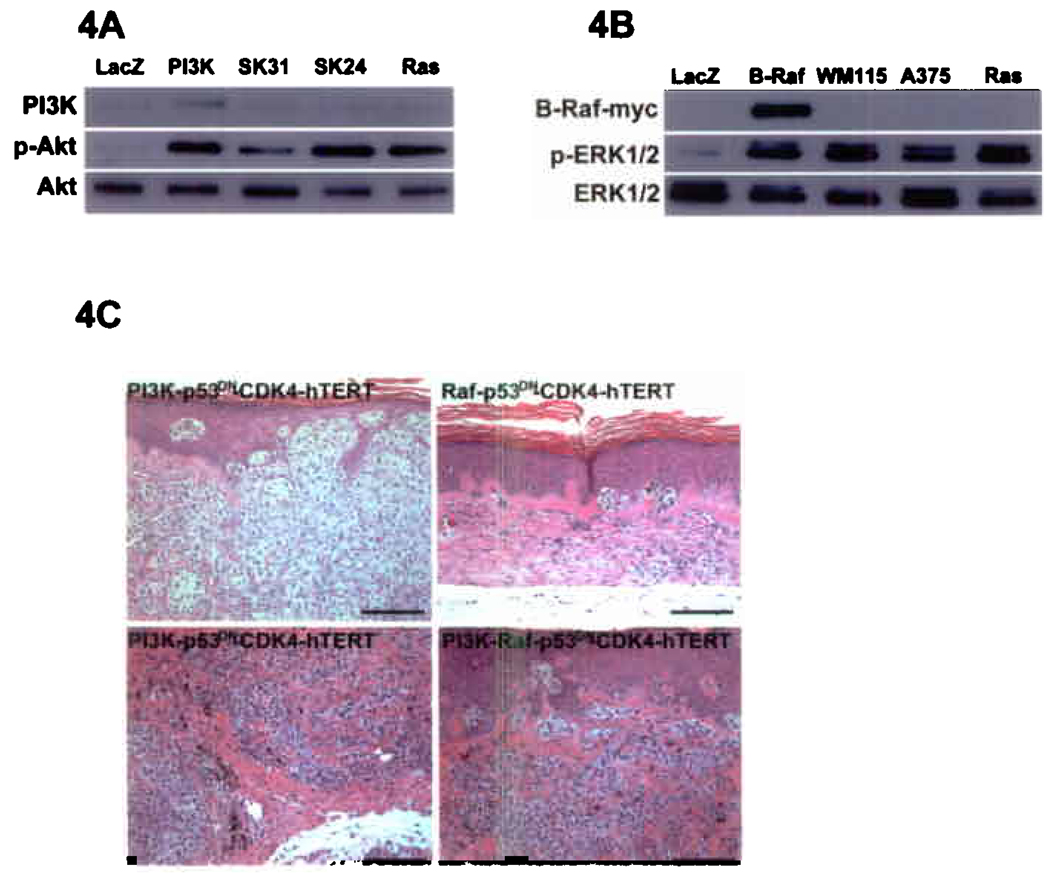
PI3K and Raf initiate major effector pathways that are implicated independently of Ras in human melanoma. Although genetic and biochemical evidence supports induction of both pathways in a substantial proportion of human melanomas, their relative functional effects on tumorigenesis are unknown. Co-expression of active PI3K p110α with CDK4, p53DN and hTERT produced invasive melanocytic neoplasia indistinguishable from that seen with oncogenic Ras (Fig. 4A, C). In contrast, B-RafV599E, the active mutant most frequently found in melanoma, failed to induce anything more than very mild junctional melanocytic nesting in this context, despite the fact that it achieved comparable ERK1/2 activation to oncogenic Ras (Fig. 4B, C). Levels of effector activation by both PI3K and B-Raf were comparable to those seen in genetically characterized melanoma cell lines with PTEN loss and endogenous mutant B-RafV599E expression, respectively (Fig. 4A, B), confirming signaling induction at magnitudes that occur in at least some melanoma cells. Co-expression of active B-Raf with PI3K p110α failed to accelerate or increase the severity of melanocytic neoplasia over the course of 12 weeks. These data indicate that activation of PI3K, but not Raf, can replace Ras to induce invasive human melanocytic neoplasia.
