Abstract
Background
Early biomarkers of skeletal muscle anabolism will facilitate the development of therapies for sarcopenia and frailty.
Methods and results
We examined plasma type III collagen N-terminal propeptide (P3NP), skeletal muscle protein fractional synthesis rate, and gene and protein expression profiles to identify testosterone-induced changes in muscle anabolism. Two placebo-controlled studies enrolled community-dwelling men (study 1, 60–75 years; study 2, 18–40 years) with low to normal testosterone levels. Men were randomized to lower dose (study 1, 100 mg; study 2, 200 mg) or higher dose (study 1, 300 mg; study 2, 600 mg) single intramuscular testosterone or saline injection. After 1 week, testosterone acutely increased plasma P3NP levels in a dose-dependent manner and altered the expression of several skeletal muscle transcripts and proteins. Though not statistically significant, mixed muscle protein fractional synthesis rate tended to increase (1.08-fold with 100 mg testosterone, 1.12-fold with 300 mg testosterone). Testosterone exposure also increased skeletal muscle expression of the collagen type III gene that encodes P3NP.
Conclusion
P3NP is a potentially useful early biomarker for muscle anabolic therapy. Skeletal muscle protein and RNA profiling are useful tools for the discovery of novel muscle anabolic biomarkers.
Keywords: RNA microarray profiling, Proteomics, Testosterone, Mass spectrometry, Sarcopenia, Frailty
Introduction
Muscle size and strength deteriorate dramatically with increasing age. Sarcopenia can be defined operationally as muscle mass two standard deviations below that of young healthy controls. The prevalence of sarcopenia in individuals older than 80 years of age exceeds 50%. Exercise training can improve sarcopenia, but there are currently no marketed treatments available for sarcopenia [1–3]. Loss of muscle mass and function threatens the independence and quality of life of the elderly and may compromise their ability to survive critical illness [4]. Further understanding of the pathophysiology of sarcopenia and the development of more sensitive and specific genotyping and phenotyping tools will enable us to develop targeted therapeutic agents to treat this disorder.
A challenge in developing drug therapies for sarcopenia is the long treatment duration required for assessing clinical efficacy. Currently, we assess clinical efficacy by quantifying changes in lean body mass using dual-energy X-ray absorptiometry and changes in muscle function by using bilateral leg maximum voluntary force production, but measurable improvement in these endpoints requires at least 12 weeks of follow-up. We hypothesized that short-term testosterone exposure would induce several candidate early predictive biomarkers of muscle anabolism that could be identified using skeletal muscle gene and protein expression profiling platforms. Testosterone enanthate was used because it is a potent anabolic agent known to improve muscle size and strength [5, 6]. Sarcopenia can affect women, but to minimize heterogeneity, only men were enrolled given there are clear data regarding the effects of testosterone in men [5–7].
Several existing candidate biomarkers for muscle anabolism were examined. Collagen type III is the characteristic collagen of parenchymal organs and soft connective tissues such as muscle and skin. Collagen III is synthesized from larger procollagen type III with cleavage of peptide fragments from the N- and C-terminal ends. Procollagen type III N-terminal peptide (P3NP) is released into the circulation during collagen III synthesis and increases when skeletal muscle remodeling is activated [8]. Skeletal muscle protein fractional synthesis rate (FSR) increases with muscle protein anabolism and can be assessed by measuring the rate of incorporation of stable isotope-labeled phenylalanine into skeletal muscle proteins. Muscle RNA microarray and protein expression profiling are newer high-throughput approaches that can be used to examine transcript and protein expression changes and can be useful for discovering new candidate biomarkers for myoanabolism.
Materials and methods
The study protocols were approved by the institutional review boards at the participating study centers. All participants provided verbal and written informed consent before undergoing any study procedures. Patients were recruited at Celerion (formerly MDS Pharma Services), 22-24 Lisburn Road, Belfast, Northern Ireland, BY9 6AD. The same ethics committee approved both studies, the Plymouth Independent Ethics Committee, ITTC Building 2, Room 107, Tamar Science Park, Derriford, Plymouth, Devon, PL6 8BX.
