Abstract
Previous studies have found that common genetic variants on chromosome 8q24 are associated with the risk of developing colorectal neoplasia. We conducted a hospital-based case-control study, including 435 cases and 788 unrelated controls to investigate the associations between common variants on 8q24 and the risk of colorectal cancer in a Chinese population. We also evaluated the association of rs6983267 with colorectal neoplasia in the published literature via a meta-analysis study. We found that rs6983267 was significantly associated with the risk of colorectal cancer in the Chinese population, with an adjusted odds-ratio (OR) for the GT heterozygotes and GG homozygotes of 1.30 (95% CI = 0.98–1.71, P = 0.069) and 1.66 (95% CI = 1.18–2.34, P = 0.004), respectively, compared to the TT homozygotes, with a P-trend value of 0.003. No association was found for the other three loci (rs16901979, rs1447295 and rs7837688). In the meta-analysis of the published genetic association studies, the rs6983267 variant was found to be associated with an increased risk of colorectal neoplasia. The heterozygous GT carriers showed a 20% increased risk of colorectal neoplasia (OR = 1.20, 95% CI = 1.16–1.25; random effects model) with a summary OR for homozygous GG carriers of 1.39 (95% CI = 1.32–1.48; random effects model) compared to the TT genotype carriers. We found no significant differences between the association of rs6983267 and colorectal cancer and colorectal adenomas. In summary, our study confirms that the variant rs6983267 is a risk factor for colorectal neoplasia in various populations, including the Chinese population.
Introduction
Colorectal cancer (CRC) is the third most common cancer and the fourth most frequent cause of cancer death worldwide [1]. The lifetime risk in Western European and North American populations is around 5% [2]. Among the risk factors and causes for CRC, inherited genetic factors account for approximately 35% of the disease etiology [2]. Many genetic factors, such as mutations of critical genes (e.g. APC, MLH1, MSH2, TGFBR2 and SMAD4) have been identified [3]. However, many factors that increase the susceptibility to CRC, but with low penetrance need to be explored. In the past few years, several genome-wide association studies (GWAS) have identified novel loci that are associated with CRC risk, such as variants on 11q23 [2], 8q24 [4], [5], [6], 10p14 [7], 8q23.3 [7], 15q13.3 (HMPS) [8] and SMAD7 [9]. However, the extent to which these genetic factors contribute to the disease has not been established.
Among these loci, variants on 8q24 have shown strong evidence of an association with the risk of CRC in different populations. Haiman et al genotyped six variants in 1,124 individuals with invasive CRC and 4,573 controls that had been previously identified as having an underlying risk for prostate cancer due to alterations on 8q24 [10] and found that one variant, rs6983267, was also significantly associated with colorectal cancer [4]. Similarly, Tomlinson et al conducted a genome-wide association study of 550,000 tag SNPs in 930 familial colorectal cancer patients and 960 controls and found that rs6983267 had the strongest association with CRC risk [5]. Analyses based on 1,477 individuals with colorectal adenoma and 2,136 controls suggested the possibility that this locus is involved in tumor initiation rather than progression [5]. SNP rs10505477, which maps about 5.86 kb centromeric to rs6983267 and has a high linkage disequilibrium with rs6983267, was also found to be associated with CRC risk, and has been implicated across many cohort and case-control studies [5], [11], [12], [13]. It has also been reported that five different haplotype blocks at the 8q24 region have been identified, and only those loci located between the 128.47 Mb and the 128.54 Mb region (e.g. rs6983267, rs10808556 and rs10505477) were associated with CRC risk. The variations in the other regions were found to be associated with prostate, breast and ovarian cancers [14].
The constantly improving standard of living in China has brought about extensive changes to the lifestyle and diet of the average Chinese citizen over the past three decades, and has led to a corresponding increase in the incidence and mortality of CRC in China [15]. The steady increase in the incidence and mortality of CRC has made the disease one of the leading causes of death in China. However, few studies have examined the genetic factors that influence the risk of CRC in the Chinese population. We herein report the results of a case-control study that we performed to investigate the allelic variants on 8q24 and their effect on CRC risk in a Chinese population. To further investigate the association of variant rs6983267 with the CRC risk, a systemic review of the literature published about the locus was undertaken, and a meta-analysis was conducted.
Materials and Methods
Study populations
Most of the participants in the study have been described previously [16]. In brief, a total of 478 CRC patients and 838 controls aged between 30 and 80 years old were enrolled between 2001 and 2003 from three hospitals (Xi'nan Hospital, Xinqiao Hospital and Daping Hospital) in Chongqing, China. The subjects were genetically unrelated ethnic Han Chinese from Chongqing and the surrounding regions served by these hospitals, including parts of Sichuan, Yunnan and Guizhou provinces in southwest China, which are adjacent to Chongqing. The recruitment followed the Japan, Korea and China Colorectal Cancer Collaboration Group guidelines. All patients had been histopathologically diagnosed with primary CRC within the past 6 months, and had not received any treatment. The medical records of the patients were thoroughly checked and for those who were suffering from ileocecal junction tumors or anal canal tumors and those with any of the following conditions: i. recurrence of CRC; ii. familial adenomatous polyposis (FAP); iii. hereditary nonpolyposis colorectal cancer (HNPCC); iv. other tumors; v. severe digestive tract diseases lasting over 2 years; vi. diabetes, fatty liver, hepatic cirrhosis, metabolic syndrome or severe cardiovascular diseases were excluded from the study. One or two controls matched to each eligible case based on age (±5 years), sex and residence were selected. The controls were recruited from non-CRC patients seen in the Departments of General Surgery, Orthopedics or Trauma who were admitted for trauma, bone fractures, appendicitis, arthritis, or varicose veins. Patients with tumors, severe digestive tract diseases lasting over 2 years, diabetes, fatty liver, hepatic cirrhosis, metabolic syndrome or severe cardiovascular diseases were also excluded from the control group.
