Abstract
Hybridization plays a central role in plant evolution, but its overall importance in fungi is unknown. New plant pathogens are thought to arise by hybridization between formerly separated fungal species. Evolution of hybrid plant pathogens from non-pathogenic ancestors in the fungal-like protist Phytophthora has been demonstrated, but in fungi, the most important group of plant pathogens, there are few well-characterized examples of hybrids. We focused our attention on the hybrid and plant pathogen Verticillium longisporum, the causal agent of the Verticillium wilt disease in crucifer crops. In order to address questions related to the evolutionary origin of V. longisporum, we used phylogenetic analyses of seven nuclear loci and a dataset of 203 isolates of V. longisporum, V. dahliae and related species. We confirmed that V. longisporum was diploid, and originated three different times, involving four different lineages and three different parental species. All hybrids shared a common parent, species A1, that hybridized respectively with species D1, V. dahliae lineage D2 and V. dahliae lineage D3, to give rise to three different lineages of V. longisporum. Species A1 and species D1 constituted as yet unknown taxa. Verticillium longisporum likely originated recently, as each V. longisporum lineage was genetically homogenous, and comprised species A1 alleles that were identical across lineages.
Introduction
Hybridization plays a central role in plant evolution. More than 10% of currently existing species might be hybrids [1], which includes many of our most important crops such as wheat, oat, cotton, coffee and canola [2]. Hybrids often exhibit new or extreme phenotypes with respect to their parents, and hybrid vigor or heterosis in cultivated crops is widely exploited.
Whereas hybridization in plants has been well studied, relatively little is known about hybrids in fungi. Historically, there have been few reports of fungal hybrids inferred from credible morphological and genetic evidence [3], [4], [5], [6]. The advent of molecular phylogenetics has greatly improved our ability to detect hybrids [7], and has resulted in increased numbers of documented examples [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19].
The consequences of hybridization at the molecular level in fungi are unknown, but in plants they include both genomic and epigenetic changes resulting in the alteration of gene expression and the evolution of new phenotypes [1], [2], [20], [21]. In natural fungal hybrids, both morphological and life style changes have been observed. Hybrids may display intermediate parental morphology [3], [4], [5], [9] and fermentation properties [10], a switch from pathogen to mutualist [22], an increase in virulence [3], a decrease in fitness [13] and a new host range with respect to their parents [3]. Some of these changes are also observed in artificial hybrids [16], [23].
Most known cases of fungal hybrids involve plant pathogens. It has been suggested that new plant pathogens with increased virulence and extended host range could arise through hybridization between formerly geographically separated species, brought into contact through human activities such as global trade [24], [25].
In fungi, there are only a limited number of examples of natural hybrid pathogens with increased virulence and extended host range. These include Melampsora x columbiana G. Newc., a hybrid between M. medusae and M. occidentalis that became the dominant rust on hybrid poplar Populus trichocarpa x P. deltoides in the Pacific Northwest in 1998 [3], as well as V. longisporum, a pathogen of crucifers. Molecular evidence suggests that V. longisporum is an allopolyploid, possibly between V. dahliae and a species related to V. albo-atrum [26]. Verticillium longisporum is the cause of Verticillium wilt of oilseed rape, a major disease in Europe [27] that is unknown in North America [28]. Even though V. dahliae, a relative of V. longisporum, is at times isolated from oilseed rape, only V. longisporum is capable of causing disease [29]. Both V. longisporum and V. dahliae survive as microsclerotia in the soil, which facilitates pathogen dispersal through water, farm equipment and personnel [30].
Laying the foundations for future studies on pathogenicity, virulence and host specificity in V. longisporum, we employed a phylogenetic approach using 203 isolates of V. longisporum, V. dahliae, V. albo-atrum, V. nubilium and V. tricorpus, and seven different loci, to elucidate the parental species and hybridization events involved in the evolution of V. longisporum.
Results
Allelic diversity in species of Verticillium
The typical ascomycete life cycle is dominated by the haploid state, but Verticillium longisporum is diploid, its nuclei contain approximately twice the amount of DNA as the closely related V. dahliae [31], [32], [33]. We found that all V. longisporum strains contained two alleles at each of the eight loci examined, except at the nuclear ribosomal internal transcribed spacer region (ITS) which contained a single allele, and at the beta-tubulin locus which was represented through an additional, paralogous copy in some isolates of V. dahliae and V. longisporum. These conclusions were derived from sequencing 225 PCR product clones from 12 different strains and all eight loci (Table S1), and PCR screening of V. longisporum strains using allele specific primers (Figures S1, S2, S3), followed by DNA sequencing and phylognetic analyses. All loci in V. albo-atrum, V. nubilum and V. tricorpus contained only a single allele, as inferred by the absence of polymorphic positions in directly sequenced PCR products.
Mating type idiomorphs in V. longisporum and other species
Mating polarity in ascomycetes is determined by the mating type locus that contains alleles differing in gene content referred to as idiomorphs [34]. Idiomorphs are designated by ‘MAT’ followed by a series of numbers that reflect idiomorph gene content. Two strains are sexually compatible if they carry either a MAT1-1 or a MAT1-2 idiomorph [35]. We found that V. longisporum strains contained two MAT1-1 idiomorphs based on cloning and DNA sequencing (Table S1), and failed to detect MAT1-2 using idiomorph-specific PCR screens (Figure S4). DNA sequence data obtained from V. longisporum strains PD342, PD348 and PD356 showed that the V. longisporum MAT1-1 idiomorph contained MAT1-1-1 approximately 1.5 kb upstream of MAT1-1-2 (GenBank Accessions HQ415004 - HQ415009), as in the closely related V. albo-atrum (Klosterman et al., accepted for publication in PLoS Pathogens pending revisions).
The majority of the V. dahliae strains carried a MAT1-2 idiomorph, only 11 strains were MAT1-1 (Figure S4). DNA sequence data of the region spanning from MAT1-1-1 to MAT1-1-2 was obtained for the V. dahliae MAT1-1 strains PD404, PD585 and PD617 (GenBank Accessions HQ415012, HQ415014, HQ415025), confirming the presence of MAT1-1-1 and MAT1-1-2 arranged as in V. longisporum.
Verticillium albo-atrum strains were found to be either MAT1-1 or MAT1-2, the mating types for the remaining species could not be determined (Figure S4).
Preliminary phylogenetic analyses
Phylogenetic analyses based on the ITS region were used to investigate the relationships of all strains included in this study (Table S2). An alignment comprising 203 taxa and 493 characters was generated and analyzed using parsimony, resulting in one most parsimonious tree (Figure 1). The most parsimonious tree contained five different clades corresponding to the five different species included in these analyses, Verticillium tricorpus, V. nubilum, V. albo-atrum, V. dahliae and V. longisporum. The exceptions were V. longisporum strains PD589, PD614, PD687 and PD715, which grouped with V. dahliae (Figure 1). For additional details on the ITS analyses, see Table 1. The alignment was submitted to TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S11128), the DNA sequences were deposited in GenBank. (GenBank Accessions HQ206718 - HQ206920).
Figure 1. Evolutionary origins of the diploid hybrid Verticillium longisporum based on phylogenetic inference from ribosomal internal transcribed spacer (ITS) data comprising 203 taxa and 493 characters.
Shown is the single, most parsimonious tree, 20 steps in length. Isolates are represented by a strain identifier. Hosts and geographic origins are given. Bootstrap supports above 70% are shown by the branches. Species delimitations are indicated by vertical bars on the right. The isolates fell into five different groups corresponding to the five species of Verticillium. The exceptions were four isolates of V. longisporum, V. longisporum strains PD589, PD614, PD687 and PD715, which grouped with V. dahliae.
Table 1. Statistics of the ITS, ACT, EF, GPD, OX and TS single locus datasets and the combined five-locus dataset and respective most parsimonious trees.
| Haplotypes | Characters | Variable characters | Pars info characters | MPTs: number/steps | CI/RIb | Clades >70% support | |
| ITS | 5 | 493 | 20 (4%)a | 6 (1%)a | 1/120 | 1.000/1.000 | 1 |
| ACT | 7 | 532 | 171 (32%) | 53 (10%) | 1/199 | 0.956/0.904 | 7 |
| EF | 11 | 600 | 251 (42%) | 70 (12%) | 6/318 | 0.947/0.832 | 9 |
| GPD | 9 | 678 | 131 (19%) | 44 (6%) | 1/165 | 0.909/0.741 | 8 |
| OX | 9 | 606 | 172 (28%) | 64 (11%) | 1/214 | 0.953/0.870 | 7 |
| TS | 10 | 591 | 177 (30%) | 52 (9%) | 35/243 | 0.930/0.754 | 8 |
| Combined ACT, EF, GPD, OX, TS | 21 | 3007 | 902 (30%) | 283 (9%) | 6/1152 | 0.932/0.832 | 14 |
Percentages refer to the proportions of variable and parsimony informative characters in each dataset.
CI: consistency index; RI: retention index.
Phylogenetic analyses based on five protein coding genes
From each of the five clades in the ITS tree (Fig. 1), isolates were chosen according to host and geographic origin for sequencing of intron-rich portions of the five protein coding genes actin (ACT), elongation factor 1-alpha (EF), glyceraldehyde-3-phosphate dehydrogenase (GPD), mitochondrial oxaloacetate transport protein (OX) and tryptophan synthase (TS). Within V. dahliae, all isolates with the rare MAT1-1 idiomorph were also selected, in V. longisporum all isolates grouping with V. dahliae, as well as all V. longisporum isolates with the D2 and D3 alleles which were less abundant than the D1 allele (see below for information on allelic diversity). In total, 73 taxa were chosen, which included one strain of V. tricorpus and V. nubilum, two strains of V. albo-atrum, 47 strains of V. dahliae and 22 V. longisporum strains. Each V. longisporum strain was represented by two alleles. The resulting five datasets ACT, EF, GPD, OX and TS were first analyzed separately using parsimony to examine congruence. Most parsimonious trees from the five analyses are shown in Figures S5, S6, S7, S8, S9. The topologies of all most parsimonious trees were identical except within the clade containing the V. dahliae isolates, suggesting incomplete sorting of ancestral polymorphisms or sexual recombination [36]. For more details on the single dataset analyses, see Table 1.