Study design
Both trials were randomized, placebo-controlled, single-blind single-dose studies. Participants were blinded to treatment; the investigators and sponsor were not blinded for safety reasons. The primary objective of study 1 was to evaluate the utility of the in vivo quantitation of skeletal muscle protein FSR as an early biomarker for testosterone-induced muscle protein anabolism. Twenty-two healthy 60- to 75-year-old men were randomized to one of three groups: low-dose (100 mg, n = 6) or high-dose (300 mg, n = 8) intramuscular (IM) testosterone enanthate or IM saline injection (n = 8). Intramuscular injection was performed on the gluteus maximus muscle. FSR was measured before (baseline) and 7 days (1 week) after testosterone/saline injection. The primary objective of study 2 was to evaluate testosterone-induced alterations in muscle protein expression profiles using mass spectrometry-based high-throughput comparative proteomics profiling. Twenty-eight healthy 18- to 40-year-old men were randomized to one of three groups: low-dose (200 mg, n = 9), high-dose (600 mg, n = 10) IM testosterone enanthate or IM saline injection (n = 9). Vastus lateralis muscle and blood samples were obtained after an overnight fast at baseline (before the testosterone/saline injection, day 0) and 7 days after the single testosterone/saline injection (day 7). Muscle and blood samples were processed as above.
Serum P3NP concentration and skeletal muscle mRNA microarray profiling were examined in both studies.
Participants
Healthy community-dwelling men between the ages of 60 and 75 years (study 1) with low–normal testosterone levels (9.7–17.3 nmol/L) or 18 and 40 years (study 2) with low–normal testosterone levels (9.7–24.6 nmol/L) and prostate-specific antigen ≤ 4.0 ng/mL were enrolled. Because older men may experience more side effects from higher doses of testosterone [6], the 600-mg testosterone dose was only administered to younger men (18–40 years old) in study 2. Participants were nonsmokers, with body mass index of 21–30 kg/m [2] and stable weight for 3 months by self-report. Participants agreed to follow the meat-free controlled protein diet and avoid strenuous physical activity during the study. Exclusion criteria included treatment with androgenic steroids, growth hormone (GH), GH secretagogues, or other anabolic agents in the previous 12 months; oral, inhaled, or nasal glucocorticoids; inflammatory modulators such as ibuprofen, etoricoxib, or celecoxib; and blood anticoagulants or high doses of aspirin.
Testosterone concentration
Serum total testosterone was quantified using immunoassay (Roche Diagnostics, Indianapolis, IN, USA). Free testosterone was quantified using the Coat-A-Count Free Testosterone radioimmunoassay kit (Siemens Medical Solutions Diagnostics, Deerfield, IL, USA).
Procollagen III N-terminal peptide
Serum P3NP concentration was quantified using a commercial equilibrium radioimmunoassay (reference range 2.3–6.4 ng/mL, sensitivity 0.8 ng/mL; UniQ PIIINP RIA kit, Orion Diagnostica, Finland) [9].
Assessment of muscle protein fractional synthetic rate
A primed (3 μmol/kg) 12-h constant intravenous infusion (3 μmol kg−1 h−1) of 13C6-phenylalanine (13C6-phe; dissolved in sterile saline; ~95% 13C6, Cambridge Isotope Laboratories Inc., Andover, MA, USA) was used to quantify the in vivo rate of phenylalanine incorporation into mixed skeletal muscle proteins [10, 11]. Using a Bergström needle biopsy procedure, vastus lateralis muscle samples (~100 mg) were obtained 2.5–3 and 11.5–12 h (from the contralateral thigh) after the 13C6-phe infusion started. Muscle samples were immediately cleaned of adipose and connective tissues, partitioned, and frozen (−80°C) for later analysis. Blood samples were obtained before starting, at 2–3 h, and at 11–12 h during the 13C6-phe infusion. Serum or plasma was separated from cells, partitioned, and frozen for later analysis. After completing the baseline 13C6-phe infusion, participants received a single testosterone enanthate or saline injection (IM 100 or 300 mg testosterone or IM placebo). The procedure for quantifying in vivo FSR was repeated 7 days after the single testosterone/saline injection.
Plasma and muscle tissue free pool amino acids were converted to their N-heptafluorobutyryl n-propyl ester chemical derivatives [12] and 13C6-phe (m/z 383 and 389) enrichment (tracer/tracee ratio, TTR) [13] was quantified using capillary gas chromatography–negative chemical ionization–mass spectrometry (GC-NCI-MS; Agilent 6890N gas chromatograph and Agilent 5973N mass selective detector, Palo Alto, CA, USA) [14, 15]. The GC-NCI-MS instrument response was calibrated using gravimetric standards of known 13C6-phe enrichment. Mixed muscle protein 13C6-phe enrichment (TTR) was quantified using gas chromatography–combustion–isotope ratio mass spectrometry (Finnigan Delta+XL-IRMS, Bremen, Germany) [16, 17].
Comparative proteomics
Muscle proteins were extracted from frozen muscle samples (20–32 mg) using lysis buffer with protease inhibitor cocktail (EMD Bioscience, San Diego, CA, USA). Fourier transform mass spectrometry data were analyzed using Rosetta Biosoftware Elucidator® System 3.1 (Seattle, WA, USA) [18]. Further details on proteomics methods are available in a prior publication [19].
