Abstract
The notion that some adult diseases may have their origins in utero has recently captured scientists’ attention. Some of these effects persist across generations and may involve epigenetic mechanisms. Epigenetic modifications, DNA methylation together with covalent modifications of histones, alter chromatin density and accessibility of DNA to cellular machinery, modulating the transcriptional potential of the underlying DNA sequence. Here, we will discuss the different epigenetic modifications and their potential role in and contribution to renal disease development.
Keywords: Epigenetics, Intrauterine environment, Histone modification, DNA methylation
Introduction
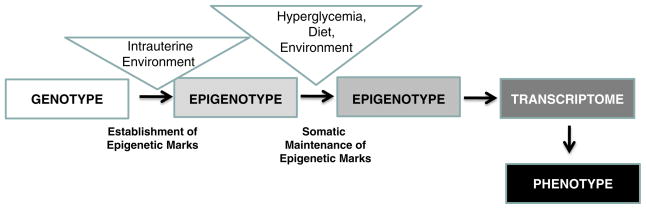
During normal development, somatic cells descending from a single progenitor and containing a similar genotype differentiate to acquire diverse biological functions by expressing and repressing different sets of genes. During development, the genome is partitioned into active and inactive segments by establishing epigenetic marks (Fig. 1). This process occurs via modification of genetic material while the nucleotide sequence remains untouched. Later, epigenetic marks are maintained through cell division in order to retain cell identity. A classic example of epigenetics is genomic imprinting, a phenomenon in which the father and mother contribute different epigenetic patterns for specific genomic loci in their germ cells that are responsible for the clinical phenotype in certain pediatric diseases [1]. For example, Angelman and Prader Willi syndromes are both caused by partial deletion of chromosome 15q. However, they have different phenotypes; Angelman syndrome is characterized by severe mental retardation, absence of speech, microcephaly, facial dysmorphism, seizures, neonatal hypotonia, ataxic movements, and inappropriate laughter while patients with Prader-Willi syndrome are characterized by hypotonia, short stature, hyperphagia, obesity, behavioral issues, small hands and feet, hypogonadism, and mild mental retardation. The particular phenotype that ensues depends on whether the mutation is inherited maternally or paternally [2, 3]. Paternal inheritance of a deletion of this region is associated with Prader-Willi syndrome, whereas maternal inheritance of the same deletion is associated with Angelman syndrome.
Fig. 1.
The phenotype (or disease) development depends on genetic and epigenetic factors. During normal development, somatic cells descending from a single progenitor and containing a similar genotype will differentiate to acquire diverse biological functions by expressing and repressing different sets of genes via establishing new epigenetic marks. While the genotype of an individual does not change, hyperglycemia, uremia, different dietary and environmental factors might change the epigenome of cells leading to differences in gene and protein expression. Differences in the epigenotype might be responsible for the development of a differing (including disease) phenotype. While the genotype is stable, there is a more dynamic link between environmental factors and phenotype development
Recent discoveries also indicate that epigenetic reprogramming can facilitate the dedifferentiation of cells back to pluripotent stem cells [4–6]. Environmental factors can also alter the epigenome and thereby the cellular phenotype. Although the role of epigenetics in cancer development is fairly well established, the evidence is now emerging that epigenetic changes play a role in renal disease and kidney development as well [7, 8].
The case for the role of epigenetics in kidney disease
While studying Swedish boys who were exposed to famine during preadolescence, Pembrey et al. observed that paternal (not maternal) grandsons were less likely to die of cardiovascular disease [9, 10]. The opposite effect was observed for females: only paternal (not maternal) granddaughters of women who experienced famine while in the womb lived shorter lives on average, suggesting trans-generational epigenetic inheritance.