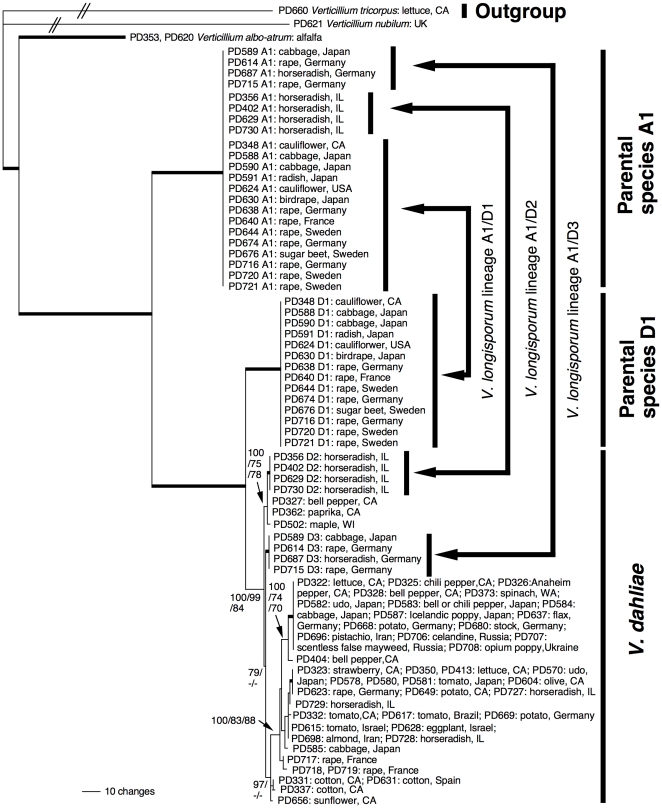
Since the single dataset analyses did not uncover conflicts between datasets except within V. dahliae, the five datasets were concatenated into one alignment containing 95 taxa and 3007 characters, and analyzed using Bayesian inference, parsimony, and maximum likelihood. Bayesian inference and maximum likelihood analyses implemented a GTR+G model of DNA sequence evolution, the most appropriate model as determined by the Akaike Information Criterion in Modeltest. The Bayesian consensus tree is shown in Figure 2, rooted with V. tricorpus based on a study by Pantou et al. [37]. Phylogenetic analyses identified four different alleles in V. longisporum, alleles A1, D1, D2 and D3. As diploids, each V. longisporum isolate contained two alleles at each locus, allele A1 was present in all V. longisporum isolates, in addition to allele D1, D2 and D3, respectively. The four alleles represented four different lineages; there was no variation within lineages. Lineage A1 contained alleles A1 of all 22 V. longisporum isolates. Lineage D1 comprised alleles D1 of 14 V. longisporum isolates, lineage D2 contained alleles D2 of four V. longisporum isolates, and lineage D3 contained alleles D3 of the four remaining V. longisporum isolates. Verticillium longisporum lineage D2 was most closely related to the three V. dahliae strains PD327, PD362 and PD502, whereas V. longisporum lineage D3 was sister group to the remaining V. dahliae isolates. Verticillium longisporum lineages D2 and D3, as well as the V. dahliae isolates were sister group to V. longisporum lineage D1. The V. longisporum lineages D1, D2, D3 and the V. dahliae isolates were the sister group to V. longisporum lineage A1. The two V. albo-atrum isolates were monophyletic, and formed the sister group to the V. longisporum lineages plus the V. dahliae isolates. The V. albo-atrum and V. dahliae isolates, together with the V. longisporum lineages, were sister to V. tricorpus plus V. nubilum.
Figure 2. Evolutionary origins of the diploid hybrid Verticillium longisporum.
Shown is a Bayesian consensus tree inferred from a combined ACT, EF, GPD, OX, TS dataset (95 taxa, 3007 characters). Isolates are represented by a strain identifier, in the case of V. longisporum followed by allele designations A1, D1, D2 or D3. Hosts and geographic origins are given. Species delimitations are indicated by vertical bars on the right. Values by the branches are Bayesian posterior probabilities, parsimony and likelihood bootstrap supports, in that order. Additional support values within V. dahliae are listed in Table S3. Branches in bold have maximal support in all analyses. The branches leading to V. tricorpus and V. nubilum are not to scale. Each V. longisporum isolate has two alleles that group in four different clades corresponding to three different species. One allele of each V. longisporum isolate groups in species A1, the second allele clusters in species D1, V. dahliae lineage D2 or V. dahliae lineage D3. Verticillium longisporum alleles D2 are most closely related to the free-living haploid V. dahliae strains PD327, PD362 and PD502. None of the other V. longisporum alleles group with any haploid parental isolates. Arrows on the right show the three inferred hybridization events between species A1 and species D1, V. dahliae lineage D2 or V. dahliae lineage D3, respectively, to give rise to the three lineages of V. longisporum, V. longisporum lineage A1/D1, lineage A1/D2 and lineage A1/D3. Of the three parental species, only V. dahliae is known.
To determine which clades represented phylogenetic species, we applied the Genealogical Concordance Phylogenetic Species Recognition concept according to which well supported, terminal clades in multigene phylogenies are recognized as phylogenetic species [38]. We thus identified the following phylogenetic species: The clade with the two V. albo-atrum isolates, V. longisporum lineage A1, V. longisporum lineage D1 and the monophyletic group with V. longisporum lineages D2 and D3 as well as the V. dahliae isolates. The V. longisporum lineages D2 and D3 did not correspond to separate phylogenetic species, since they were part of a monophyletic group together with the V. dahliae isolates, within which there were topological conflicts between the single gene trees (Figures S5, S6, S7, S8, S9), likely indicative of sexual recombination [38]. Close affinity of V. longisporum lineages D2 and D3 with the V. dahliae isolates was supported by morphological evidence. Verticillium dahliae strains PD327, PD362 and PD502, the closest relatives of V. longisporum lineage D2, were morphologically indistinguishable from the remaining V. dahliae isolates. Thus, haploid isolates of the V. longisporum lineages D2 and D3 would be expected to belong to the morphological species V. dahliae. No morphological species corresponding to V. longisporum lineages A1 and D1 are known. Our analyses included representatives of all four haploid Verticillium morphological species [39], none of which grouped with V. longisporum lineages A1 and D1. The phylogenetic species containing the V. albo-atrum isolates corresponded to the morphological species V. albo-atrum.
The ITS tree shown in Figure 1 is not directly comparable to the Bayesian consensus tree based on five protein coding loci shown in Figure 2, since all V. longisporum isolates only carried a single ITS allele. Verticillium longisporum ITS alleles fell into two groups, correlating with the V. longisporum lineages D1, D2 and D3. Isolates of Verticillium longisporum lineages D1 and D2 had identical ITS alleles, forming a sister group to the V. dahliae clade in a polytomy with the V. albo-atrum isolates (Figure 1). ITS alleles of V. longisporum lineage D3 were identical to the ITS alleles of the majority of V. dahliae isolates.
The phylogenetic analyses using parsimony and maximum likelihood supported the results from Bayesian inference on the combined five-locus dataset. Parsimony analyses yielded six most parsimonious trees of 1152 steps each, differing from each other and the tree in Figure 2 by poorly supported branches within V. dahliae. See Figure 2 for parsimony branch supports, and Table 1 for additional details in the parsimony analyses. The most likely tree had a –ln likelihood score of 8851.96, and differed from the tree in Figure 2 and the most parsimonious trees by poorly supported branches within V. dahliae (see Figure 2 for likelihood branch supports). See Table S3 for support values within V. dahliae not presented in Figure 2. The five-locus alignment was submitted to TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S11128), and the DNA sequences were deposited in GenBank (Accessions: ACT: HQ206921 - HQ207015; EF: HQ414624 - HQ414718, GPD: HQ414719 - HQ414813, OX: HQ414814 - HQ414908, TS: HQ414909 - HQ415003).
Phylogenetic analyses and taxon selection for the MAT1-1 tree
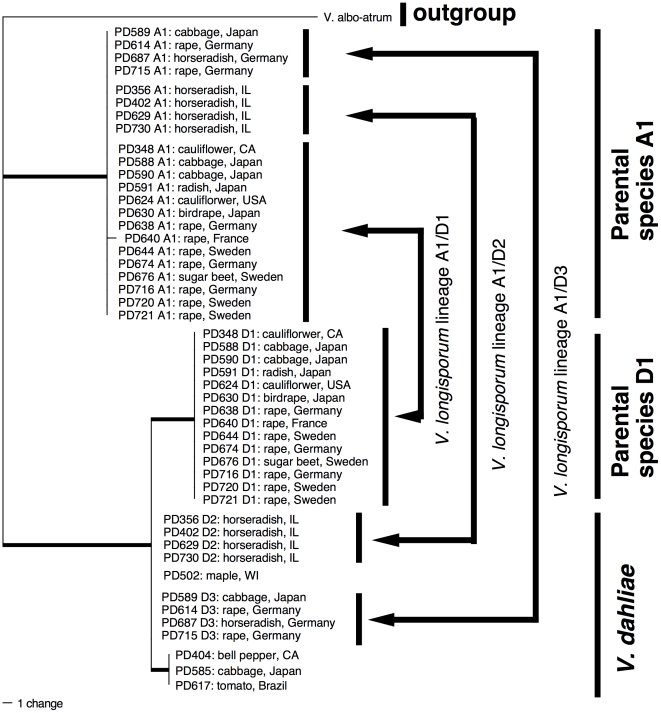
Results from phylogenetic analyses based on the MAT1-1 region (Figure 3) were congruent with results from the tree derived from five loci (Figure 2), except that V. dahliae lineages D2 and D3 were not resolved. All V. longisporum isolates in Figure 2 were included in the MAT1-1 analyses along with the four V. dahliae strains PD404, PD502, PD585 and PD617, and the homologous region from the genome sequence of V. albo-atrum strain retrieved from the Broad Institute website. The V. dahliae strains were randomly chosen among the V. dahliae strains with MAT1-1 with the exception of strain PD502, which was included because it was the only V. dahliae MAT1-1 strain sharing a most recent common ancestor with a V. longisporum group. The MAT dataset consisted of 45 taxa and 420 characters from the intergenic spacer region between MAT1-1-1 and MAT1-1-2 (GenBank Accessions HQ415004 - HQ415049). The alignment was submitted to TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S11128) and analyzed using Bayesian inference, parsimony, and maximum likelihood. Both the Bayesian consensus tree shown in Figure 3, the most parsimonious tree (73 steps in length) and the most likely tree (–ln likelihood = 922.64) had identical topologies (see Figure 3 for branch support values). The K81 model of DNA sequence evolution was used for likelihood and Bayesian analyses as determined by Modeltest.