RNA microarray expression analysis
Because study 1 was primarily designed to assess FSR, muscle samples from two time points (after 3 and 12 h of 13C6-phe infusion) were available from the baseline and day 7 infusion studies. These samples provided enough material for two gene expression analyses that were used to compare transcript levels between baseline and 7 days after testosterone/saline injection. During study 2, one muscle sample was obtained at baseline and at day 7 and was used to compare the gene expression profiles between baseline and 7 days after testosterone/saline injection. Tissue was stored in RNAlater (Ambion, Inc., Austin, TX, USA) and RNA purified using TRIzol. Sample purification, amplification, labeling, and microarray processing were performed by Rosetta Inpharmatics. Cell files were loaded into Rosetta Biosoftware Resolver® System 7.2. Expression summaries were calculated using Robust Multichip Analysis via Affymetrix Power Tool with only perfect match (PM) probes included. MAS 5.0 p values were calculated with both PM and MM (mismatch) probes included.
Statistical methods
The assessments of P3NP, total/free testosterone, and FSR were performed after log transformation using an unequal variance ANCOVA model with a treatment term and baseline as covariate. No deviations from normality were observed. Heterogeneity of variance reduced after log transformation. The primary comparisons were based on the mean (fold) changes from baseline between two testosterone doses and placebo. A difference (expected to be >0) was considered significant if its one-sided p value was <0.05. To adjust for multiple comparisons within an endpoint, a closed stepwise testing procedure was used, starting with (a) high-dose testosterone vs. placebo and, if significant, then (b) low-dose testosterone vs. placebo. Spearman rank correlation analysis of the endpoints with serum total and free testosterone was performed. To further assess whether serum testosterone levels predicted P3NP, regression models including the terms study, dose, and with or without testosterone were fitted to the combined data from both studies. “Dose” is in the logarithmic scale of the active doses and set to 0 for placebo. The proportion of treatment effect on change in P3NP explained by serum testosterone was derived from the regression models.
Proteomic data were analyzed using a linear mixed model. Briefly, for each of the 65,152 features with liquid chromatography retention times between 10 and 45 min, the 0 intensities were replaced with half the smallest positive intensity measured for that feature. A linear mixed model was fitted for each feature using a log intensity scale, with the six treatment period combinations as fixed effects and subjects as random effects.
For gene and protein expression, two-sided p values and 95% CIs were derived. The false discovery rate (FDR) defined as the proportion of significant changes that are false positives was used to assess significance. Candidate biomarkers significant on their own (univariate analysis) between the placebo and testosterone groups were identified with 10% FDR. Significant group changes in gene expression from baseline were identified using standard ANOVA on each study set at two-sided p < 0.05. Gene expression changes from baseline that followed a significant trend were identified using the Jonckheere–Terpstra ordered trend test [20, 21]. The intersection of the ANOVA gene set and the Jonckheere–Terpstra trending gene set were used to select gene probe sets for assessing association with testosterone treatment in muscle.
For P3NP, total/free testosterone and FSR data were analyzed using SAS 9.1 (SAS Institute Inc., Cary, NC, USA). The RNA statistical analysis software used was MatLab (The Mathworks, Natick, MA, USA). The proteomic statistical analysis software used was R-2.7.0. (R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria; 2010).
Results
Participant demographics and baseline characteristics
In study 1, 22 participants (eight in the placebo arm, six in the 100 mg, and eight in the 300 mg testosterone arms) were randomly assigned and completed treatment. One placebo participant had a muscle biopsy that was inadequate for FSR measurement. In study 2, 28 participants (nine in placebo, nine in 300 mg, and ten in 600 mg testosterone groups) were randomly assigned; 27 completed the study and one participant withdrew from the study following a reaction (10-cm area of induration) at the testosterone injection site. Baseline testosterone levels were similar between the study groups (Table 1). For study 1, baseline mean (±SD) total testosterone was 14.6 ± 2.7 nmol/L (range 10.5–19.5 nmol/L). For study 2, baseline mean total testosterone was 18.6 ± 4.5 nmol/L (range 8.7–29.7 nmol/L). Baseline P3NP levels were similar between groups. In study 1, baseline mean P3NP level was 3.2 ± 0.7 ng/L (range 1.8–4.6 ng/L) and in study 2 was 3.7 ± 0.7 ng/L (range 2.4–5.6 ng/L).
Table 1.