A 5-mL peripheral venous blood sample was obtained from each subject after written informed consent was obtained. Each participant was personally interviewed by trained interviewers to complete a Semi-quantitative Food Frequency Questionnaire (SQFFQ), which collected demographic information and information about dietary, and smoking habits (current smoker, former or never smoker), alcohol use (more or less than 15 g/day, according to the recommended level of daily alcohol consumption suggested by the China Health Care Association) and other lifestyle factors [16]. After quality control procedures were completed for both SQFFQ and DNA samples, a total of 435 patients and 788 controls of Han ethnicity were finally included in the study.
Ethics statement
The study was approved by all of the ethics committees of the participating hospitals (“Ethics Committee of Xi'nan Hosipital”, “Ethics Committee of Xinqiao Hosipital” and “Ethics Committee of Daping Hosipital”). All of the samples were collected with a written informed consent provided by the participants, and all protocols were approved by the human research ethics committees of the participating hospitals.
Selection of SNPs on 8q24
Genome-wide association studies and gene-based candidate studies have confirmed that chromosome 8q24 is a susceptibility region for CRC. It has also been shown that specific loci on 8q24 are associated with specific cancers [14]. Among these loci are two SNPs, rs6983267 and rs10505477, which were separately found to be associated with CRC risk [5], [17]. These two SNPs are in high linkage disequilibrium (LD) in the Chinese population (r2 = 0.95) according to the Hapmap database, and they were also found to be associated with an increased risk of prostate, breast and ovarian cancers in many population studies [4], [18], [19]. We chose to further study the association of rs6983267 with colorectal cancer in the Chinese population. Another 3 SNPs, rs16901979, rs1447295 and rs7837688, which were previously found to be associated with prostate cancer, were also selected to evaluate their association with colorectal cancer risk in the Chinese population [14], [20].
DNA isolation and genotyping
Genomic DNA was extracted from 2.5-mL of whole blood with a Promega DNA Purification Wizard kit according to the manufacturer's instructions. The genotyping methods have been reported previously [16]. Three SNPs (rs16901979, rs1447295 and rs7837688) were genotyped using the Applied Biosystems SNPLex system (Applied Biosystems Incorporated, California, USA) together with 45 other loci including previously reported loci on CYP2E1 [16]. Loci were submitted online to ABI Inc for probe design and synthesis. The OLA (oligonucleotide ligation assay), purification, and PCR reactions were performed on an Eppendorf 5333 Mastercycler, and allele inspection was performed on an ABI 3130xl Gene Analyzer. The SNP information was collected using Data Collection Software version 3.0, and data were analyzed by the GeneMapper Software version 4.0. Another locus (rs6983267) was genotyped using a TaqMan® SNP Genotyping Assay (Applied Biosystems Incorporated, California, USA) on a 7900HT Fast Real-Time PCR System (Applied Biosystems Incorporated, California, USA). A total of 10% of the samples were randomly selected for duplication for these loci to assess the reproducibility of the genotyping calls and a more than 99% concordance rate was found.
Meta-analysis of the rs6983267 locus in subjects with colorectal neoplasia
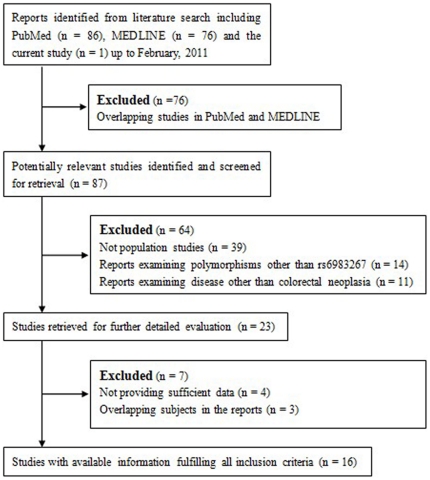
In order to explore the association between rs6983267 and the risk of colorectal neoplasia, a meta-analysis was conducted. We performed a comprehensive and systematic bibliographic search updated to February of 2011 based on the MEDLINE and PubMed databases. We used the terms “8q24” and “rs6983267” in combination with “colorectal neoplasia”, “colorectal cancer” or “colorectal adenoma” to search the database to identify the studies regarding the association between the rs6983267 polymorphism and the risk of colorectal cancer. References were also checked to identify any missing studies. The details of the studies were thoroughly examined in order to exclude potentially overlapping data. If the same participants were used in different papers, only the largest and most complete study was included here. The studies included were those that provided data about rs6983267 and the risk of colorectal cancer or colorectal adenomas and could be case-control, cohort or cross-sectional studies that were reported in English. The studies included should also provide sufficient data about the frequency of the genotypes. Individual authors were also contacted for further data when the criteria were not met. The flow chart that tracks the selection process for the studies and the reasons for exclusion is presented in Figure 1.