Figure 4.
Capacity of active PI3K and B-Raf to recapitulate Ras-driven human melanocytic neoplasia. (A) Expression and function of active PI3K p110α-CAAX (PI3K) in primary human melanocytes. Levels of phosphorylated active Akt in melanocytes expressing either active PI3K or N-Ras are compared to SK-Mel-24 (SK24) and SK-Mel-31 (SK31) melanoma cells with known activation of PI3K via PTEN deletion. (B) Expression and function of active B-RafV599E. Levels of phosphorylated active ERK1/2 in melanocytes expressing either active B-Raf or N-Ras are compared to WM115 and A375 melanoma cells with known mutation of endogenous BRAF leading to expression of the active B-RafV599E mutant. (C) Human tissue regenerated with melanocytes expressing either p110α-CAAX, B-RafV599E or both in combination with CDK4, p53DN and hTERT at 12 weeks post-grafting. Note deeply invasive melanocytic neoplasia in presence of active PI3K and only mild junctional hyperplasia seen with active B-Raf. The lower left panel represents a more deeply invasive region of the tumor shown in the upper left panel; scale bars=100µm.
Both active Ras and PI3K along with Rb and p53 inhibition as well as hTERT expression are sufficient to induce invasive human melanocytic neoplasia. B-Raf, although currently a subject of intense interest in melanoma, lacked the oncogenic potency of either Ras or PI3K in the human skin tissue context studied here. Because mutants are introduced nearly simultaneously, however, the present strategy does not exclude other potential outcomes that might occur with different sequences or timing of mutational events. Interestingly, BRAF mutations may occur at a higher frequency in benign nevi than in melanoma, while the opposite is reported for NRAS4, underscoring the importance of future studies of B-Raf functional impacts. In contrast, evidence for PI3K pathway induction occurs more frequently in melanoma than in benign nevi7,8, consistent with our findings that activating PI3K pathway elements may represent a more potent oncogenic stimulus.
The observation that interference with either Rb or p53 pathway function is oncogenic in the context of co-expression of Ras and hTERT is consistent with observations of human melanomas harboring CDKN2A mutations that selectively ablate the transcript encoding either p16INK4A or ARF as well as with murine data indicating that Ras can increase melanoma frequency in concert with deficiency of either protein18,23,24. Although no short-term growth differences were noted among gene combination groups in vitro, both Rb and p53 pathways are known to inhibit uncontrolled progression through the G1 phase of the cell cycle. Thus, long-term inhibition of the function of either protein in vivo may facilitate tumor growth via diminished G1 restraints. p53, however, exerts a variety of tumor-suppressive impacts and understanding its precise role in this context requires further study. hTERT co-expression, while dispensable for early stages of melanocytic hyperplasia, was required to induce progressively invasive melanocytic neoplasia. Cessation of mitotic activity in the absence of hTERT co-expression did not accompany this finding, consistent with its reported effects on tumorigenesis beyond promoting immortalization25,26. These data assess the oncogenic capacity of specific genetic alterations observed in human melanoma and identify a minimal set of genetic changes sufficient to induce this neoplasm in human skin tissue. Use of genetically engineered human tissue may thus provide a general means of testing the oncogenic potency of genetic alterations identified in other human tumors and of developing models for assessment of new therapeutics aimed at such genetic targets.
Methods
Cell Growth
Normal human keratinocytes and normal human melanocytes were isolated from human skin, including adult abdominal and breast skin as well as neonatal foreskin from a total of approximately 10 unrelated patients; cells from all tissues used produced similar results, with no donor-linked differences observed. Keratinocytes were grown in SFM (GIBCO/BRL, Gaithersburg, MD), while all melanocytes were grown in medium154 (Cascade Biologics, Portland, OR) containing phorbol ester; the latter was necessary for cell growth in vitro but not sufficient for subsequent tumorigenesis. Melanoma cell lines A375, SK-Mel-24, SK-Mel-31, WM115, and SK-Mel-2 were purchased from ATCC (Manassas, VA). Cells were grown in DMEM (GIBCO/BRL) supplemented with 10% (A375, SK-Mel-2, SK-Mel-24, WM115) or 15% (SK-Mel-31) fetal bovine serum (Omega Scientific, Tarzana, CA). Giemsa banding was performed at the Stanford Cytogenetics Laboratory (Stanford, CA).