Baseline characteristics for treatment groups
| Placebo | Low-dose testosteronea | High-dose testosteroneb | |
|---|---|---|---|
| Study 1 | |||
| No. of subjects | 8 | 6 | 8 |
| Age (years) | 67.5 (4.1) | 62.3 (2.3) | 64.6 (3.9) |
| Weight (kg) | 79.8 (8.5) | 76.3 (7.0) | 85.8 (7.7) |
| Body mass index (kg/m2) | 26.7 (2.2) | 27.0 (1.9) | 27.1 (1.0) |
| Free testosterone (pg/mL) | 8.79 (2.01) | 8.92 (1.53) | 10.16 (1.74) |
| Total testosterone (nmol/L) | 13.64 (2.87) | 14.53 (2.52) | 15.51 (2.73) |
| Plasma P3NP (ng/L) | 3.33 (0.95) | 3.08 (0.55) | 3.13 (0.64) |
| Study 2 | |||
| No. of subjects | 9 | 9 | 10 |
| Age (years) | 30.0 (6.1) | 26.2 (6.3) | 30.1 (5.1) |
| Weight (kg) | 81.7 (9.2) | 83.7 (11.2) | 81.6 (11.9) |
| Body mass index (kg/m2) | 25.0 (2.2) | 26.7 (2.6) | 24.9 (2.3) |
| Free testosterone (pg/mL) | 17.14 (2.84) | 16.09 (3.78) | 17.80 (3.74) |
| Total testosterone (nmol/L) | 19.79 (3.00) | 16.26 (4.49) | 19.50 (5.10) |
| Plasma P3NP (ng/L) | 3.51 (0.76) | 3.69 (0.80) | 3.82 (0.66) |
Mean (standard deviation)
aFor study 1, low = testosterone 100 mg. For study 2, low = testosterone 200 mg.
bFor study 1, high = testosterone 300 mg. For study 2, high = testosterone 600 mg.
Changes in testosterone levels
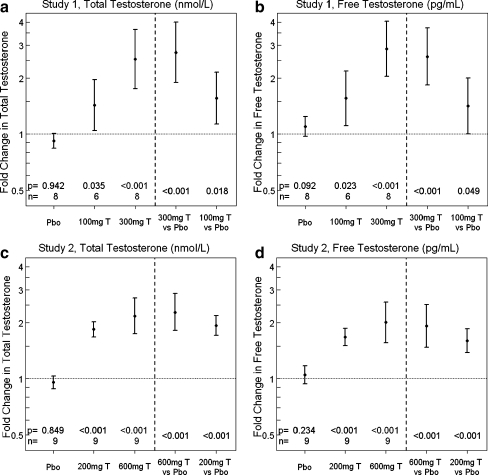
In both studies, total (Fig. 1a, c) and free testosterone levels (Fig. 1b, d) increased 7 days after IM testosterone injection in a dose-dependent manner in comparison to saline injection. As expected, for each dose administered, there was considerable inter-participant variability in the total and free testosterone levels. In study 1, total testosterone levels increased 1.43-fold from baseline in the 100 mg IM, 2.53-fold in the 300 mg IM, and 0.9-fold in the saline IM group (Fig. 1a). In study 2, total testosterone levels increased 1.84-fold in the 200 mg IM, 2.18-fold in the 600 mg IM, and 0.95-fold in the saline IM group (Fig. 1c).
Fig. 1.
Testosterone administration significantly increased both serum total and free testosterone levels in both young and older men after 1-week treatment. Serum total (a) and free testosterone (b) increased significantly with 300 and 100 mg testosterone IM compared with placebo in study 1 involving healthy 60- to 75-year-old men. Serum total (c) and free testosterone (d) increased significantly with 600 and 200 mg testosterone IM compared with placebo in study 2 involving healthy 18- to 40-year-old men. Y-axis is in log scale. Values of p, sample size (n), mean and 90% confidence intervals are shown
Seven of nine 600-mg samples (day 7) had testosterone levels above the upper limit of quantitation for the assay. Two samples from the 600-mg group (day 7) had testosterone levels near the pretreatment baseline values. One possible explanation for the absence of an increase in serum testosterone in these two participants would be a dosing error, but this was not reported nor verified to have occurred. Another possible explanation is that testosterone clearance rate was extraordinarily rapid in these two participants. As expected, there were strong positive correlations between changes in free and total testosterone within and between studies, with Spearman’s correlation ranging from 0.82 to 0.92, p < 0.001.