Figure 1. The flow chart for the selection of studies and specific reasons for exclusion of studies from the meta-analysis.
For each study, the following information was recorded: first author, publication year, study design, study location, study population/ethnicity, total number and sources of cases and controls, sub-group and disease categories, total number of cases and controls, and the allele frequency in the study. When there were sub-group studies described in the paper (such as those stratified by ethnicity, study stage, etc), they were considered individually. Pooled allelic effects were estimated both under a fixed effects model and a random effects model, and the pooled OR with its 95% CIs was used as the summary measurement.
Statistical analysis
The prevalence of each of the alleles was measured in cases and controls. The χ2 test (for categorical variables) and Student's t-test (for continuous variables) were used to evaluate the differences in demographic characteristics and selected variables. Hardy-Weinberg Equilibrium (HWE) was assessed by the χ2 test (1 degree of freedom (d.f)). The common homozygote was used as the reference to calculate the genotype-specific odds ratio (OR) and its 95% confidence intervals (CI) with or without adjustment for age, sex, smoking status and alcohol use under the unconditional Logistic regression statistic model. The statistical powers of the study were calculated using the Power software under the assuming of two sided test with alpha level is 0.05 with the log-additive genetic model [21], [22].
For meta-analysis, the pooled OR and its 95% CI were calculated using the standard inverse variance weighting method for the fixed effects model and the DerSimonian-Laird method for the random-effects model. Heterogeneity between studies was assessed using the Cochrane Q-test in combination with the I2 statistic. Publication bias was graphically represented by funnel plotting and was assessed both by Egger's linear regression [23] and Begg's rank correlation tests [24]. Statistical analyses were undertaken using the R Software with the SNPassoc and Meta packages (http://www.r-project.org/).
Results
The characteristics of cases and controls are given in Table 1. There were more patients with a daily average alcohol intake of more than 15 g/day (P = 0.011) in the neoplasia patients, as has been indicated previously [15]. In addition, the cases were slightly older than the controls (P = 0.017). Other potential confounders were not significantly different between the cases and controls (Table 1). The genotyping results of the selected SNPs in the CRC cases and controls are shown in Table 2. The overall call rate of each SNP was more than 98%, and no SNP deviated from Hardy-Weinberg equilibrium (P>0.05, Table 2).
Table 1. The characteristics of the participants from a Chinese population.
| Variance | Controls (N = 788) | Cases (N = 435) | P-value |
| Age (years)a | 51.73±11.29 | 53.49±12.88 | 0.017 |
| Sex | 0.972 | ||
| female | 351 (44.5%) | 195 (44.8%) | |
| male | 437 (55.5%) | 240 (55.2%) | |
| Smoking status | 0.547 | ||
| former and never | 488 (61.9%) | 261 (60.0%) | |
| current | 300 (38.1%) | 174 (40.0%) | |
| Alcohol use (>15 g/day) | 0.011 | ||
| yes | 152 (19.3%) | 112 (25.7%) | |
| no | 636 (80.7%) | 323 (74.3%) |
Age was presented as the mean ± SD (years).
Table 2. An association study of 8q24 loci and colorectal cancer risk.
| SNP | Genotype | Controls(N %) | Cases(N %) | Crude OR(95% CI) | Adjusted ORa(95% CI) | P valuea | P-trenda | Call rate | HWE-testP-value |
| rs16901979 | CC | 403 (51.5) | 219 (50.7) | 1 | 1 | 99.3% | 0.66 | ||
| AC | 321 (41.0) | 171 (39.6) | 0.98 (0.76–1.26) | 0.95 (0.74–1.22) | 0.706 | ||||
| AA | 58 (7.4) | 42 (9.7) | 1.33 (0.87–2.05) | 1.30 (0.84–2.00) | 0.243 | 0.550 | |||
| rs6983267 | TT | 256 (32.6) | 111 (25.8) | 1 | 1 | 99.4% | 0.61 | ||
| GT | 392 (49.9) | 219 (50.9) | 1.29 (0.98–1.70) | 1.30 (0.98–1.71) | 0.069 | ||||
| GG | 138 (17.6) | 100 (23.3) | 1.67 (1.19–2.35) | 1.66 (1.18–2.34) | 0.004 | 0.003 | |||
| rs1447295 | CC | 567 (72.5) | 294 (67.9) | 1 | 1 | 99.3% | 0.39 | ||
| AC | 202 (25.8) | 127 (29.3) | 1.21 (0.93–1.58) | 1.22 (0.94–1.59) | 0.140 | ||||
| AA | 13 (1.7) | 12 (2.8) | 1.78 (0.80–3.95) | 1.73 (0.78–3.87) | 0.180 | 0.062 | |||
| rs7837688 | GG | 587 (75.8) | 308 (71.0) | 1 | 1 | 98.8% | 1.00 | ||
| TG | 175 (22.6) | 117 (27.0) | 1.27 (0.97–1.67) | 1.26 (0.95–1.65) | 0.103 | ||||
| TT | 12 (1.6) | 9 (2.1) | 1.43 (0.60–3.43) | 1.38 (0.57–3.32) | 0.479 | 0.089 |
Adjusted for age, sex, alcohol use and smoking status.