Figure 3. Evolutionary origins of the diploid hybrid Verticillium longisporum based on partial MAT1-1 idiomorph data.
Shown is a Bayesian consensus tree inferred from non-coding region between MAT1-1-1 and MAT1-1-2 (45 taxa, 420 characters). Taxa included were all V. longisporum strains used in Fig. 2, as well as a selection of V. dahliae MAT1-1 isolates. Isolates are represented by a strain identifier, in the case of V. longisporum followed by allele designations A1, D1, D2 or D3. Hosts and geographic origins are given. Species delimitations are indicated by vertical bars on the right. Values by the branches are Bayesian posterior probabilities, parsimony and likelihood bootstrap supports, in that order. Branches in bold have maximal support in all analyses. Each V. longisporum isolate has two alleles that group in three different clades corresponding to three different species. One allele of each V. longisporum isolate groups in species A1, the second allele clusters in species D1 or in V. dahliae lineages D2/D3. Unlike in the five-locus tree in Figure 2, V. dahliae lineages D2 and D3 are not differentiated, otherwise results are congruent. Arrows on the right show the three inferred hybridization events between species A1 and species D1, V. dahliae lineage D2 or V. dahliae lineage D3, respectively, to give rise to the three lineages of V. longisporum, V. longisporum lineage A1/D1, lineage A1/D2 and lineage A1/D3. The V. albo-atrum sequence was retrieved from the BROAD website from supercontig 1_2.
Detection of a beta-tubulin paralog in V. dahliae and V. longisporum
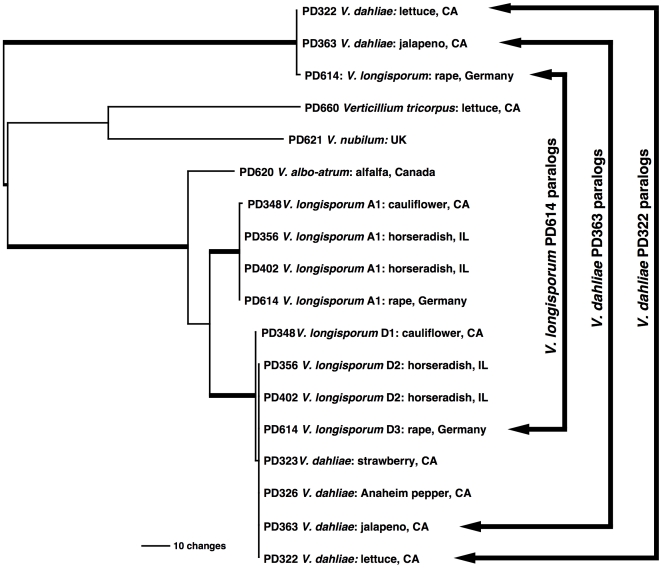
Beta-tubulin (TUB) has frequently been used in phylogenetic studies of fungi, including Verticillium [40], [41], [42], [43]. We did not use the TUB in this study, as initial PCR screens followed by cloning and phylogenetic analyses showed that V. dahliae strains PD322 and PD363, as well as V. longisporum strain PD614 harbored a TUB paralog. Phylogenetic analyses involving the TUB paralogs were performed on an 18 taxa, 666 character dataset using parsimony, resulting in two most parsimonious trees of 399 steps each (CI = 0.912; RI = 0.934) that were topologically identical on a 70% bootstrap support level. The taxa included in the TUB analyses represented all species and major lineages found in the phylogenetic analyses based on the combined, five-locus dataset (Figure 2). The most parsimonious TUB tree illustrated in Figure 4 showed that the three paralogs formed the sister group to the remaining taxa, and thus arose by gene duplication before speciation of V. tricorpus, V. nubilum, V. albo-atrum, V. longisporum and V. dahliae. No TUB paralogs were detected in any other taxa included in the tree, no other taxa included in this study were screened for TUB paralogs. The alignment was submitted to TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S11128), and the DNA sequences were deposited in GenBank (Accessions JF343442 - JF343459).
Figure 4. Presence of beta-tubulin paralogs in V. dahliae and V. longisporum isolates.
Shown is a most parsimonious tree from a 18 taxa, 666 character dataset, 399 steps in length. The taxa selected represent the major clades from the five-locus tree in Figure 2. Isolates are represented by a strain identifier. Hosts and geographic origins are given. Arrows in the right mark the pairs of paralogs present in V. dahliae strains PD322 and PD363 as well as V. longisporum strain PD614. The tree suggests that the paralogs originated before speciation in Verticillium. No paralogs were found in any of the other taxa included in the tree, but not all taxa used in this study were investigated for the presence of beta-tubulin paralogs.
Discussion
Our results suggest that Verticillium longisporum is an allodiploid hybrid that originated at least three different times in independent hybridization events involving four different parental lineages representing three different species.
Verticillium longisporum is diploid
Unlike the majority of ascomycetes, Verticillium longisporum is a stable diploid [6]. Its nuclei contain approximately twice as much DNA as the closely related V. dahliae [31], [32], [33], its conidia (asexual spores) are uninucleate and almost twice the size of V. dahliae conidia [44], [45]. Artificial diploids of V. dahliae have similar conidia sizes as V. longisporum [46], and artificial haploids of V. longisporum have similar conidia to wild type V. dahliae [6]. Auxotrophic mutants of V. longisporum are more challenging to obtain than in V. dahliae [6], [47], [48]. Our data are in agreement with V. longisporum being diploid. Verticillium dahliae, a close relative of V. longisporum, has six or seven chromosomes [49], [50]. We assayed six non-ribosomal loci that in V. dahliae are on at least five different chromosomes, and found two alleles at each locus (Figures 2 &3). Since all our strains were derived from single conidia that are uninucleate [31], [44], [45], this suggested that V. longisporum nuclei are diploid. All V. longisporum strains had just one ITS allele, consistent with the homogenizing selection pressure exerted on ribosomal gene repeats by concerted evolution. All other Verticillium species assayed here had just one allele at each locus, which is the norm in ascomycete fungi which are haploid.
Verticillium longisporum has three different parental species
Phylogenetic analyses demonstrated that V. longisporum had three different parental species, species A1, species D1 and V. dahliae. Each species corresponded to a monophyletic group with maximal statistical support. Species A1 and species D1 did not correspond to known Verticillium species. We included representatives of all five Verticillium species [39], none of which clustered with species A1 and species D1. Blast searches at GenBank with V. longisporum alleles A1 and D1 did not return any 100% matches other than to V. longisporum sequences.
It was previously suggested that parents of V. longisporum included V. dahliae, V. albo-atrum and/or close relatives of the two [26], [40], [41], [45]. Our data demonstrated that the parents were V. dahliae and its close relative, species D1, as well as the more divergent species A1, sister taxon to species D1 plus V. dahliae (Figure 2). Species D1 differed from V. dahliae by 1.4% of all sequenced sites, and species A1 and V. dahliae differed by 4.4%. The phylogenetic topology together with the fixed genetic differences suggested that the three species have been distinct entities for a considerable amount of time.
Collins et al. [40] suggested that one of the V. longisporum parents was a diploid strain of V. dahliae, based on the presence of two beta-tubulin alleles in some strains of V. dahliae and V. longisporum. Our phylogenetic anlayses showed that V. dahliae and V. longisporum strains may contain a beta-tubulin paralog (Fig. 4). No other loci we examined in V. dahliae or in any other Verticillium species besides V. longisporum contained more than one allele. Thus, a more likely scenario for the presence of an additional beta-tubulin copy in some V. dahliae and V. longisporum strains is ancient duplication with subsequent differential losses during speciation in Verticillium.
Verticillium longisporum originated at least three times
How often V. longisporum originated is unclear. Collins et al. [40] suggested two or possibly three different hybridization events. Using some of the same strains as Collins et al. [40], we demonstrated that V. longisporum originated at least three different times given our taxon sampling (Figure 2). Species A1 was involved in each hybridization event, hybridizing with species D1 resulting in V. longisporum lineage A1/D1, with V. dahliae lineage D2 resulting in V. longisporum lineage A1/D2, and with V. dahliae lineage D3 resulting in V. longisporum lineage A1/D3. This conclusion is derived from the observation that each V. longisporum strain contained a species A allele, as well as a species D1, V. dahliae lineage D2 or V. dahliae lineage D3 allele, at each non-ribosomal locus examined (Figure 2). The ITS locus was monomorphic in all V. longisporum strains, likely due to concerted evolution. The V. longisporum lineages A1/D1 and A1/D2 contained the ITS region of species A1, and all isolates of V. longisporum lineage A1/D3 contained V. dahliae ITS regions (Figure 1).