Epidemiologic data shows an association between low birth weight and subsequent adult hypertension, diabetes, cardiovascular disease and chronic kidney disease (CKD) [11]. A direct correlation of low birth weight (LBW) with microalbuminuria has been reported in type 1 diabetics and in older nondiabetic persons [12]. LBW also directly correlates with albuminuria in type 2 diabetic Pima Indians, and with CKD progression [12]. Animal studies and indirect evidence from human studies support the hypothesis that LBW (a marker of adverse intrauterine circumstances) is associated with a congenital deficit in nephron number [12]. In humans, the final endowment of nephrons depends on the intrauterine environment and gestational age at birth as no new nephrons are formed after birth. Growth-retarded stillbirth and liveborn infants with intra-uterine growth retardation (IUGR) demonstrate a reduced nephron number compared to control infants with birth weights appropriate for gestational age [13]. In adults, autopsy studies also reveal a significant correlation between birth weight, nephron number, and mean arterial pressure [14]. Normal birth weight, however, does not always indicate normal nephron number. Oligomeganephronia, a type of renal hypoplasia that results from arrested development of the metanephric blastema at 14 to 20 weeks of gestation, is characterized by a reduced nephron number and marked compensatory glomerular and tubular hypertrophy [15]. Recently, Hodgin et al. reported six cases of focal segmental glomerulosclerosis (FSGS) where the clinical and pathological findings were most consistent with the secondary form of FSGS in whom a history of prematurity and very low birth weight were the only identifiable risk factors for secondary FSGS [16]. These studies highlight the connection between the intrauterine environment, nephron number, and renal disease development.
Important factors influencing fetal development are malnutrition and uteroplacental insufficiency. Animal models of fetal growth restriction can be produced by maternal caloric or protein restriction, change in sodium or iron intake, placental embolization or surgical reduction of blood flow, and exposure to corticosteroids. These interventions often result in a marked reduction of nephron number, kidney function, and increased susceptibility for hypertension. Molecular mechanisms that have been implicated include inhibition of the renin-angiotensin system in utero, reduced circulating insulin-like growth factor 1, and increased apoptosis. In animal models, the reduction in nephron number is associated with compensatory glomerular hypertrophy leading to the development of hypertension and glomerulosclerosis, which in turn initiate a vicious cycle leading to further nephron loss and ultimately CKD development [11]. While the low nephron number hypothesis has been developed by Barker and Brenner in the early 1960s, the underlying molecular mechanism of “intrauterine programming” and its implication on later life CKD is not yet known [12]. As epigenetic marks are established during development, we can speculate that epigenetic differences potentially play a role in this process (Fig. 1).
In addition to “fetal programming”, studies indicate that similar “programming” might occur during adulthood as well. Data from the Epidemiology of Diabetes Interventions and Complications (EDIC) study, which followed 1,441 patients with type 1 diabetes after they completed the Diabetes Control and Complication Trial (DCCT), shows that early chronic exposure to a moderately high level of hyperglycemia has prolonged effects on diabetic complications during subsequent periods of improved glycemia, a phenomenon termed “metabolic memory” [17, 18]. For example, atherosclerotic changes not present at the end of DCCT appeared subsequently in the previously higher HbA1c group, followed by a twofold increase in myocardial infarction, stroke, and cardiovascular death (Fig. 1). These changes occurred despite the fact that hemoglobinA1c (HbA1c) levels were identical in the two groups during the length of the study. Recent in vitro studies suggest that epigenetic differences and changes in histone modification occurring during periods of hyperglycemia might play a role in “hyperglycemic memory” [19, 20]. However, it should be noted that the “metabolic memory” concept might present a risk of focusing attention on interventions associated with glycemic control, while diverting attention away from other modifiable risk factors, i.e. high blood pressure and hyperlipidemia [21].
Cell type specification, kidney development, and epigenetics
The adult kidney is composed of more than 20 different cell types; different specialized epithelial, endothelial, and stromal cells [22]. Local signaling molecules (including bone morphogenetic proteins, Wnt proteins) and DNA-binding proteins, such as Pax2/8 and Osr1 (odd skipped related), play key roles in kidney development [23]. During cell type specification in the kidney, the genome is partitioned into transcriptionally active (eurochromatin) and inactive areas (heterochromatin). This process is achieved by establishing specific epigenetic marks. Much remains to be learned about the different epigenetic modifications in the kidney. Recent studies, however, indicate that Pax2 plays an important role in determining epigenetic marks during renal development. Pax2 interacts with the adaptor protein PTIP and participates in promoting histone H3K4 methylation at kidney-specific loci in response to inductive signals [24]. Trimethylation of H3K4 promotes nucleosome remodeling, which in turn attracts the RNA polymerase II complex, commencing gene activation. These results suggest that Pax2/8 functions to provide locus and tissue specificity for epigenetic cues to restrict the developmental potential of the intermediate mesoderm to the renal lineage. In addition to the Pax2/PTIP axis, there are likely many other players awaiting discovery.