Statistically significant associations were obtained only for the previously reported SNP rs6983267 (Table 2). Compared to the TT homozygotes, the GT heterozygotes showed a marginally increased risk of colorectal cancer (adjusted OR = 1.30, 95% CI = 0.98–1.71; P = 0.069). However, the GG homozygotes showed a significant, 66% increased risk of colorectal cancer (adjusted OR = 1.66, 95% CI = 1.18–2.34; P = 0.004). The locus was also found to be associated with a gene-dose response relationship for an increased risk for CRC (P-trend = 0.003). The statistical power for the association of SNP rs6983267 and CCR in our study was 0.839 for the observed OR. We found no statistically significant interactions between rs6983267 and age, sex, smoking status or alcohol use. No other locus was found to be significantly associated with the CRC risk (Table 2). However, the statistical powers for the three loci were relatively lower (0.290 for rs16901979, 0.702 for 1447295 and 0.316 for rs7837688 for the observed ORs).
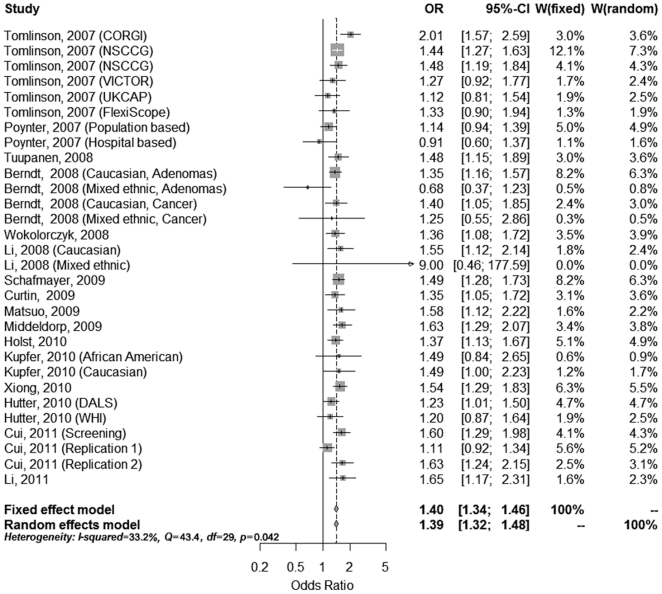
For the meta-analysis regarding the association between colorectal cancer risk and the variant rs6983267, we identified twenty-three reports regarding the association between rs6983267 and colorectal neoplasia. After detailed evaluation, sixteen studies including a total of 36,761 cases and 38,901 controls were used for the meta-analysis (Table 3) [5], [11], [12], [13], [19], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34]. Four studies lacked sufficient data about the genotype frequency [4], [14], [35], [36] and three studies contained overlapping participants with the included reports [37], [38], [39], and were therefore excluded from the analysis (Figure 1). For rs6983267, the summary odds ratio (SOR) was 1.20 (95% = 1.16–1.25) under the fixed effects model and the random effects model for the GT heterozygotes compared to the TT homozygotes (Figure 2). There was no significant heterogeneity between the studies (Q = 21.77, df = 29, P = 0.829; I2 = 0%), and no evidence of a significant publication bias was found (Begg's test, P = 0.630; Egger's test, P = 0.465). For the GG homozygotes, we noted that there was a significant increase in colorectal neoplasia, with the SOR of 1.40 (95% CI = 1.33–1.46) under the fixed effects model and 1.39 (95% CI = 1.32–1.48) under the random effects model compared to the TT carriers (Figure 3). However, significant heterogeneity was found among these studies (Q = 43.39, df = 29, P = 0.042; I2 = 33.2%). However, there still no significant publication bias (Begg's test, P = 0.762; Egger's test, P = 0.783). Sensitivity analyses were also performed by removing the individual studies sequentially, and we found that none individual studies dramatically affected the overall polled ORs.
Table 3. Studies included in the meta-analysis of the association of rs6983267 with colorectal neoplasia.