It was previously known that V. longisporum was genetically diverse. We obtained some groupings similar to those reported earlier, the V. longisporum lineage A1/D1 corresponded to Zeise and von Tiedemann's [51] group lsp and Clewes et al. 's [26] group alpha. The V. longisporum lineage A1/D3 corresponded to group lsp* and group beta of Zeise and von Tiedemann [51], and Clewes et al. [26], respectively. The group beta-gamma of Clewes et al. [26] corresponded to V. longisporum lineage A1/D2. Karapapa et al. [45] found another group, called recombinants which we could not confirm. Verticillium longisporum strain PD674, one of the recombinants, was a member of V. longisporum lineage A1/D1 (Figure 2).
Multiple evolutionary origins of hybrids in plants are well documented [52]. A fungal example includes the poplar rust Melampsora x columbiana that has evolved multiple times in different geographic areas [3].
Verticillium longisporum likely did not originate through parasexual processes
It has been suggested that V. longisporum originated by parasexual recombination [45], which involves hyphal fusion, karyogamy, mitotic recombination and chromosome loss to restore the haploid state [53]. A parasexual origin is supported by the observation that nuclei of V. longisporum contain approximately double the DNA of V. dahliae [31], [44]. But there is considerable variation in DNA content between nuclei of different isolates, suggesting differences in chromosome content between isolates. This is exemplified by two isolates included in our study, V. longisporum strains PD721 and PD644, whose respective nuclear DNA contents were 0.05 and 0.075 pg DNA/nucleus [33]. Also, parasexual recombination has been observed in the laboratory within and between species of Verticillium [46], [54], making it an attractive hypothesis to account for the evolution of V. longisporum. However, our data do not support parasexual recombination. All V. longisporum strains we examined had two alleles at all six non-ribosomal loci (Figures 2 & 3), which suggested they are stable and have not reverted to a haploid state. This was despite the fact that 18 of our strains were isolated more than 13 years ago, the oldest 51 years ago (Table S2). Genetically stable hybrids in Verticillium have also been found in some laboratory experiments [55], but stability varies between parental strains even within a single species [54]. The variability of nuclear DNA content in V. longisporum isolates is similar to what is known from other fungi [56], including V. dahliae [49], [50]. Thus, the evidence at hand does not suggest that V. longisporum originated through parasexual recombination. Hyphal fusion between strains of different species, followed by nuclear fusion leading to the creation of a stable, diploid nucleus, seems more plausible.
An alternative explanation for the origin of V. longisporum is sexual recombination without separation of homologous chromosomes. However, this seems less likely. In ascomycetes, sexual recombination is initiated when two individuals of opposite mating types fuse. We showed that all V. longisporum isolates lacked the mating type MAT1-2 (Figure S4), but carried two idiomorphs (alleles) of the other mating type, MAT1-1, one from each parent (Figure 3). In addition, no sexual state has ever been detected in any species of Verticillium, including V. dahliae [57].
Verticillium longisporum originated on unusual host for both parents
Compared with animals and plants, not much is known about the evolutionary origins of fungal hybrids. One of the best-studied groups of fungal hybrids includes Neotyphodium, a genus of grass endophytes. Neotyphodium hybrids often have different hosts than their parents [22], [58]. This suggested that hybridizations occurred on unusual hosts for both parents, increasing the chances for the hybrid to outcompete its parents [58]. A similar scenario might be playing out in Verticillium [59]. Nothing is known about the life styles of two of the V. longisporum parents, species A1 and species D1, none of which has ever been found. Species A1 and species D1 might be associated with non-agricultural hosts or live as saprobes which would explain why they have been overlooked so far, or they might be extinct. The life style of species A1 and species D1 might be similar to V. nubilum, another potentially saprobic species in Verticillium. Verticillium nubilum was originally isolated from potato where it failed to induce disease symptoms [60]. Verticillium dahliae, the third parent, has a wide host range and infects many different crops [30], which in general excludes crucifers with some exceptions [61], [62]. The host range of V. longisporum is centered on crucifers [61], [63], but infection of a wide diversity of hosts has been demonstrated in greenhouse studies [48], [64], [65], suggesting that V. longisporum could have originated on many different hosts. We considered one host in detail, horseradish in Illinois where both V. longisporum and V. dahliae are pathogenic and are isolated from diseased plants [62]. We found that all V. longisporum isolates from horseradish in Illinois belonged to V. longisporum lineage A1/D2. None of the three V. dahliae isolates (strains PD727, PD728, PD729) from the same host and location were in contention as parents of V. longisporum, as they grouped in the main clade of V. dahliae, distantly related to the V. longisporum parents V. dahliae lineages D2 and D3 (Figure 2). Similarly, all other V. dahliae isolates (strains PD584, PD585, PD623, PD717, PD718, PD719) from typical V. longisporum hosts (cabbage, oilseed rape) grouped in the main clade of V. dahliae (Figure 2).
Further support that V. longisporum originated on unusual hosts for both parents comes from V. dahliae strains PD327, PD362 and PD502, which are part of V. dahliae lineage D2, the only V. longisporum ancestor for which free-living, haploid strains are known (Figure 2). Verticillium dahliae strains PD327, PD362 and PD502 were isolated from pepper in California and maple in Wisconsin, neither of which is a host of V. longisporum. Pathogenicity tests of one of the isolates, V. dahliae strain PD327, on a variety of different hosts showed that V. dahliae strain PD327 was more virulent on its original host than on the V. longisporum hosts cabbage and cauliflower [65]. However, more research is needed to conclusively identify the sites of hybridization in the ancestry of V. longisporum.
Verticillium longisporum, a new hybrid fungal pathogen with an expanded host range
Hybridization can lead to the evolution of new phenotypes. Verticillium longisporum has larger conidia than any other species of Verticillium, and it also is exceptionally virulent on oilseed rape in Europe and Japan [28], where it may severely reduce yields. In comparative studies of the infection process in oilseed rape, V. longisporum was more virulent than V. dahliae, one of its parental species. Both V. longisporum and V. dahliae entered the plant by penetrating the roots, but only V. longisporum spread to the shoot and induced disease symptoms, including stunted growth, chlorosis and discoloration of the vessels in older leaves [29]. The differences in virulence suggest a particular adaptation to oilseed rape of V. longisporum relative to V. dahliae, which possibly includes suppression of or resistance to the host defense response [29]. The differences observed between the infection process of V. dahliae and V. longisporum at the microscopic level agreed with the fact that V. longisporum is the most important Verticillium wilt-inducing pathogen of oilseed rape and other crucifer crops such as cabbage and cauliflower [48], suggesting that the increased virulence of V. longisporum might be related to its hybrid origin.
In Phytophthora, a group of fungal-like protists, there are examples of new plant pathogens arising through hybridization between different species [66], [67]. In fungi, a major group of plant pathogens, reports of hybrids are rare [25], and in the few known examples, the virulence and host ranges of the hybrids is generally equal or reduced relative to their parents, or unknown [4], [5], [8], [9], [10], [11], [12], [13], [14], [15], [68], [69]. The only example, to our knowledge, with increased virulence of a hybrid is the example of the poplar rust fungus Melampsora x columbiana that became the dominant species on poplar in the Pacific Northwest in 1998 by replacing one of its parents [3]. Another example for increased virulence of a fungal hybrid might be the human pathogen Cryptococcus neoformans, where hybrids between different lineages of the fungus occur in nature. These hybrids were artificially recreated, and were shown to have increased virulence in an animal model [16].
Verticillium longisporum might be one of the first examples of a hybrid fungal pathogen with a new host range. The strains of V. longisporum used in our study were isolated from various diseased agricultural crops, including oilseed rape where V. longisporum is more virulent than any of its parents, presumably due to its allodiploid status. In plants, phenotypic variation in polyploids might be due to changes in gene expression, altered regulatory interactions, and rapid genetic and epigenetic changes [2]. In fungi, little is known about the molecular consequences of polyploidy.
Potential phenotypic differences between the V. longisporum hybrid lineages
There is evidence that the three V. longisporum lineages differ phenotypically in their host range and pathogenicity. Zeise and von Tiedemann [63] found that the isolates of their V. longisporum group lsp* were avirulent on oilseed rape, whereas all isolates from group lsp caused disease. We included representatives of both groups, and showed that V. longisporum groups lsp* and lsp corresponded to V. longisporum lineages A1/D3 and A1/D1, respectively. Interestingly, one parent of the oilseed rape pathogen V. longisporum lineage A1/D3 is V. dahliae, which is not pathogenic on oilseed rape [29].
Another lineage of V. longisporum is lineage A1/D2, only known from horseradish in Illinois [62]. Whether V. longisporum lineage A1/D2 causes disease on oilseed rape is not known. Considerable acreage of oilseed rape is grown in relative proximity to Illinois, in Kansas, Oklahoma, Minnesota and North Dakota (http://www.uscanola.com/, accessed on June 17, 2010), but Verticillium wilt disease on oilseed rape has never been reported from North America [28], despite the fact that no commercially grown oilseed rape varieties are resistant to V. longisporum [28]. This suggests that V. longisporum lineage A1/D2, which is confined to horseradish in Illinois, might not be pathogenic on oilseed rape. However, the agent of Verticillium wilt of oilseed rape, V. longisporum lineage A1/D1, is present in North America as a pathogen of cauliflower in California [48], and is pathogenic on oilseed rape in greenhouse assays [63].
Verticillium longisporum originated recently
The three lineages of V. longisporum are genetically homogenous. Based on the seven different loci examined, there is no variation within the lineages except for one substitution in MAT1-1 in one of the isolates (Figure 2, Figure 3). This suggests a recent origin of all three lineages, which is further supported by the fact that the alleles of species A1, a parent of all three V. longisporum lineages, are all identical regardless of lineage. Brasier [24] proposed that movement of fungi away from where they initially evolved might allow them to come into contact with other species, hybridize and evolve into new pathogens. We don't know whether this applies to V. longisporum. However, the lack of genetic variation suggests recent origin, conceivably facilitated through human activities.