Gene Transfer
The pLZRS retroviral backbone27 was used to express all the genes studied, with sequence verification performed in all cases by sequencing both vector strands. The human N-RasG12V coding sequence was purchased from the Guthrie cDNA Resource Center (Sayre, PA) and subcloned into the BamHI-XhoI sites of pLZRS. Coding sequences of active PI3K p110α-CAAX were subcloned into the BamHI site of pLZRS (courtesy of T. Cai). Human Myc-tagged B-RafV599E cDNA (gift of R. Marais) was subcloned into the XmnI-XbaI sites of pENTR1A (Invitrogen, Carlsbad, CA) to make pENTR1A-B-Raf. To remove the 3’ UTR, pENTR1A-B-Raf was used as a PCR template with the following primers: TGTCTCCAGATCTCAGTAAGG (forward) and ACTCTCTAG ATTCAGTGGACAGGAAACGC (reverse). The resulting product was digested with BglII/XbaI and used to replace the original BglII-XbaI fragment of pENTR1A-B-Raf to make pENTR1A-B-Raf-noUTR. The B-RafV599E coding sequence was then transferred into pLZRS containing a GATEWAY (Invitrogen) destination site (pLZRS-GATEWAY). Vectors for H-RasG12V, CDK4R24C, p53R248W, and hTERT have been described previously21. Extracellular matrix (ECM)-coated plates were made by lysing confluent keratinocytes in 34mM ammonium hydroxide and rinsing away cell debris with PBS (GIBCO/BRL). Melanocytes were seeded on ECM plates and transduced by multiplex serial gene transfer (MSGT) at multiplicities of infection of 10 to 15 with retroviral vectors using low-speed centrifugation at intervals of 12–24h21,28; no drug selection was used in any experiment.
Animal studies
48–72 hours after MSGT, transduced melanocytes were combined with non-transduced keratinocytes on devitalized human dermal substrate at a ratio of 1:5 (melanocyte: keratinocyte), which has been proposed as the physiological ratio of epidermal melanocytes to basal keratinocytes29. 7 days after seeding, tissue was grafted on CB.17 scid/scid mice as described21. Prior to seeding, expression of all introduced genes was confirmed in melanocytes by immunoblotting as noted below. All studies were done with multiple replicates in separate rounds of experiments (n=5–25 animals/group); both N-RasG12V and H-RasG12V were tested in parallel and produced melanocytic neoplasms with identical features. For B-Raf studies, 25 independent grafts in 5 rounds of experiments were performed to confirm observed results. Human skin tissue was analyzed initially at 4–12 weeks with matched tissue from all the different experimental groups harvested at each time point. Findings in individual grafts were consistent for each gene set studied. For hTERT progression studies, skin tissue was harvested at approximately 4, 12 and 24 weeks after grafting. At the time of skin tissue harvesting, animals were sacrificed and visceral organs examined for metastases, clinically and histologically. Tumor burden in skin was estimated to be 5–10×107 melanocytic cells at 24 weeks via cross-sectional cell counting of tissue sections and calculation of tumor volumes. Human skin tissue containing A375 melanoma cells was regenerated and analyzed as above, with A375 cells used in place of genetically engineered melanocytes.