| Study(First author, year) | Country and ethnicity | Study design | Sub-group study and disease category | Cases | Controls | Ref |
| Tomlinson, 2007 | UK, Caucasian | Population based case-control | CORGI, cancer | 620 | 960 | [5] |
| UK, Caucasian | Population based case-control | CORGI, adenomas | 407 | |||
| UK, Caucasian | Nested case-control | NSCCG, cancer | 4361 | 3752 | ||
| UK, Caucasian | Nested case-control | NSCCG, cancer | 1901 | 1079 | ||
| UK, Caucasian | Nested case-control | VICTOR, cancer | 1072 | 415 | ||
| UK, Caucasian | Nested case-control | UKCAP, adenomas | 607 | 765 | ||
| UK, Caucasian | Population based case-control | FlexiScope, adenomas | 463 | 411 | ||
| Poynter, 2007 | USA and Canada, Mixed ethnic | Population based case-control | Cancer | 1339 | 2191 | [11] |
| USA and Canada, Mixed ethnic | Hospital based case-control | Cancer | 288 | 502 | ||
| Berndt, 2008 | USA, Caucasian | Nested case-control | Adenomas | 2569 | 2779 | [12] |
| USA, Mixed ethnic | Nested case-control | Adenomas | 151 | 194 | ||
| USA, Caucasian | Nested case-control | Cancer | 538 | 1644 | ||
| USA, Mixed ethnic | Nested case-control | Cancer | 71 | 165 | ||
| Tuupanen, 2008 | Finland, Caucasian | Population based case-control | Cancer | 996 | 1012 | [13] |
| Wokolorczyk, 2008 | Poland, Caucasian | Hospital based case-control | Cancer | 779 | 1910 | [19] |
| Li, 2008 | USA, Caucasian | Population based case-control | Cancer | 527 | 679 | [25] |
| USA, Mixed ethnic | Population based case-control | Cancer | 34 | 42 | ||
| Curtin, 2009 | UK and USA, Caucasian | Population based case-control | Cancer | 1069 | 1040 | [26] |
| Matsuo, 2009 | Japan, Japanese | Hospital based case-control | Cancer | 481 | 962 | [27] |
| Schafmayer, 2009 | Germany, Caucasian | Population based case-control | Cancer | 2712 | 2713 | [28] |
| Middeldorp, 2009 | Netherlands, Caucasian | Population based case-control | Cancer | 995 | 1340 | [29] |
| Kupfer, 2009 | USA, African American | Hospital based case-control | Cancer | 1795 | 2378 | [30] |
| USA, Caucasian | Hospital based case-control | Cancer | 399 | 367 | ||
| Holst, 2010 | Sweden, Caucasian | Population based case-control | Cancer | 1786 | 1749 | [31] |
| Xiong, 2010 | China, Chinese Han | Hospital based case-control | Cancer | 2124 | 2124 | [32] |
| Hutter, 2010 | USA, Caucasian | Population based case-control | Cancer | 1461 | 1813 | [33] |
| USA, Caucasian | Nested case-control | Cancer | 614 | 633 | ||
| Cui, 2011 | Japan, Japanese | Population based case-control | Screening stage, cancer | 1583 | 1898 | [34] |
| Japan, Japanese | Population based case-control | Replication 1,cancer | 3099 | 1777 | ||
| Japan, Japanese | Population based case-control | Replication 2, cancer | 1485 | 819 | ||
| Li, 2011 | China, Chinese Han | Hospital based case-control | Cancer | 435 | 788 |
Figure 2. A forest plot of the association of colorectal neoplasia with rs6983267 heterozygosity (G/T vs. T/T).
Figure 3. A forest plot of the association of colorectal neoplasia with rs6983267 homozygosity (G/G vs. T/T).
To determine whether the association between variant rs6983267 and colorectal neoplasia could differ for colorectal adenomas and colorectal cancer, we examined the association between the variant and colorectal cancer and the variant and colorectal adenomas. Among the fourteen identified reports, two examined the association between the variant and colorectal adenomas, with a total of 4,197 cases and 5,109 controls [5], [12]. All of the reports studied the association between rs6983267 and colorectal cancer, with a pool of 32,564 total cases and 34,752 controls. From the meta-analysis, we found that the SORs for GT heterozygotes and GG homozygotes compared to the TT homozygotes were similar in magnitude to those found for colorectal adenomas and total colorectal cancer (Table 4). In the subgroup analysis by ethnicity, we found that the variant rs6983267 was associated with an increased risk of colorectal neoplasia both in the Asian populations (9,207 cases and 8,368 controls) and the Caucasians (23,818 cases and 25,021 controls), although there was a slight different of the pooled OR between the Asian populations and for the Caucasians (Table 4).
Table 4. The results of the meta-analysis of the association of rs6983267 with colorectal neoplasia.