The geographic locations where V. longisporum originated are uncertain. Lineage A1/D2 is restricted to Illinois where it might have originated; lineage A1/D3 is confined to Europe and Japan; and lineage A1/D1 occurs in Europe, Japan and North America. Population genetics studies might shed light on the centers of origins of these two lineages, as might finding all V. longisporum parents, which, with the exception of V. dahliae, are yet unknown.
Summary and further research
Our data demonstrate that V. longisporum originated three different times, involving three different parental species. Further work will focus on identification and characterization of all parental species, which will provide the foundations for further research into the effects of hybridization on the parental genomes of V. longisporum, including the basis of pathogenicity and virulence. This work will allow for the correct identification and differentiation of the V. longisporum lineages which is important for disease management and enforcement of quarantine regulations.
Materials and Methods
Taxon sampling and origins of fungal strains
The isolates included in this study were identified based on morphological characters, and selected to represent all five species of Verticillium [39]. A total of 203 isolates were used, 154 isolates of V. dahliae, 42 isolates of V. longisporum, five isolates of V. albo-atrum, and one isolate each of V. tricorpus and V. nubilum (Table S2). The isolates were from 43 different hosts, from 16 countries and four continents.
DNA extraction, PCR amplification for direct sequencing and DNA sequencing conditions
Fungi were grown on cellophane membranes placed on PDA plates. Mycelia were peeled off, freeze-dried and ground using liquid nitrogen. DNA was extracted with the FastDNA Kit in conjunction with the FastPrep Instrument (MP Biomedicals, Irvine, CA). Buffer CLS-Y was used, and grinding intensity was set to 4.5 for 30 seconds. PCRs were performed using GoTaq Colorless Master Mix (Promega Corp., Madison, WI, USA) in a 25 µl reaction volume according to the manufacturer's instructions. The PCR program consisted of a 2 min initial denaturation step at 94°C, 32 cycles of 10 sec at 94°C, 20 sec at the primer pair dependent annealing temperature, and 1 min at 72°C, followed by a final extension of 7 min at 72°C. PCR products were purified by sodium acetate precipitation [70]. DNA sequences were determined at the UC Davis DNA Sequencing Facility, using ABI BigDye Terminator v3.1 Cycle Sequencing chemistry on an ABI 3730 Capillary Electrophoresis Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Loci used for phylogenetic analyses and primer design
Eight loci were PCR amplified and sequenced with primer pairs designed based on conserved regions in the V. dahliae and V. albo-atrum genomes accessed on the BROAD Institute website (http://www.broadinstitute.org/annotation/genome/verticillium_dahliae/MultiHome.html, accessed February 10, 2009), and the V. dahliae MAT1-1 sequence AB469828 retrieved from GenBank. Some primers were taken from the literature. The ribosomal internal transcribed spacer region was amplified with ITS1-F [71] and ITS4, and sequenced with ITS5 and ITS4 [72]. Parts of the following protein coding genes were PCR amplified and sequenced: actin (ACT) with primer pair VActF/VActR, elongation factor 1-alpha (EF) with VEFf/VEFr, glyceraldehyde-3-phosphate dehydrogenase (GPD) with primer pair VGPDf2/VGPDr, mitochondrial oxaloacetate transport protein (OX) with VOx3f/VOx2r, tryptophan synthase (TS) with VTs3f/VTs3r and beta-tubulin (TUB) with VTubF2/VTubR. Parts of the MAT1-1 idiomorph spanning from MAT1-1-1 to MAT1-1-2 were amplified with Alf/MAT12r, and sequenced with various primers. For details on loci, primer sequences, PCR conditions, and sequencing primers used, see Table S4, Table S5 and Table S6.
For V. longisporum, allele specific primers were designed for six non-ribosomal loci (ACT, EF, GPD, OX, TS, MAT) based on cloned PCR products obtained with the primer pairs mentioned above. For ACT, the allele specific primers were ActFa1, ActF2d1, and ActF2d2, paired with reverse primer VActR; for EF, EFfa1, EFfd1 and EFfd2, paired with VEFr; for GPD, GPDfa1, GPDfd1, and GPDfd2, paired with VGPDr; for OX, OxFa1, OxFd1 and OxFd2 paired with VOxR; for TS, TsFa1, TsF2d1 and TsFd2, paired with VTs2r; and for the MAT1-1 idiomorph, the primer pairs MATa1f/MATa1r and MATdf/MATdr. For more details on loci and PCR conditions, see Table S7.
MAT screening
All isolates were screened for presence of the mating type idiomorphs MAT1-1 and MAT1-2 by PCR, using primers designed based on the sequenced V. albo-atrum and V. dahliae strains which were MAT1-1 and MAT1-2, respectively. The primer pair Alf/MAT11r targeted parts of MAT1-1-1, whereas HMG21f/MAT21r targeted parts of MAT1-2-1. For primer sequences and PCR conditions, see Table S5 and Table S6.
Cloning of mixed PCR products in Verticillium longisporum
Direct sequencing of PCR products for all but the ITS region failed in V. longisporum, indicating the presence of a mixed template population. To determine the number of alleles present at each locus, PCR products were cloned. To avoid formation of chimera PCR products due to the presence of mixed template, the following guidelines proposed by Beser et al. [73] were implemented. As compared to standard PCR conditions, the denaturation temperature, elongation time, and primer concentrations were increased, and the number of PCR cycles was decreased, resulting in the following PCR protocol. PCR reactant concentrations 0.8 mM for each dNTP and 0.8 µM for each primer, in a 25 µl reaction volume containing 10 or 100 ng of template DNA. To reduce PCR-induced substitutions, Easy-A High-Fidelity PCR cloning Enzyme or PfuUltra High-Fidelity DNA polymerase (Agilent Technologies, Stratagene Products Division, La Jolla, CA) were used according to the manufacturer's instruction, and the PCR program consisted of a 3 min initial denaturation at 94°C, followed by 22 cycles of 1 min at 96°C, 1 min at the primer pair dependent annealing temperature, and 4.5 min at 72°C, and followed by a final extension of 7 min at 72°C. For primer specific annealing temperatures, see Table S6. PCR products were purified with the DNA Clean & Concentrator-5 Kit (Zymo Research Corp., Orange, CA) according to the manufacturer's instructions, with an additional washing step using 80% ethanol, and eluted in a final volume of 6 µl. In case of multiple PCR bands, target bands were purified using a Zymoclean Gel DNA Recovery Kit (Zymo Research Corp., Orange, CA) following the manufacturer's instructions. PCR products were ligated to a pGEM-T Vector and cloned using the JM109 High Efficiency Competent Cells (Promega Corp., Madison, WI, USA) following the manufacturer's instructions. Transformant colonies were screened for the presence of insert by PCR with the standard M13f/M13r primers. PCR products of expected lengths were sequenced with the initial PCR primers.
Phylogenetic analyses
Four different datasets were analyzed, including the ITS dataset, a combined five-locus dataset consisting of concatenated ITS, ACT, GPD, EF, OX and TS datasets, a MAT1-1 dataset, as well as a TUB dataset. The MAT1-1 outgroup sequence of V. albo-atrum was retrieved from the V. albo-atrum genome sequence from the BROAD Institute website (http://www.broadinstitute.org/annotation/genome/verticillium_dahliae/MultiHome.html, accessed February 10, 2009).
Three different algorithms were used. The ITS and TUB datasets were analyzed under the maximum parsimony criterion using PAUP v.4.0b 10 [74]. The combined, five-locus and the MAT1-1 datasets were analyzed using parsimony, maximum likelihood as implemented in PAUP v.4.0b 10 [74], as well as MrBayes v3.0b4 [75] implementing a Bayesian approach to inferring phylogenies.
Most parsimonious trees were inferred using 30 random addition replicates; otherwise, default settings were used, including treating insertion/deletion gaps as missing data. Bootstrap support values were based on 500 replicates. Maximum likelihood analyses were performed using default settings and 30 random addition replicates, and bootstrap supports were based on 500 replicates. Bayesian analyses were performed with default settings, running four chains over 10 million generations and sampling each 100th tree. The first 1000 of the 10,000 saved trees were discarded and the consensus tree was based on the remaining 9,000 trees. Maximum likelihood and Bayesian analyses implemented an optimal model of DNA sequence evolution determined using Modeltest 3.7 [76]. All analyses were run with a single representative of each haplotype to speed up analyses.
Supporting Information
PCR gels documenting the presence of allele A1 in all 42 isolates of Verticillium longisporum using allele A1 specific primers. Loci are indicated on the left. Numbers in the top row refer to V. longisporum isolates as they appear in Table S2, except for strains PD356, PD402, PD629, PD730, PD589, PD614, PD687 and PD715 which are in lanes 35–42. ‘L’ indicates DNA size standards (arrowhead = 500 bp), ‘C’ PCR negative controls. For names of loci, details on primers and PCR conditions, see text.
(TIF)
PCR gels documenting the distribution of allele D1 in all 42 isolates of Verticillium longisporum using allele D1 specific primers. Allele D1 was absent in V. longisporum strains PD356, PD402, PD629, PD730, PD589, PD614, PD687 and PD715, corresponding to lanes 35 – 42. The bands for locus TS in lanes 35 – 42 were due to non-specific amplification of alleles D2 or D3 by allele D1 specific primers as confirmed by DNA sequencing. Numbers in the top row refer to V. longisporum isolates as they appear in Table S2, except for strains PD356, PD402, PD629, PD730, PD589, PD614, PD687 and PD715 which are in lanes 35–42. ‘L’ indicates DNA size standards (arrowhead = 500 bp), ‘C’ PCR negative controls. For names of loci, details on primers and PCR conditions, see text.