DNA methylation and CKD
In eukaryotes, DNA methylation entails the covalent addition of methyl groups at the 5-position of cytosines [25, 26]. This methylation occurs specifically on cytosines followed by guanines (CpG dinucleotides). In the human genome, CpG dinucleotides are generally concentrated in regions called CpG islands, which are preferentially located in gene promoter areas. Some physiological processes require DNA methylation of CpG islands [27, 28], these include silencing of imprinted genes, where only one allele should be expressed in a progenitor-of-origin manner (i.e. only paternal or maternal expression) and X chromosome inactivation in women, a mechanism for dose compensation [29]. CpG dinucleotides not embedded in CpG islands are mainly located in repetitive or centromeric sequences while some are distributed in gene or intergenic regions. DNA methylation may affect the transcription of genes in two ways; first, the methylation of DNA may itself physically impede the binding of transcriptional proteins to the gene, and second, (likely more important) methylated DNA may be bound by proteins known as methyl-CpG-binding domain proteins (MBDs) [30]. MBD proteins recruit additional proteins to the locus, such as histone deacetylases and other chromatin remodeling proteins that can modify histones to form compact, inactive chromatin, termed silent chromatin. This link between DNA methylation and chromatin structure appears to be very important. In particular, loss of methyl-CpG-binding protein 2 (MeCP2) has been implicated in Rett syndrome; and methyl-CpG-binding domain protein 2 (MBD2) mediates the transcriptional silencing of hyper-methylated genes in cancer [31, 32].
Maintenance methylation activity is necessary to preserve DNA methylation after every cellular DNA replication cycle. Without DNA methyltransferase (DNMT), the replication machinery itself would produce unmethylated daughter strands leading to passive demethylation over time. DNMT1 is the proposed maintenance methyltransferase responsible for copying DNA methylation patterns to daughter strands during DNA replication. Mouse models with both copies of DNMT1 deleted are embryonic lethal at approximately day 9, due to the requirement of DNMT1 activity for development in mammalian cells. It is thought that DNMT3a and DNMT3b are the de novo methyltransferases that set up DNA methylation patterns early in development.
Since many tumor suppressor genes are silenced by DNA methylation during carcinogenesis, there have been attempts to re-express these genes by inhibiting DNMTs. 5-Aza-2′-deoxycytidine (decitabine) is a nucleoside analog that inhibits DNMTs by trapping them in a covalent complex on DNA by preventing the β-elimination step of catalysis, resulting in the enzymes’ degradation. However, for decitabine to be active, it must be incorporated into the genome of the cell, which can cause mutations in the daughter cells if the cell does not die. In addition, decitabine’s therapeutic potential is undermined by its toxicity to bone marrow.
A report from the Zeisberg group indicates that DNA methylation patterns of kidney fibroblasts differ between control and diseased kidneys [33]. The differential DNA methylation appears to control fibroblast proliferation and contribute to the development of fibrosis. Mice treated with 5′ azacytidine, where DNA methylation was inhibited, were protected from renal fibrosis, indicating that increased DNA methylation contributes to fibrosis development. This is the first report to support a direct role of DNA methylation in CKD development.
The methylation differences might even be established during development. Einstein et al. reported that IUGR subjects (compared to controls) possessed a distinctive number of consistent differences in methylation near genes involved in processes critical to stem cell function, including cell cycle and cellular maintenance [34]. A locus that emerged consistently in their pathway analysis was HNF4A, a gene already implicated in type 2 diabetes (T2DM), but not previously demonstrated to undergo epigenetic dysregulation in response to IUGR. HNF4A is best known as a monogenic cause for the autosomal dominant form of maturity onset diabetes of the young (MODY). They found differences in DNA methylation targeted to one of the HNF4A promoters, supporting a model of isoform variation of the gene being related to susceptibility to T2DM.