| Category | Genotype(Cases/Controls) | Fixed effects model | Random effects model | Q value/df | P value for Q test | I2 | P value of Begg's test | P value of Egger's test | |
| Disease | Neoplasia | TT (8,334/10,088) | 1 | 1 | |||||
| GT (17,583/18,400) | 1.20 (1.16–1.25) | 1.20 (1.16–1.25) | 21.77/29 | 0.829 | 0% | 0.630 | 0.465 | ||
| GG (10,770/10,372) | 1.40 (1.33–1.46) | 1.39 (1.32–1.48) | 43.39/29 | 0.042 | 33.2% | 0.762 | 0.783 | ||
| Adenomas only | TT (862/1,253) | 1 | 1 | ||||||
| GT (2,110/2,538) | 1.20 (1.08–1.34) | 1.21 (1.04–1.41) | 6.10/4 | 0.192 | 34.4% | 1.000 | 0.868 | ||
| GG (1,225/1,318) | 1.36 (1.21–1.53) | 1.32 (1.00–1.74) | 15.09/4 | 0.005 | 73.5% | 0.624 | 0.712 | ||
| Carcinomas only | TT (7,537/9,024) | 1 | 1 | ||||||
| GT (15,471/16,335) | 1.20 (1.15–1.25) | 1.20 (1.15–1.25) | 16.09/25 | 0.912 | 0% | 0.843 | 0.647 | ||
| GG (9,478/9,356) | 1.41 (1.34–1.48) | 1.41 (1.33–1.49) | 31.46/25 | 0.174 | 20.5% | 0.708 | 0.584 | ||
| Ethnic group | Caucasian | TT (4,670/6,041) | 1 | 1 | |||||
| GT (11,953/12,566) | 1.21 (1.16–1.27) | 1.21 (1.16–1.27) | 11.24/17 | 0.844 | 0% | 0.791 | 0.591 | ||
| GG (7,195/6,414) | 1.42 (1.35–1.49) | 1.42 (1.35–1.49) | 16.37/17 | 0.498 | 0% | 0.677 | 0.745 | ||
| Asian | TT (3,276/3,338) | 1 | 1 | ||||||
| GT (4,382/3,898) | 1.18 (1.10–1.26) | 1.18 (1.10–1.26) | 1.46/5 | 0.918 | 0% | 0.573 | 0.371 | ||
| GG (1,533/1,131) | 1.45 (1.32–1.59) | 1.48 (1.28–1.70) | 10.66/5 | 0.059 | 53.1% | 0.573 | 0.369 | ||
Discussion
Variants on 8q24 (position 128.14 Mb to 128.62 Mb) were found to be associated with various types of cancer. It was reported by Ghoussaini et al that there are five different haplotype blocks within this region. The rs6983267 locus and its highly correlated locus, rs10505477 (r2 = 0.95 in CHB population for Hapmap database), which are located between a 128.47 and 128.54 Mb region, have been found to be associated with prostate, colorectal and ovarian cancers [14]. Loci outside this region were also found to be associated with prostate or breast cancers, but not colorectal cancer [14]. Our study also found a significant association between rs6983267 and colorectal cancer in the Chinese population. However, no significant association for the other three loci, which are located outside the 128.47 and 128.54 Mb region, was identified in the Chinese population. As indicated in our previous study and in other studies, alcohol use is a risk factor for colorectal cancer. In the stratification analysis, we found a trend toward a stronger association in the participants without a drinking habit (<15 g/d), however, we found no statistically significant interactions between the variant rs6983267 and alcohol use. We also did not find any significant interactions between rs6983267 CRC risk and other potential confounders. These findings were consistent with other studies indicating that the association between rs6983267 and colorectal cancer may not modified by the age at diagnosis [11], [12], [25], [27], sex [12], [25], [27], family history of CRC [11], [12], [25], [27], smoking status [27], or alcohol use [27]. However, the current study is a hospital based case-control study and the number of participants was relatively small, which may have led to spurious results. Whether the association between rs6983267 and colorectal neoplasia was affected by the confounders needs further investigation.
In the meta-analysis of rs6983267 and colorectal neoplasia, sixteen eligible papers that examined the risk of colorectal carcinoma, including two that also reported the association between the locus and colorectal adenomas risk were examined. We found that the GT heterozygotes had a 20% (95% CI = 1.16–1.25; random effects model) and the GG homozygotes showed a 39% (95% CI = 1.32–1.48; random effects model) increased risk of colorectal neoplasia. The SORs determined for colorectal neoplasia were similar in magnitude to those observed for colorectal cancer and colorectal adenomas (Table 4). These findings indicated that rs6983267 may play a similar role in the etiology of both colorectal cancer and colorectal adenomas. Although the frequency for the risk allele G was different in different populations (0.487 in CEU, 0.394 in CHB and 0.301 in JPT according to Hapmap database), we also found that the strength of the association between rs6983267 and colorectal neoplasia were similar between the Caucasian and Asian populations (Table 4), which was consistent with a previous meta-analysis study conducted by Hutter et al concerning the association between this locus and CRC risk [33].
Although the variant rs6983267 had been found to be associated with an increased susceptibility to colorectal neoplasia, whether the rs6983267 SNP is the causal locus is still uncertain. Several recently published studies have shown that rs6983267 is located within a transcriptional enhancer region and affects a binding site for TCF4 (also called TCF7L2), a transcription factor which is activated in most CRCs. TCF4 interacts with β-catenin to activate the transcription of Wnt target genes, which are part of a key pathway involved in CRC initiation [40], [41]. It has been found that the rs6983267 G allele has an approximately 1.5-fold stronger enhancer activity and increased affinity for TCF4 compared to the T allele. And Pomerantz et al. demonstrated that a DNA fragment region containing the variant shows a long-range physical interaction with the MYC promoter located ∼330 kb downstream using a chromatin conformation capture (3C) technique [40]. These findings indicate the biological mechanism(s) that potentially underlie the role of this non-protein-coding risk variant, and indicated that rs6983267 may be a causal variant for the susceptibility to CRC.