(TIF)
PCR gels documenting the distribution of alleles D2 and D3 in all 42 isolates of Verticillium longisporum using a primer set specific to alleles D2 and D3. Alleles D2 and D3 were absent in all but V. longisporum strains PD356, PD402, PD629, PD730, PD589, PD614, PD687 and PD715, corresponding to lanes 35 – 42. The bands for locus OX in lanes 1 – 34 were due to non-specific amplification of allele D1 as confirmed by DNA sequencing. Numbers in the top row refer to V. longisporum isolates as they appear in Table S2, except for strains PD356, PD402, PD629, PD730, PD589, PD614, PD687 and PD715 which are in lanes 35–42. ‘L’ indicates DNA size standards (arrowhead = 500 bp), ‘C’ PCR negative controls. For names of loci, details on primers and PCR conditions, see text.
(TIF)
PCR gels documenting the distribution of MAT1-1 and MAT1-2 idiomorphs in all 203 Verticillium isolates using idiomorph specific primers. Each of the four panes shows presence and absence of MAT1-1 and MAT1-2 in a subset of isolates. The order of the isolates is as in Table S2, isolate numbers are given above the panes for isolates near the DNA size standards (arrowheads = 500 bp). ‘C’ stands for PCR negative control. Each isolate amplifies for either MAT1-1 or MAT1-2, except the three isolates marked by asterisks corresponding to V. nubilum strain PD621, V. tricorpus strain PD660 and V. dahliae strain PD707, respectively, which failed to amplify for either primer set.
(TIF)
Evolutionary origins of the diploid hybrid Verticillium longisporum based on phylogenetic inference from the ACT dataset comprising 95 taxa and 532 characters. Shown is the single, most parsimonious tree, 199 steps in length. Isolates are represented by a strain identifier, V. longisporum identifiers are followed by an allele designation. Hosts and geographic origins are given. Branches with 100% bootstrap support are in bold, other support values above 70% are given by the branches.
(TIF)
Evolutionary origins of the diploid hybrid Verticillium longisporum based on phylogenetic inference from the EF dataset comprising 95 taxa and 600 characters. Shown is one most parsimonious tree, 318 steps in length. Isolates are represented by a strain identifier, V. longisporum identifiers are followed by an allele designation. Hosts and geographic origins are given. Branches with 100% bootstrap support are in bold, other support values above 70% are given by the branches.
(TIF)
Evolutionary origins of the diploid hybrid Verticillium longisporum based on phylogenetic inference from the GPD dataset comprising 95 taxa and 678 characters. Shown is the single, most parsimonious tree, 165 steps in length. Isolates are represented by a strain identifier, V. longisporum identifiers are followed by an allele designation. Hosts and geographic origins are given. Branches with 100% bootstrap support are in bold, other support values above 70% are given by the branches.
(TIF)
Evolutionary origins of the diploid hybrid Verticillium longisporum based on phylogenetic inference from the OX dataset comprising 95 taxa and 606 characters. Shown is the single, most parsimonious tree, 214 steps in length. Isolates are represented by a strain identifier, V. longisporum identifiers are followed by an allele designation. Hosts and geographic origins are given. Branches with 100% bootstrap support are in bold, other support values above 70% are given by the branches.
(TIF)
Evolutionary origins of the diploid hybrid Verticillium longisporum based on phylogenetic inference from the TS dataset comprising 95 taxa and 591 characters. Shown is one most parsimonious tree, 243 steps in length. Isolates are represented by a strain identifier, V. longisporum identifiers are followed by an allele designation. Hosts and geographic origins are given. Branches with 100% bootstrap support are in bold, other support values above 70% are given by the branches.
(TIF)
Numbers of PCR product clones sequenced for each Verticillium longisporum strain at each locus. For details on strains, see Table S2.
(DOC)
Fungal isolates used; given are the strain identifiers used in this study, additional strain identifiers, the host scientific and common names, the location and date of collection, the source, the number of loci sequenced, as well as the mating type.
(DOC)
Loci used for phylogenetic analyses; details of chromosomal locations, lengths of amplicons, introns and intergenic spacers, and locus IDs are given with respect to the sequenced genome of V. dahliae strain PD322 on the Broad Institute website ( http://www.broadinstitute.org/annotation/genome/verticillium_dahliae/MultiHome.html , accessed February 10, 2009).
(DOC)
The DNA sequences of all primers used are listed by primer name in ascending alphanumeric order. Primer sequences are given 5′->3′.
(DOC)
PCR conditions used in this study. For all loci, forward and reverse PCR primers, annealing temperature, annealing temperature for cloning, and expected product length in V. dahliae strain PD322 are given. For details on PCR conditions, see text.
(DOC)
Loci and conditions used for V. longisporum allele-specific PCR amplifications. For each locus and allele, forward and reverse primers, annealing temperature, amplicon length with respect to V. dahliae strain PD322, numbers of introns targeted and total intron lengths are given. For details on PCR conditions, see text.
(DOC)
Acknowledgments
We thank the following individuals for providing strains that were crucial for the successful completion of this study. A. von Tiedemann, K. Zeise, University of Göttingen; C. Dixelius, University of Agricultural Sciences, Uppsala; D. Barbara, University of Warwick; G. Stanosz, University of Wisconsin; L. du Toit, Washington State University; M. Typas, University of Athens; M. Babadoost, University of Illinois; E. L. Gasich, P. Gannibal, All-Russian Institute of Plant Protection, St. Petersburg; R. Zare, Iranian Research Institute of Plant Protection, Tehran; T. Gordon, J. Duniway, S. Koike, G. Miyao, R. Wilson, UC Davis; T. Usami, Chiba University; H. Sakai, T. Shiraishi, Department of Agriculture, Gunma Prefecture; B. W. Pennypacker, Pennsylvania State University; D. Fravel, USDA-ARS; D. Lawn, United Genetics Seeds Company; K. Johnson, M. Powelson, M. Putnam, Oregon State University; National Institute of Agrobiological Sciences, Japan, for providing cultures free of charge. G. I. Craig, C. Rosa, UC Davis, helped with cloning. Z. K. Atallah, UC Davis, provided suggestions that improved the manuscript. J. Jenkins, E. Gutierrez, A. Felt provided expert technical assistance. We thank two anonymous reviewers for helpful comments and suggestions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Financial support was provided by the California Leafy Greens Board (2008-10, 2008-11, 2009-10, and 2009-11), United States Department of Agriculture-National Institute of Food and Agriculture (USDA-NIFA) (2007-51100-03846), and the Hatch funds allocated to University of California Agricultural Experiment Station Project CA-D*-PPA-5681-H. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hegarty MJ, Hiscock SJ. Hybrid speciation in plants: new insights from molecular studies. New Phytol. 2005;165:411–423. doi: 10.1111/j.1469-8137.2004.01253.x. [DOI] [PubMed] [Google Scholar]
- 2.Osborn TC, Pires JC, Birchler JA, Auger DL, Chen ZJ, et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003;19:141–147. doi: 10.1016/s0168-9525(03)00015-5. [DOI] [PubMed] [Google Scholar]
- 3.Newcombe G, Stirling B, McDonald S, Bradshaw HD. Melampsora x columbiana, a natural hybrid of M. medusae and M. occidentalis. Mycol Res. 2000;104:261–274. [Google Scholar]
- 4.Flor HH. Heterothallism and hybridization in Tilletia tritici and T. laevis. J Agr Res. 1932;44:49–58. [Google Scholar]
- 5.Spiers AG, Hopcroft DH. Comparative studies of the poplar rusts Melampsora medusae, M. larici-populina and their interspecific hybrid M. medusae-populina Mycol Res. 1994;98:889–903. [Google Scholar]
- 6.Ingram R. Verticillium dahliae var. longisporum, a stable diploid. Trans Br Mycol Soc. 1968;51:339–341. [Google Scholar]
- 7.Schardl CL, Craven KD. Interspecific hybridization in plant-associated fungi and oomycetes: a review. Mol Ecol. 2003;12:2861–2873. doi: 10.1046/j.1365-294x.2003.01965.x. [DOI] [PubMed] [Google Scholar]
- 8.Gonthier P, Nicolotti G, Linzer R, Guglielmo F, Garbelotto M. Invasion of European pine stands by a North American forest pathogen and its hybridization with a native interfertile taxon. Mol Ecol. 2007;16:1389–1400. doi: 10.1111/j.1365-294X.2007.03250.x. [DOI] [PubMed] [Google Scholar]
- 9.Joly DL, Langor DW, Hamelin RC. Molecular and morphological evidence for interspecific hybridization between Cronartium ribicola and C. comandrae on Pinus flexilis in southwestern Alberta. Plant Dis. 2006;90:1552–1552. doi: 10.1094/PD-90-1552A. [DOI] [PubMed] [Google Scholar]
- 10.Masneuf I, Hansen J, Groth C, Piskur J, Dubourdieu D. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl Environ Microbiol. 1998;64:3887–3892. doi: 10.1128/aem.64.10.3887-3892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Donnell K, Cigelnik E, Nirenberg HI. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia. 1998;90:465–493. [Google Scholar]
- 12.Morin L, Van Der Merwe M, Hartley D, Muller P. Putative natural hybrid between Puccinia lagenophorae and an unknown rust fungus on Senecio madagascariensis in KwaZulu-Natal, South Africa. Mycol Res. 2009;113:725–736. doi: 10.1016/j.mycres.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Brasier CM, Kirk SA, Pipe ND, Buck KW. Rare interspecific hybrids in natural populations of the Dutch elm disease pathogens Ophiostoma ulmi and O. novo-ulmi. Mycol Res. 1998;102:45–57. [Google Scholar]