Gene and protein expression analysis
For immunoblotting, 15µg of whole-cell protein extract was loaded per lane, and rabbit antibodies specific for Ras, p53, p21 (Santa Cruz Biotechnology, Santa Cruz, CA), phospho-ERK1/2, total ERK1/2, phospho-Rb, phospho-Akt, and total Akt (Cell Signaling Technology, Beverly, MA), PERP (gift of L. Attardi) and mouse antibodies specific for the Myc-tag, total Rb (Santa Cruz Biotechnology), PI3K p110α (BD Transduction Laboratories, San Diego, CA), CDK4 (US Biological, Swampscott, MA), hypo-phosphorylated Rb (BD Pharmingen, San Diego, CA) and β-actin (Sigma, St. Louis, MO) were used. Telomerase activation was measured with the Trapeze ELISA Telomerase Detection Kit (Chemicon, Temecula, CA). Relative telomerase activity is calculated as the (A450-A690 sample/A450-A690 manufacturer’s control), with data expressed relative to LacZ control, which was assigned a value of 1.0. p53 reporter assays were performed with the internally controlled Dual-Luciferase Reporter Assay System (Promega, Madison, WI) using lucifererase reporter constructs containing a multimer of wild-type or mutant p53 binding sites30; basal levels of reporter activity were consistently observed with cells in culture. TUNEL staining was performed using the in situ death detection kit (Roche Applied Science, Indianapolis, IN). For RT-PCR, RNA was extracted from cells and dissected tumor specimens using Trizol reagent (Invitrogen) then further purified using the RNeasy Protect Mini Kit (Qiagen, Valencia, CA) and reverse transcribed and amplified using Superscript 1-step with Platinum Taq (Invitrogen). The following primers were used: forward primer from LZRS retrovector TGGATACACGCCGCCCACGTG, reverse primers for Ras TCCTCCTGGCCGGCGGTA, p53 ACGTGCAAGTCACAGACTTG, CDK4 GTCTA CATGCTCAAACACCAG. For hTERT the forward vector primer CTTACACAGTCCTGCT GAC was used with the reverse primer GCCGCACGAACGTGGCCA; GAPDH mRNA-specific primers were ATCACCATCTTCCAGGAG and GTGAGCTTCCCGTTCAGC. Immunohistochemistry was performed on paraffin-embedded tissue sections, using mouse antibodies specific for pan-cytokeratin, HMB45, and Melan-A and rabbit antibodies specific for S100 and Ki-67 (Dako, Carpenteria, CA); melanin bleaching was performed to assist visualization of immunostaining. Immunofluorescence was performed on tissue cryosections and cells, using mouse antibodies against Ras, p53 (Oncogene Research Products, Boston, MA), CDK4 (US Biological), and HMB45 (Abcam, Cambridge, MA), Melan-A/MART-1 (Labvision, Fremont, CA); rat antibodies were used against the vascular marker CD31 (PECAM, BD Pharmingen) and rabbit antibodies used for β-galactosidase (MP Biomedicals, Aurora, OH) and Ki-67 (Labvision, Fremont, CA). Sections were counterstained with rabbit antibody against Collagen VII (Calbiochem, San Diego, CA) and Hoechst 33342 (Molecular Probes, Eugene, OR).
Acknowledgments
Supported by the US Veterans Affairs Office of Research and Development, by National Institutes of Arthritis and Musculoskeletal and Skin Diseases, US National Institutes of Health. We thank D. Felsher, A. Oro, S. Artandi, H.Y. Chang, S. Swetter, H. Lee, Z. Siprashvili, F. Scholl, J. Reuter, T. Ridky, and P. Dumesic for pre-submission review and helpful discussions, T. Cai for PI3K reagents, B. Vogelstein and K. Kinzler for p53 reporter constructs, M. Mihm, S. Kohler and J. Harvell for expert pathologic evaluation, S. Tao for technical support, L. Attardi for PERP antibody and R. Marais for B-RafV599E and helpful advice. Y.C. is a Howard Hughes Medical Institute predoctoral fellow and A.E.A. was supported by a training grant from the NIAMS, NIH and a Warren-Whitman-Richardson Fellowship.