There are several potential limitations to the current study. First, for a hospital-based case-control study, subjects were recruited based on their outcome (with CRC or without CRC) rather than their exposure. The potential CRC risks for the controls are immeasurable, and may have caused a cause bias in studying the etiology of the disease. It could also have led to spurious results in the gene/environmental interaction studies. Second, the sample size was relatively small. We cannot exclude the possibility there is an association between rs16901979, rs1447295 and rs7837688 and colorectal cancer in the Chinese population due to the inadequate statistical power of our study (0.290 for rs16901979, 0.702 for 1447295 and 0.316 for rs7837688, respectively), although many other studies have also reported no significant association for these loci with CRC in other populations [12], [14], [26], [28]. Accordingly, the null interaction between the confounders (alcohol use, smoking status, sex, age) and rs6983267 may also be due to the low power of small sample size. Third, there are at least five different cancer susceptibility regions on the 8q24 “desert” as reported by Ghoussaini et al [14]. Only the loci in the third region such as rs6983267 and rs10505477, were found to be significantly associated with CRC, and evidence from the in vitro experiment indicated that rs6983267 may be the causal locus, although we cannot exclude the possibility that other functional variants which are highly correlated with rs6983267 also lead to an increased risk of CRC. This study evaluated the representative loci from different regions on 8q24 and their association with CRC risk in an Asian (Han Chinese), which has been not well studied for these variants so far. It is valuable to conduct a fine-mapping study for the third region to identify other potential causal variants that are associated with CRC risk in a larger Chinese population.
In summary, we observed a statistically significant association between SNP rs6983267 on 8q24 and the risk of colorectal cancer in a Chinese population. Moreover, a meta-analysis of previous studies conducted in different populations confirmed the association between this locus and colorectal neoplasia risk in different populations and ethnic groups. These results provide a more complete picture of the role of this polymorphism in the risk of colorectal neoplasia, and may give genetic insight into possible strategies for prevention of colorectal neoplasia.
Acknowledgments
We thank Dr Zhibin Hu for helpful discussion, Xiaoguang Li for technical assistance and Dr. Elizabeth R. Rayburn (Magic City Medical Communications) for assistance in editing this manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by grants from One Hundred Talents Program of the Chinese Academy of Sciences, the National Nature Science Foundation (30870513, 30800933, 31070680 and 91029715), the Ministry of Science and Technology of China (2007CB947100), the Science and Technology Commission of Shanghai Municipality (08391910800, 10391902100), National Science and Technology Major Project “Key New Drug Creation and Manufacturing Program” (2009ZX09102-114, 2009ZX09301-011), the Grant-in Aid for Scientific Research on Special Priority Areas of Cancer from the Ministry of Education, Culture, Sports, Science and Technology of Japan (12670383), and grants from the Major International (Regional) Joint Research Projects (30320140461), the Science and Technology Commission of Xuhui District of Shanghai Municipality (RCT201001, CRC2010002), Director Foundation of INS (20090101), the Food Safety Research Center and Key Laboratory of Nutrition and Metabolism of INS, SIBS, CAS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Y, Pasche B. TGF-beta signaling alterations and susceptibility to colorectal cancer. Hum Mol Genet . 2007;16 (Spec No 1):R14–20. doi: 10.1093/hmg/ddl486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 6.Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 7.Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 8.Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40:26–28. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 9.Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 10.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poynter JN, Figueiredo JC, Conti DV, Kennedy K, Gallinger S, et al. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: results from the Colon Cancer Family Registry. Cancer Res. 2007;67:11128–11132. doi: 10.1158/0008-5472.CAN-07-3239. [DOI] [PubMed] [Google Scholar]
- 12.Berndt SI, Potter JD, Hazra A, Yeager M, Thomas G, et al. Pooled analysis of genetic variation at chromosome 8q24 and colorectal neoplasia risk. Hum Mol Genet. 2008;17:2665–2672. doi: 10.1093/hmg/ddn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuupanen S, Niittymaki I, Nousiainen K, Vanharanta S, Mecklin JP, et al. Allelic imbalance at rs6983267 suggests selection of the risk allele in somatic colorectal tumor evolution. Cancer Res. 2008;68:14–17. doi: 10.1158/0008-5472.CAN-07-5766. [DOI] [PubMed] [Google Scholar]
- 14.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sung JJ, Lau JY, Goh KL, Leung WK. Increasing incidence of colorectal cancer in Asia: implications for screening. Lancet Oncol. 2005;6:871–876. doi: 10.1016/S1470-2045(05)70422-8. [DOI] [PubMed] [Google Scholar]