- 14.O'Donnell K, Ward TJ, Geiser DM, Kistler HC, Aoki T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet Biol. 2004;41:600–623. doi: 10.1016/j.fgb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Staats M, van Baarlen P, van Kan JAL. Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity Mol Biol Evol. 2005;22:333–346. doi: 10.1093/molbev/msi020. [DOI] [PubMed] [Google Scholar]
- 16.Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, et al. Alpha AD alpha hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 2007;3:e186. doi: 10.1371/journal.pgen.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai H-F, Liu J-S, Staben C, Christensen MJ, Latch GCM, et al. Evolutionary diversification of fungal endophytes of tall fescue grass by hybridization with Epichloë species. Proc Natl Acad Sci USA. 1994;91:2542–2546. doi: 10.1073/pnas.91.7.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moon CD, Miles CO, Jarlfors U, Schardl CL. The evolutionary origins of three new Neotyphodium endophyte species from grasses indigenous to the Southern Hemisphere. Mycologia. 2002;94:694–711. doi: 10.1080/15572536.2003.11833197. [DOI] [PubMed] [Google Scholar]
- 19.Moon CD, Guillaumin J-J, Ravel C, Li C, Craven KD, et al. New Neotyphodium endophyte species from the grass tribes Stipeae and Meliceae. Mycologia. 2007;99:895–905. doi: 10.3852/mycologia.99.6.895. [DOI] [PubMed] [Google Scholar]
- 20.Paun O, Fay MF, Soltis DE, Chase MW. Genetic and epigenetic alterations after hybridization and genome doubling. Taxon. 2007;56:649–656. [PMC free article] [PubMed] [Google Scholar]
- 21.Ma X-F, Gustafson JP. Allopolyploidization-accommodated genomic sequence changes in Triticale. Ann Bot. 2008;101:825–832. doi: 10.1093/aob/mcm331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moon CD, Craven KD, Leuchtmann A, Clement SL, Schardl CL. Prevalence of interspecific hybrids amongst asexual fungal endophytes of grasses. Mol Ecol. 2004;13:1455–1467. doi: 10.1111/j.1365-294X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez-Bello MA, Chilvers MI, Akamatsu H, Peever TL. Host specificity of Ascochyta spp. infecting legumes of the Viciae and Cicerae tribes and pathogenicity of an interspecific hybrid. Phytopathology. 2006;96:1148–1156. doi: 10.1094/PHYTO-96-1148. [DOI] [PubMed] [Google Scholar]
- 24.Brasier CM. The rise of the hybrid fungi. Nature. 2000;405:134–135. doi: 10.1038/35012193. [DOI] [PubMed] [Google Scholar]
- 25.Brasier CM. Rapid evolution of introduced plant pathogens via interspecific hybridization. BioScience. 2001;51:123–133. [Google Scholar]
- 26.Clewes E, Edwards SG, Barbara DJ. Direct molecular evidence supports long-spored microsclerotial isolates of Verticillium from crucifers being interspecific hybrids. Plant Pathol. 2008;57:1047–1057. [Google Scholar]
- 27.Heale JB. Diversification and speciation in Verticillium - an overview. In: Tjamos EC, Rowe RC, Heale JB, Fravel DR, editors. Advances in Verticillium research and disease management. St. Paul, MN: APS Press; 2000. pp. 1–14. Proceedings of the Seventh International Verticillium Symposium, Cape Souinion, Athens, Greece, October 1997 ed. [Google Scholar]
- 28.Heale J, Karapapa VK. The Verticillium threat to Canada's major oilseed crop: Canola. Can J Plant Pathol. 1999;21:1–7. [Google Scholar]
- 29.Eynck C, Koopmann B, Grunewaldt-Stoecker G, Karlovsky P, von Tiedemann A. Differential interactions of Verticillium longisporum and V. dahliae with Brassica napus detected with molecular and histological techniques. Eur J Plant Pathol. 2007;118:259–272. [Google Scholar]
- 30.Pegg GF, Brady BL. Wallingford, Oxon, UK: CABI Publishing; 2002. Verticillium wilts.552 [Google Scholar]
- 31.Typas MA, Heale JB. DNA content of germinating spores, individual hyphal cells and resting structure cells of Verticillium spp. measured by microdensitometry. J Gen Microbiol. 1980;121:231–242. [Google Scholar]
- 32.Jackson CW, Heale JB. Relationship between DNA content and spore volume in sixteen isolates of Verticillium lecanii and two new diploids of V. dahliae ( = V. dahliae var. longisporum Stark). Journal of General Microbiology. 1985;131:3229–3236. [Google Scholar]
- 33.Steventon LA, Fahleson J, Hu Q, Dixelius C. Identification of the causal agent of Verticillium wilt of winter oilseed rape in Sweden, V. longisporum. Mycol Res. 2002;106:570–578. [Google Scholar]
- 34.Metzenberg RL, Glass NL. Mating type and mating strategies in Neurospora. BioEssays. 1990;12:53–59. doi: 10.1002/bies.950120202. [DOI] [PubMed] [Google Scholar]
- 35.Turgeon BG, Yoder OC. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet Biol. 2000;31:1–5. doi: 10.1006/fgbi.2000.1227. [DOI] [PubMed] [Google Scholar]
- 36.Taylor JW, Geiser DM, Burt A, Koufopanou V. The evolutionary biology and population genetics underlying fungal strain typing. Clin Microbiol Rev. 1999;12:126–146. doi: 10.1128/cmr.12.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pantou MP, Strunnikova OK, Shakhnazarova VY, Vishnevskaya NA, Papalouka VG, et al. Molecular and immunochemical phylogeny of Verticillium species. Mycol Res. 2005;109:889–902. doi: 10.1017/s0953756205003345. [DOI] [PubMed] [Google Scholar]
- 38.Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- 39.Barbara DJ, Clewes E. Plant pathogenic Verticillium species: how many of them are there? Molecular Plant Pathology. 2003;4:297–305. doi: 10.1046/j.1364-3703.2003.00172.x. [DOI] [PubMed] [Google Scholar]
- 40.Collins A, Okoli CA, Morton A, Parry D, Edwards SG, et al. Isolates of Verticillium dahliae pathogenic to crucifers are of at least three distinct molecular types. Phytopathology. 2003;93:364–376. doi: 10.1094/PHYTO.2003.93.3.364. [DOI] [PubMed] [Google Scholar]
- 41.Collado-Romero M, Jiménez-Díaz RM, Mercado-Blanco J. DNA sequence analysis of conserved genes reveals hybridization events that increase genetic diversity in Verticillium dahliae Fungal Biology. 2010;114:209–218. doi: 10.1016/j.funbio.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Collado-Romero M, Mercado-Blanco J, Olivares-García C, Jiménez-Díaz RM. Phylogenetic analysis of Verticillium dahliae vegetative compatibility groups. Phytopathology. 2008;98:1019–1028. doi: 10.1094/PHYTO-98-9-1019. [DOI] [PubMed] [Google Scholar]
- 43.Qin QM, Vallad GE, Wu BM, Subbarao KV. Phylogenetic analyses of phytopathogenic Isolates of Verticillium spp. Phytopathology. 2006;96:582–592. doi: 10.1094/PHYTO-96-0582. [DOI] [PubMed] [Google Scholar]
- 44.Stark C. Das Auftreten der Verticillium-Tracheomykosen in Hamburger Gartenbaukulturen. Gartenbauwissenschaft. 1961;26:493–528. [Google Scholar]
- 45.Karapapa VK, Bainbridge BW, Heale JB. Morphological and molecular characterization of Verticillium longisporum comb. nov., pathogenic to oilseed rape. Mycol Res. 1997;101:1281–1294. [Google Scholar]
- 46.Hastie AC. The parasexual cycle in Verticillium albo-atrum. Genetical Research. 1964;5:305–315. [Google Scholar]
- 47.Puhalla JE, Hummel M. Vegetative compatibility groups within Verticillium dahliae. Phytopathology. 1983;73:1305–1308. [Google Scholar]
- 48.Subbarao KV, Chassot A, Gordon TR, Hubbard JC, Bonello P, et al. Genetic relationships and cross pathogenicities of Verticillium dahliae isolates from cauliflower and other crops. Phytopathology. 1995;85:1105–1112. [Google Scholar]
- 49.Pantou MP, Typas MA. Electrophoretic karyotype and gene mapping of the vascular wilt fungus Verticillium dahliae. FEMS Microbiol Lett. 2005;245:213–220. doi: 10.1016/j.femsle.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 50.Usami T, Fukaya M, Amemiya Y. Electrophoretic karyotyping and mapping of pathotype-specific DNA sequences in Japanese isolates of Verticillium dahliae. Journal of General Plant Pathology. 2008;74:61–65. [Google Scholar]
- 51.Zeise K, von Tiedemann A. Application of RAPD-PCR for virulence type analysis within Verticillium dahliae and V. longisporum. Phytopathol Z. 2002;150:557–563. [Google Scholar]
- 52.Soltis DE, Soltis PS. Molecular data and the dynamic nature of polyploidy. Crit Rev Plant Sci. 1993;12:243–273. [Google Scholar]
- 53.Caten CE. Parasexual processes in fungi. In: Gull K, Oliver SG, editors. The Fungal Nucleus. Cambridge: Cambridge University Press; 1981. pp. 191–214. [Google Scholar]
- 54.Hastie AC. Hybridization of Verticillium albo-atrum and Verticillium dahliae. Trans Br Mycol Soc. 1973;60:511–523. [Google Scholar]
- 55.Clarkson JM, Heale JB. Heterokaryon compatibility and genetic recombination within a host plant between hop wilt isolates of Verticillium albo-atrum. Plant Pathol. 1985;34:129–138. [Google Scholar]
- 56.Zolan ME. Chromosome-length polymorphism in Fungi. Microbiol Rev. 1995;59:686–698. doi: 10.1128/mr.59.4.686-698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Usami T, Itoh M, Amemiya Y. Asexual fungus Verticillium dahliae is potentially heterothallic Journal of General Plant Pathology. 2009;75:422–427. [Google Scholar]
- 58.Selosse MA, Schardl CL. Fungal endophytes of grasses: hybrids rescued by vertical transmission? An evolutionary perspective. New Phytol. 2007;173:452–458. doi: 10.1111/j.1469-8137.2007.01978.x. [DOI] [PubMed] [Google Scholar]
- 59.Clewes E, Barbara DJ. Two allopolyploid ascomycete fungal plant pathogens were not rescued by vertical transmission. New Phytol. 2008;177:583–585. doi: 10.1111/j.1469-8137.2007.02327.x. [DOI] [PubMed] [Google Scholar]