References
- 1.Gilchrest BA, Eller MS, Geller AC, Yaar M. The pathogenesis of melanoma induced by ultraviolet radiation. N Engl J Med. 1999;340:1341–1348. doi: 10.1056/NEJM199904293401707. [DOI] [PubMed] [Google Scholar]
- 2.Schaffer JV, Rigel DS, Kopf AW, Bolognia JL. Cutaneous melanoma--past, present, and future. J Am Acad Dermatol. 2004;51:S65–S69. doi: 10.1016/j.jaad.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 3.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 4.Pollock PM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 5.Rodolfo M, Daniotti M, Vallacchi V. Genetic progression of metastatic melanoma. Cancer Lett. 2004;214:133–147. doi: 10.1016/j.canlet.2004.06.049. [DOI] [PubMed] [Google Scholar]
- 6.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004;122:337–341. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene. 2003;22:3113–3122. doi: 10.1038/sj.onc.1206451. [DOI] [PubMed] [Google Scholar]
- 8.Stahl JM, et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64:7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 9.Kamb A, et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet. 1994;8:23–26. doi: 10.1038/ng0994-22. [DOI] [PubMed] [Google Scholar]
- 10.Hussussian CJ, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8:15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 11.Hayward NK. Genetics of melanoma predisposition. Oncogene. 2003;22:3053–3062. doi: 10.1038/sj.onc.1206445. [DOI] [PubMed] [Google Scholar]
- 12.Wolfel T, et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 13.Albino AP, et al. Mutation and expression of the p53 gene in human malignant melanoma. Melanoma Res. 1994;4:35–45. doi: 10.1097/00008390-199402000-00006. [DOI] [PubMed] [Google Scholar]
- 14.Weitzman JB, Yaniv M. Rebuilding the road to cancer. Nature. 1999;400:401–402. doi: 10.1038/22637. [DOI] [PubMed] [Google Scholar]
- 15.Hamad NM, et al. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16:2045–2057. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimura EK, et al. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416:854–860. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- 17.Sharpless NE, Kannan K, Xu J, Bosenberg MW, Chin L. Both products of the mouse Ink4a/Arf locus suppress melanoma formation in vivo. Oncogene. 2003;22:5055–5059. doi: 10.1038/sj.onc.1206809. [DOI] [PubMed] [Google Scholar]
- 18.Kannan K, et al. Components of the Rb pathway are critical targets of UV mutagenesis in a murine melanoma model. Proc Natl Acad Sci U S A. 2003;100:1221–1225. doi: 10.1073/pnas.0336397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beer S, et al. Developmental Context Determines Latency of MYC-Induced Tumorigenesis. PLoS Biol. 2004;2:e332. doi: 10.1371/journal.pbio.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dajee M, et al. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- 21.Lazarov M, et al. CDK4 coexpression with Ras generates malignant human epidermal tumorigenesis. Nat Med. 2002;8:1105–1114. doi: 10.1038/nm779. [DOI] [PubMed] [Google Scholar]
- 22.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 23.Brookes S, Rowe J, Gutierrez Del Arroyo A, Bond J, Peters G. Contribution of p16(INK4a) to replicative senescence of human fibroblasts. Exp Cell Res. 2004;298:549–559. doi: 10.1016/j.yexcr.2004.04.035. [DOI] [PubMed] [Google Scholar]
- 24.Randerson-Moor JA, et al. A germline deletion of p14(ARF) but not CDKN2A in a melanoma-neural system tumour syndrome family. Hum Mol Genet. 2001;10:55–62. doi: 10.1093/hmg/10.1.55. [DOI] [PubMed] [Google Scholar]
- 25.Stewart SA, et al. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc Natl Acad Sci U S A. 2002;99:12606–12611. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang S, DePinho RA. Telomerase extracurricular activities. Proc Natl Acad Sci U S A. 2002;99:12520–12522. doi: 10.1073/pnas.212514699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinsella TM, Nolan GP. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 28.Deng H, Choate KA, Lin Q, Khavari PA. High efficiency gene transfer and pharmacologic selection of genetically engineered human keratinocytes. BioTechniques. 1998;25:274–280. doi: 10.2144/98252gt02. [DOI] [PubMed] [Google Scholar]
- 29.Hsu M, Andl T, Li G, Meinkoth JL, Herlyn M. Cadherin repertoire determines partner-specific gap junctional communication during melanoma progression. J Cell Sci. 2000;113(Pt 9):1535–1542. doi: 10.1242/jcs.113.9.1535. [DOI] [PubMed] [Google Scholar]
- 30.Kern SE, et al. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]