- 16.Yang H, Zhou Y, Zhou Z, Liu J, Yuan X, et al. A novel polymorphism rs1329149 of CYP2E1 and a known polymorphism rs671 of ALDH2 of alcohol metabolizing enzymes are associated with colorectal cancer in a southwestern Chinese population. Cancer Epidemiol Biomarkers Prev. 2009;18:2522–2527. doi: 10.1158/1055-9965.EPI-09-0398. [DOI] [PubMed] [Google Scholar]
- 17.Gruber SB, Moreno V, Rozek LS, Rennerts HS, Lejbkowicz F, et al. Genetic variation in 8q24 associated with risk of colorectal cancer. Cancer Biol Ther. 2007;6:1143–1147. doi: 10.4161/cbt.6.7.4704. [DOI] [PubMed] [Google Scholar]
- 18.White KL, Sellers TA, Fridley BL, Vierkant RA, Phelan CM, et al. Variation at 8q24 and 9p24 and risk of epithelial ovarian cancer. Twin Res Hum Genet. 2010;13:43–56. doi: 10.1375/twin.13.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wokolorczyk D, Gliniewicz B, Sikorski A, Zlowocka E, Masojc B, et al. A range of cancers is associated with the rs6983267 marker on chromosome 8. Cancer Res. 2008;68:9982–9986. doi: 10.1158/0008-5472.CAN-08-1838. [DOI] [PubMed] [Google Scholar]
- 20.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Closas M, Lubin JH. Power and sample size calculations in case-control studies of gene-environment interactions: comments on different approaches. Am J Epidemiol. 1999;149:689–692. doi: 10.1093/oxfordjournals.aje.a009876. [DOI] [PubMed] [Google Scholar]
- 22.Lubin JH, Gail MH. On power and sample size for studying features of the relative odds of disease. Am J Epidemiol. 1990;131:552–566. doi: 10.1093/oxfordjournals.aje.a115530. [DOI] [PubMed] [Google Scholar]
- 23.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 25.Li L, Plummer SJ, Thompson CL, Merkulova A, Acheson LS, et al. A common 8q24 variant and the risk of colon cancer: a population-based case-control study. Cancer Epidemiol Biomarkers Prev. 2008;17:339–342. doi: 10.1158/1055-9965.EPI-07-0713. [DOI] [PubMed] [Google Scholar]
- 26.Curtin K, Lin WY, George R, Katory M, Shorto J, et al. Meta association of colorectal cancer confirms risk alleles at 8q24 and 18q21. Cancer Epidemiol Biomarkers Prev. 2009;18:616–621. doi: 10.1158/1055-9965.EPI-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuo K, Suzuki T, Ito H, Hosono S, Kawase T, et al. Association between an 8q24 locus and the risk of colorectal cancer in Japanese. BMC Cancer. 2009;9:379. doi: 10.1186/1471-2407-9-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schafmayer C, Buch S, Volzke H, von Schonfels W, Egberts JH, et al. Investigation of the colorectal cancer susceptibility region on chromosome 8q24.21 in a large German case-control sample. Int J Cancer. 2009;124:75–80. doi: 10.1002/ijc.23872. [DOI] [PubMed] [Google Scholar]
- 29.Middeldorp A, Jagmohan-Changur S, van Eijk R, Tops C, Devilee P, et al. Enrichment of low penetrance susceptibility loci in a Dutch familial colorectal cancer cohort. Cancer Epidemiol Biomarkers Prev. 2009;18:3062–3067. doi: 10.1158/1055-9965.EPI-09-0601. [DOI] [PubMed] [Google Scholar]
- 30.Kupfer SS, Anderson JR, Hooker S, Skol A, Kittles RA, et al. Genetic heterogeneity in colorectal cancer associations between African and European americans. Gastroenterology : . 2010;139:1677–1685, 1685 e1671-1678. doi: 10.1053/j.gastro.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Holst S, Picelli S, Edler D, Lenander C, Dalen J, et al. Association studies on 11 published colorectal cancer risk loci. Br J Cancer. 2010;103:575–580. doi: 10.1038/sj.bjc.6605774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong F, Wu C, Bi X, Yu D, Huang L, et al. Risk of genome-wide association study-identified genetic variants for colorectal cancer in a Chinese population. Cancer Epidemiol Biomarkers Prev. 2010;19:1855–1861. doi: 10.1158/1055-9965.EPI-10-0210. [DOI] [PubMed] [Google Scholar]
- 33.Hutter CM, Slattery ML, Duggan DJ, Muehling J, Curtin K, et al. Characterization of the association between 8q24 and colon cancer: gene-environment exploration and meta-analysis. BMC Cancer. 2010;10:670. doi: 10.1186/1471-2407-10-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui R, Okada Y, Jang SG, Ku JL, Park JG, et al. Gut [Epub ahead of print]; 2011. Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cicek MS, Slager SL, Achenbach SJ, French AJ, Blair HE, et al. Functional and clinical significance of variants localized to 8q24 in colon cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:2492–2500. doi: 10.1158/1055-9965.EPI-09-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He J, Wilkens LR, Stram DO, Kolonel LN, Henderson BE, et al. Generalizability and epidemiologic characterization of eleven colorectal cancer GWAS hits in multiple populations. Cancer Epidemiol Biomarkers Prev. 2011;20:70–81. doi: 10.1158/1055-9965.EPI-10-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pittman AM, Broderick P, Sullivan K, Fielding S, Webb E, et al. CASP8 variants D302H and -652 6N ins/del do not influence the risk of colorectal cancer in the United Kingdom population. Br J Cancer. 2008;98:1434–1436. doi: 10.1038/sj.bjc.6604314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kupfer SS, Torres JB, Hooker S, Anderson JR, Skol AD, et al. Novel single nucleotide polymorphism associations with colorectal cancer on chromosome 8q24 in African and European Americans. Carcinogenesis. 2009;30:1353–1357. doi: 10.1093/carcin/bgp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niittymaki I, Kaasinen E, Tuupanen S, Karhu A, Jarvinen H, et al. Low-penetrance susceptibility variants in familial colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:1478–1483. doi: 10.1158/1055-9965.EPI-09-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]