The histone code and kidney disease
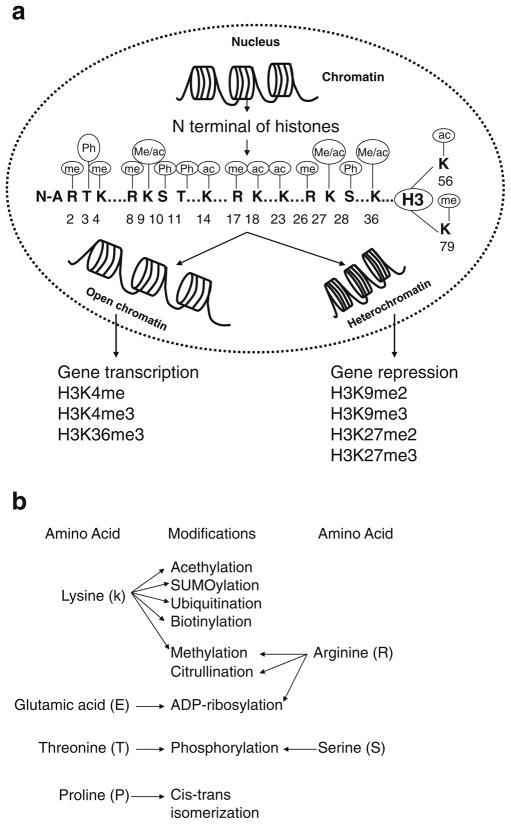
Histones are strongly basic proteins found in eukaryotic cell nuclei, which package and order DNA into structural units called nucleosomes. Histones are highly conserved and grouped into five major classes: H1/H5, H2A, H2B, H3, and H4. They fall into one of two super-classes: core histones (H2A, H2B, H3 and H4) or linker histones (H1 and H5). The nucleosome core particle, composed of two copies of each core histone, is wrapped by 147 base pairs of DNA in 1.65 left-handed super-helical turns. The N-terminal tails of the four core histone proteins H2A, H2B, H3 and H4 have biochemical sites for a range of post-translational modifications, including acetylation, methylation, O-GlcNac modification, phosphorylation, SUMOylation, ADP-ribosylation, and ubiquitination [35]. Figure 3 describes in detail the different histone tail modifications. Appropriate enzymes facilitate all of these histone modifications. Methylation and demethylation of histones are carried out by histone methyltransferases and demethylases such as the jumonji protein family. The key enzymes involved in acetylation at lysine residues include histone acetyltransferases (HATs), histone deacetylases (HDACs) [30]. The final outcome of histone modification depends not only on the identity of the modified residue but also on the site of modification, i.e. methylation of histone H3 on lysine 4 and 36 (H3K4 and H3K36) is generally associated with an “open” euchromatin structure and transcriptional activation, whereas methylation of histone H3 on lysine 9 and 27 (H3K9 and H3K27) is generally associated with a “closed” heterochromatin structure and gene silencing (Figs. 2 and 3). To make things more complex, histone lysine tails can be monomethylated, dimethylated, or trimethylated. Trimethylated H3K4 (H3k4me3) is preferentially associated with gene activation and is proposed to promote gene expression through recognition by transcription-activating effector molecules, whereas histone H3 lysine 27 trimethylation (H3K27me3) is associated with repression. It is postulated that bivalent marks identify genes that are silent but poised for transcription [23, 24] (Figs. 2 and 3).
Fig. 3.
Different histone tail modifications and their effects on gene transcription. a The different H3 histone tail modifications at the amino acid resolution level (aa 1-79) and their modifications, ac acetylation, me methylation, ph phosphorylation. Below is the most common histone modification and the effect on gene transcription i.e. repression and/or active transcription. b The different post-translational modifications of the different histone amino-acids
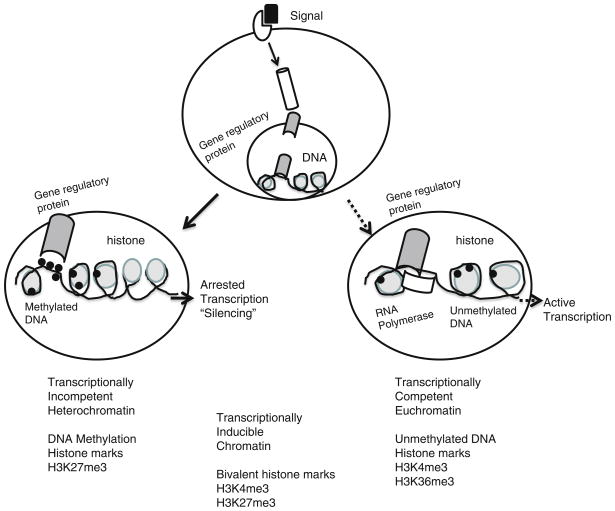
Fig. 2.