- 60.Pethybridge GH. Notes on some saprophytic species of fungi associated with diseased potato plants and tubers. Trans Br Mycol Soc. 1919;6:104–120. [Google Scholar]
- 61.Horiuchi S, Hagiwara H, Takeuchi S. Host specificity of isolates of Verticillium dahliae towards cruciferous and solanaceous hosts. In: Hornby D, editor. Biological control of soil-borne plant pathogens. Wallingford, UK: CAB International; 1990. pp. 285–298. [Google Scholar]
- 62.Babadoost M, Chen W, Bratsch AD, Eastman CE. Verticillium longisporum and Fusarium solani: two new species in the complex of internal discoloration of horseradish roots. Plant Pathol. 2004;53:669–676. [Google Scholar]
- 63.Zeise K, von Tiedemann A. Host specialization among vegetative compatibility groups of Verticillium dahliae in relation to Verticillium longisporum. Phytopathol Z. 2002;150:112–119. [Google Scholar]
- 64.Johansson A, Goud JKC, Dixelius C. Plant host range of Verticillium longisporum and microsclerotia density in Swedish soils. Eur J Plant Pathol. 2006;114:139–149. [Google Scholar]
- 65.Bhat R, Subbarao KV. Host range specificity in Verticillium dahliae. Phytopathology. 1999;89:1218–1225. doi: 10.1094/PHYTO.1999.89.12.1218. [DOI] [PubMed] [Google Scholar]
- 66.Brasier CM, Cooke DE, Duncan JM. Origin of a new Phytophthora pathogen through interspecific hybridization. Proc Natl Acad Sci U S A. 1999;96:5878–5883. doi: 10.1073/pnas.96.10.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Man in't Veld WA, de Cock AWAM, Summerbell RC. Natural hybrids of resident and introduced Phytophthora species proliferating on multiple new hosts. Eur J Plant Pathol. 2006;117:25–33. [Google Scholar]
- 68.Kauserud H, Svegarden IB, Decock C, Hallenberg N. Hybridization among cryptic species of the cellar fungus Coniophora puteana (Basidiomycota). Mol Ecol. 2007;16:389–399. doi: 10.1111/j.1365-294X.2006.03129.x. [DOI] [PubMed] [Google Scholar]
- 69.Christen AA, Bruehl GW. Hybridization of Typhula ishikariensis and T. idahoensis. Phytopathology. 1979;69:263–266. [Google Scholar]
- 70.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 71.Gardes M, Bruns TD. ITS primers with enhanced specificity of basidiomycetes: application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 72.White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols. San Diego: Academic Press, Inc; 1990. pp. 315–322. [Google Scholar]
- 73.Beser J, Hagblom P, Fernandez V. Frequent in vitro recombination in Internal transcribed spacers 1 and 2 during genotyping of Pneumocystis jirovecii. J Clin Microbiol. 2007;45:881–886. doi: 10.1128/JCM.02245-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swofford DL. Sunderland, Massachusetts: Sinauer Associates; 2002. PAUP*.Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. [Google Scholar]
- 75.Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 76.Posada D, Crandall KA. Modeltest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR gels documenting the presence of allele A1 in all 42 isolates of Verticillium longisporum using allele A1 specific primers. Loci are indicated on the left. Numbers in the top row refer to V. longisporum isolates as they appear in Table S2, except for strains PD356, PD402, PD629, PD730, PD589, PD614, PD687 and PD715 which are in lanes 35–42. ‘L’ indicates DNA size standards (arrowhead = 500 bp), ‘C’ PCR negative controls. For names of loci, details on primers and PCR conditions, see text.
(TIF)
PCR gels documenting the distribution of allele D1 in all 42 isolates of Verticillium longisporum using allele D1 specific primers. Allele D1 was absent in V. longisporum strains PD356, PD402, PD629, PD730, PD589, PD614, PD687 and PD715, corresponding to lanes 35 – 42. The bands for locus TS in lanes 35 – 42 were due to non-specific amplification of alleles D2 or D3 by allele D1 specific primers as confirmed by DNA sequencing. Numbers in the top row refer to V. longisporum isolates as they appear in Table S2, except for strains PD356, PD402, PD629, PD730, PD589, PD614, PD687 and PD715 which are in lanes 35–42. ‘L’ indicates DNA size standards (arrowhead = 500 bp), ‘C’ PCR negative controls. For names of loci, details on primers and PCR conditions, see text.
(TIF)
PCR gels documenting the distribution of alleles D2 and D3 in all 42 isolates of Verticillium longisporum using a primer set specific to alleles D2 and D3. Alleles D2 and D3 were absent in all but V. longisporum strains PD356, PD402, PD629, PD730, PD589, PD614, PD687 and PD715, corresponding to lanes 35 – 42. The bands for locus OX in lanes 1 – 34 were due to non-specific amplification of allele D1 as confirmed by DNA sequencing. Numbers in the top row refer to V. longisporum isolates as they appear in Table S2, except for strains PD356, PD402, PD629, PD730, PD589, PD614, PD687 and PD715 which are in lanes 35–42. ‘L’ indicates DNA size standards (arrowhead = 500 bp), ‘C’ PCR negative controls. For names of loci, details on primers and PCR conditions, see text.
(TIF)
PCR gels documenting the distribution of MAT1-1 and MAT1-2 idiomorphs in all 203 Verticillium isolates using idiomorph specific primers. Each of the four panes shows presence and absence of MAT1-1 and MAT1-2 in a subset of isolates. The order of the isolates is as in Table S2, isolate numbers are given above the panes for isolates near the DNA size standards (arrowheads = 500 bp). ‘C’ stands for PCR negative control. Each isolate amplifies for either MAT1-1 or MAT1-2, except the three isolates marked by asterisks corresponding to V. nubilum strain PD621, V. tricorpus strain PD660 and V. dahliae strain PD707, respectively, which failed to amplify for either primer set.
(TIF)
Evolutionary origins of the diploid hybrid Verticillium longisporum based on phylogenetic inference from the ACT dataset comprising 95 taxa and 532 characters. Shown is the single, most parsimonious tree, 199 steps in length. Isolates are represented by a strain identifier, V. longisporum identifiers are followed by an allele designation. Hosts and geographic origins are given. Branches with 100% bootstrap support are in bold, other support values above 70% are given by the branches.
(TIF)
Evolutionary origins of the diploid hybrid Verticillium longisporum based on phylogenetic inference from the EF dataset comprising 95 taxa and 600 characters. Shown is one most parsimonious tree, 318 steps in length. Isolates are represented by a strain identifier, V. longisporum identifiers are followed by an allele designation. Hosts and geographic origins are given. Branches with 100% bootstrap support are in bold, other support values above 70% are given by the branches.
(TIF)
Evolutionary origins of the diploid hybrid Verticillium longisporum based on phylogenetic inference from the GPD dataset comprising 95 taxa and 678 characters. Shown is the single, most parsimonious tree, 165 steps in length. Isolates are represented by a strain identifier, V. longisporum identifiers are followed by an allele designation. Hosts and geographic origins are given. Branches with 100% bootstrap support are in bold, other support values above 70% are given by the branches.
(TIF)
Evolutionary origins of the diploid hybrid Verticillium longisporum based on phylogenetic inference from the OX dataset comprising 95 taxa and 606 characters. Shown is the single, most parsimonious tree, 214 steps in length. Isolates are represented by a strain identifier, V. longisporum identifiers are followed by an allele designation. Hosts and geographic origins are given. Branches with 100% bootstrap support are in bold, other support values above 70% are given by the branches.
(TIF)
Evolutionary origins of the diploid hybrid Verticillium longisporum based on phylogenetic inference from the TS dataset comprising 95 taxa and 591 characters. Shown is one most parsimonious tree, 243 steps in length. Isolates are represented by a strain identifier, V. longisporum identifiers are followed by an allele designation. Hosts and geographic origins are given. Branches with 100% bootstrap support are in bold, other support values above 70% are given by the branches.
(TIF)
Numbers of PCR product clones sequenced for each Verticillium longisporum strain at each locus. For details on strains, see Table S2.
(DOC)
Fungal isolates used; given are the strain identifiers used in this study, additional strain identifiers, the host scientific and common names, the location and date of collection, the source, the number of loci sequenced, as well as the mating type.
(DOC)
Loci used for phylogenetic analyses; details of chromosomal locations, lengths of amplicons, introns and intergenic spacers, and locus IDs are given with respect to the sequenced genome of V. dahliae strain PD322 on the Broad Institute website ( http://www.broadinstitute.org/annotation/genome/verticillium_dahliae/MultiHome.html , accessed February 10, 2009).
(DOC)
The DNA sequences of all primers used are listed by primer name in ascending alphanumeric order. Primer sequences are given 5′->3′.
(DOC)
PCR conditions used in this study. For all loci, forward and reverse PCR primers, annealing temperature, annealing temperature for cloning, and expected product length in V. dahliae strain PD322 are given. For details on PCR conditions, see text.
(DOC)
Loci and conditions used for V. longisporum allele-specific PCR amplifications. For each locus and allele, forward and reverse primers, annealing temperature, amplicon length with respect to V. dahliae strain PD322, numbers of introns targeted and total intron lengths are given. For details on PCR conditions, see text.
(DOC)