The effect of different external signals depends on epigenetic modification in the target cells. The cellular response to external signals may reflect chromatin-based (epigenetic) differences superimposed on the static genetic code. In this example, the 5-regulatory region of a disease susceptibility gene is depicted in cells. In some cells, the promoter assumes an “open” chromatin architecture characterized by activated histone posttranslational marks, decreased nucleosome density, and the lack of DNA methylation. RNA polymerase II is recruited to the promoter, resulting in productive transcription. Alternatively, in other cells the genetically identical promoter assumes a “closed” chromatin configuration characterized by repressive posttranslational marks, increased nucleosome density, and prominent DNA methylation. RNA polymerase II is not recruited to the promoter, and no transcription occurs. Transcriptionally competent euchromatin is usually associated with unmethylated DNA, and trimethyl H3K4me3 and H3K36me3 marks. Transcription-ally incompetent heterochromatin is generally associated with methylated DNA and trimethyl H3K27 marks
In addition, it appears that the different histone modifications are not completely independent from each other. DNA methylation and histone modifications might function in close interplay with nucleosome remodeling and positioning complexes that bind trimethylated H3K4 and methyl CpG binding proteins and move nucleosomes on DNA by ATP-dependent mechanisms [36, 37].
The different histone modifications in the human kidney have not yet been studied in the context of CKD. However, studies performed in various mouse models of kidney disease suggest potential key differences in the “histone code”. Increased levels of renal H3K9 and H3K23 acetylation, H3K4 dimethylation and H3 phosphorylation at serine 10 were detected in mouse models of advanced diabetic nephropathy [38, 39]. The authors speculate that these changes could enhance chromatin unfolding and contribute to active gene transcription. Reports from the Natarajan group also support the role of epigenetic modification in diabetic complications [40, 41]. They found that the chromatin histone H3-lysine 4 methyltransferase, SET7/9, is a novel coactivator of NF-kappaB and that SET7/9 plays a key role in inflammation and diabetes. In addition, there are experimental examples demonstrating the influence of posttranslational histone modifications that influence renin cell identity in the kidney. Using a cultured renin linage cell system, Pentz et al. showed that renin gene expression memory is maintained in cultured cells and can be reenacted by cAMP and chromatin remodeling (histone H4 acetylation) [42]. Gomez et al. demonstrated that CREB-binding protein (CBP) and p300 (which possess histone acetyltransferase activity) are essential for renin cell identity and morphological integrity of the kidney [43].
Summary
Studies characterizing the role of epigenetic changes in human complex diseases are ongoing. The National Institute of Health is carrying out a major program initiative aimed at developing new epigenomic assays, generating reference epigenomic maps, and describing disease-specific epigenomic changes (epigenomics@nih.gov). Unlike the genetic sequence, the epigenome is under the influence of various environmental factors and might mediate the relationship between genotype and internal and external environmental factors [44, 45] (Fig. 1). Emerging evidence implicates epigenetics in CKD development, calling for new genome scale epigenomic and systems biology studies.
Acknowledgments
We thank members of the Einstein Center for Epigenomics and the Susztak lab for the discussion. Our studies are supported by NIH 1R01DK087635 to KS.
Question
Answers appear following the reference list.
-
Epigenetics (nongenetic causes of a phenotype) may involve processes of:
Genetic imprinting
Histone methylation
Cytosine methylation
RNA interference
All of the above
-
Euchromatin denotes the region in where DNA undergoes
Transcriptional activation
Demethylation
Methylation
Gene silencing
a and b
-
Active histone marks (H3K4me3 and H3K36me3) refer to:
Euchromatin
Trimethylation
Heterochromatin
a and b
All of the above
-
The epigenetic modifications play a role in following diseases and physiologic processes:
Cancer
Organ development
Fetal programming
Diabetes
All of the above
-
Differences in clinical phenotype between Prader-Willi and Angelman syndromes result from:
Histone acetylation
DNA demethylation
Expression of micro RNAs
Genetic imprinting
All of the above
Answers
c
e
d
e
d
Contributor Information
Robert Woroniecki, Department of Pediatrics, Nephrology, Albert Einstein College of Medicine, 1300 Morris Park Ave., Ullmann Bldg 619, Bronx, NY 10461, USA.
Anil Bhanudas Gaikwad, Department of Medicine, Division of Nephrology, Albert Einstein College of Medicine, 1300 Morris Park Ave., Ullmann Bldg 619, Bronx, NY 10461, USA.
Katalin Susztak, Email: Katalin.susztak@einstein.yu.edu, Department of Medicine, Division of Nephrology, Albert Einstein College of Medicine, 1300 Morris Park Ave., Ullmann Bldg 619, Bronx, NY 10461, USA, Department of Genetics, Albert Einstein College of Medicine, 1300 Morris Park Ave., Ullmann Bldg 619, Bronx, NY 10461, USA.
References
- 1.Wood AJ, Oakey RJ. Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet. 2006;2:e147. doi: 10.1371/journal.pgen.0020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knoll JH, Nicholls RD, Magenis RE, Graham JM, Jr, Lalande M, Latt SA. Angelman and Prader-Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am J Med Genet. 1989;32:285–290. doi: 10.1002/ajmg.1320320235. [DOI] [PubMed] [Google Scholar]
- 3.Knoll JH, Nicholls RD, Magenis RE, Glatt K, Graham JM, Jr, Kaplan L, Lalande M. Angelman syndrome: three molecular classes identified with chromosome 15q11q13-specific DNA markers. Am J Hum Genet. 1990;47:149–154. [PMC free article] [PubMed] [Google Scholar]
- 4.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 6.Yamanaka S. A fresh look at iPS cells. Cell. 2009;137:13–17. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 7.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 8.Dressler GR. Epigenetics, development, and the kidney. J Am Soc Nephrol. 2008;19:2060–2067. doi: 10.1681/ASN.2008010119. [DOI] [PubMed] [Google Scholar]
- 9.Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, Golding J. Sex-specific, male-line trans-generational responses in humans. Eur J Hum Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 10.Kaati G, Bygren LO, Pembrey M, Sjostrom M. Trans-generational response to nutrition, early life circumstances and longevity. Eur J Hum Genet. 2007;15:784–790. doi: 10.1038/sj.ejhg.5201832. [DOI] [PubMed] [Google Scholar]
- 11.Zandi-Nejad K, Luyckx VA, Brenner BM. Adult hypertension and kidney disease: the role of fetal programming. Hypertension. 2006;47:502–508. doi: 10.1161/01.HYP.0000198544.09909.1a. [DOI] [PubMed] [Google Scholar]
- 12.Luyckx VA, Brenner BM. Low birth weight, nephron number, and kidney disease. Kidney Int Suppl. 2005:S68–S77. doi: 10.1111/j.1523-1755.2005.09712.x. [DOI] [PubMed] [Google Scholar]
- 13.Alexander BT. Intrauterine growth restriction and reduced glomerular number: role of apoptosis. Am J Physiol Regul Integr Comp Physiol. 2003;285:R933–R934. doi: 10.1152/ajpregu.00446.2003. [DOI] [PubMed] [Google Scholar]
- 14.Hughson MD, Douglas-Denton R, Bertram JF, Hoy WE. Hypertension, glomerular number, and birth weight in African Americans and white subjects in the southeastern United States. Kidney Int. 2006;69:671–678. doi: 10.1038/sj.ki.5000041. [DOI] [PubMed] [Google Scholar]
- 15.McGraw M, Poucell S, Sweet J, Baumal R. The significance of focal segmental glomerulosclerosis in oligomeganephronia. Int J Pediatr Nephrol. 1984;5:67–72. [PubMed] [Google Scholar]
- 16.Hodgin JB, Rasoulpour M, Markowitz GS, D’Agati VD. Very low birth weight is a risk factor for secondary focal segmental glomerulosclerosis. Clin J Am Soc Nephrol. 2009;4:71–76. doi: 10.2215/CJN.01700408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group. Retinopathy and nephropathy in patients with type 1 diabetes four years after a trial of intensive therapy. The Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group. N Engl J Med. 2000;342:381–389. doi: 10.1056/NEJM200002103420603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopes-Virella MF, Carter RE, Gilbert GE, Klein RL, Jaffa M, Jenkins AJ, Lyons TJ, Garvey WT, Virella G. Risk factors related to inflammation and endothelial dysfunction in the DCCT/ EDIC cohort and their relationship with nephropathy and macro-vascular complications. Diab Care. 2008;31:2006–2012. doi: 10.2337/dc08-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, Cooper ME, Brownlee M. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205:2409–2417. doi: 10.1084/jem.20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brasacchio D, Okabe J, Tikellis C, Balcerczyk A, George P, Baker EK, Calkin AC, Brownlee M, Cooper ME, El-Osta A. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that co-exist on the lysine tail. Diabetes. 2009;58:1229–1236. doi: 10.2337/db08-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White NH, Sun W, Cleary PA, Danis RP, Davis MD, Hainsworth DP, Hubbard LD, Lachin JM, Nathan DM. Prolonged effect of intensive therapy on the risk of retinopathy complications in patients with type 1 diabetes mellitus: 10 years after the Diabetes Control and Complications Trial. Arch Ophthal. 2008;126:1707–1715. doi: 10.1001/archopht.126.12.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dressler GR. Advances in early kidney specification, development and patterning. Development. 2009;136:3863–3874. doi: 10.1242/dev.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 24.Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat Rev. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- 27.Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21:359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Agrelo R, Wutz A. X inactivation and disease. Semin Cell Dev Biol. 2009;21:194–200. doi: 10.1016/j.semcdb.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 29.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 30.Razin A. CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol Cell. 2008;29:525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 32.Lasalle JM, Yasui DH. Evolving role of MeCP2 in Rett syndrome and autism. Epigenomics. 2009;1:119–130. doi: 10.2217/epi.09.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bechtel W, McGoohan S, Zeisberg EM, Muller GA, Kalbacher H, Salant DJ, Muller CA, Kalluri R, Zeisberg M. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med. 2010;16:544–550. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Einstein F, Thompson RF, Bhagat TD, Fazzari MJ, Verma A, Barzilai N, Greally JM. Cytosine methylation dysregulation in neonates following intrauterine growth restriction. PLoS One. 2010;5:e8887. doi: 10.1371/journal.pone.0008887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marmorstein R, Trievel RC. Histone modifying enzymes: structures, mechanisms, and specificities. Biochim Biophys Acta. 2009;1789:58–68. doi: 10.1016/j.bbagrm.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 37.Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. Int J Biochem Cell Biol. 2009;41:87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Sayyed SG, Gaikwad AB, Lichtnekert J, Kulkarni O, Eulberg D, Klussmann S, Tikoo K, Anders HJ. Progressive glomerulo-sclerosis in type 2 diabetes is associated with renal histone H3K9 and H3K23 acetylation, H3K4 dimethylation and phosphorylation at serine 10. Nephrol Dial Transplant. 2010;25:1811–1817. doi: 10.1093/ndt/gfp730. [DOI] [PubMed] [Google Scholar]
- 39.Gaikwad AB, Sayyed SG, Lichtnekert J, Tikoo K, Anders HJ. Renal failure increases cardiac histone h3 acetylation, dimethylation, and phosphorylation and the induction of cardiomyopathy-related genes in type 2 diabetes. Am J Pathol. 2010;176:1079–1083. doi: 10.2353/ajpath.2010.090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Reddy MA, Miao F, Shanmugam N, Yee JK, Hawkins D, Ren B, Natarajan R. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem. 2008;283:26771–26781. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci USA. 2008;105:9047–9052. doi: 10.1073/pnas.0803623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pentz ES, Lopez ML, Kim HS, Carretero O, Smithies O, Gomez RA. Ren1d and Ren2 cooperate to preserve homeostasis: evidence from mice expressing GFP in place of Ren1d. Physiol Genomics. 2001;6:45–55. doi: 10.1152/physiolgenomics.2001.6.1.45. [DOI] [PubMed] [Google Scholar]
- 43.Gomez RA, Pentz ES, Jin X, Cordaillat M, Sequeira Lopez ML. CBP and p300 are essential for renin cell identity and morphological integrity of the kidney. Am J Physiol Heart Circ Physiol. 2009;296:H1255–H1262. doi: 10.1152/ajpheart.01266.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 45.Wilson AG. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J Periodontol. 2008;79:1514–1519. doi: 10.1902/jop.2008.080172. [DOI] [PubMed] [Google Scholar]