Procollagen III/N-terminal peptide
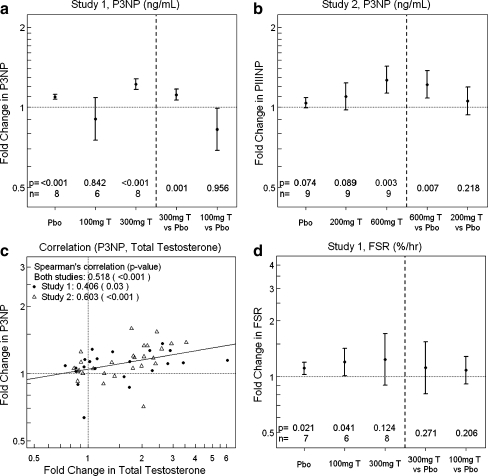
The baseline P3NP plasma concentrations were similar in the treatment groups within each study (Table 1). In study 1, higher dose (300 mg) testosterone significantly increased P3NP (1.12-fold, p = 0.001), relative to placebo, measured in healthy older men (Fig. 2a). In study 2, higher dose (600 mg) testosterone significantly increased P3NP (1.22-fold, p = 0.007), relative to placebo, measured in younger healthy men (Fig. 2b). There were moderate correlations between increases in P3NP and total testosterone within and between studies, with Spearman’s correlation ranging from 0.41 (p = 0.030) to 0.60 (p < 0.001; Fig. 2c). Significant effects of change in total testosterone (p = 0.012) and change in free testosterone (p = 0.013) on change in P3NP were observed even after adjusting for “study” and “dose” in the regression models. Change in testosterone explained a large proportion of treatment effect (PTE) on change in P3NP; the PTE estimates were over 100% for both total and free testosterone due to variability in the estimates.
Fig. 2.
Plasma P3NP changes from baseline after 1-week testosterone treatment. a Plasma P3NP concentration is significantly increased from baseline with 300 mg testosterone (T) relative to placebo (study 1). b Plasma P3NP concentration is significantly increased from baseline with 600 mg T relative to placebo (study 2). c Scatter plot of change in P3NP from baseline vs. change in total testosterone from baseline (study 1, filled circle; study 2, empty triangle). A line fitted to the points is shown (r = 0.518, p < 0.001). d Change in skeletal muscle FSR after 1-week treatment with 300 mg testosterone, 100 mg testosterone, and placebo. There were no statistically significant changes in FSR relative to placebo. Y-axis is in log scale. X-axis in log scale (c). Values of p, sample size (n), mean, and 90% confidence intervals are shown
Muscle protein FSR
The fractional synthesis rate for mixed muscle proteins was calculated using the established equation: FSR = [(increment in 13C6-phe enrichment in mixed muscle proteins from 3 to 12 h × 100)/(13C6-phe enrichment in the precursor pool × amount of time (h) between the two muscle samples)] [16, 22–25]. This approach employs a reliable estimate for 13C6-phe enrichment in the true precursor pool for muscle protein synthesis (13C6-aminoacyl-tRNA) [26, 27]. The average muscle tissue free pool 13C6-phe enrichment was used to calculate FSR. The muscle tissue free pool 13C6-phe enrichment was 61 ± 9% of the plasma 13C6-phe enrichment, so if plasma 13C6-phe enrichment were used to reflect the precursor pool enrichment, the calculated mixed muscle protein FSR would be 39% lower (Table 2).
Table 2.
13C6-phe enrichment values in various precursor pools and mixed muscle protein used to calculate muscle protein synthesis rate (FSR)
| Parameter | Placebo (n = 8) | 100 mg Testosterone (n = 6) | 300 mg Testosterone (n = 8) | |||
|---|---|---|---|---|---|---|
| Baseline | Day 7 | Baseline | Day 7 | Baseline | Day 7 | |
| Plasma 13C6-phea | 7.1 ± 0.9 | 7.2 ± 0.7 | 7.0 ± 0.8 | 7.3 ± 0.8 | 7.0 ± 0.6 | 7.2 ± 0.7 |
| Muscle Free pool 13C6-pheb | 4.3 ± 0.5 | 4.5 ± 0.8 | 4.0 ± 0.4 | 4.4 ± 0.6 | 4.4 ± 0.2 | 4.5 ± 0.7 |
| Mixed muscle protein 13C6-phec | 0.0195 ± 0.0050 | 0.0198 ± 0.0017 | 0.0184 ± 0.0035 | 0.0250 ± 0.0091 | 0.0173 ± 0.0023 | 0.0233 ± 0.0081 |
| Mixed muscle protein synthesis rate (% per hour) | 0.050 ± 0.011 | 0.052 ± 0.006 | 0.052 ± 0.010 | 0.063 ± 0.179 | 0.044 ± 0.006 | 0.060 ± 0.026 |
Mean ± SD. Mixed muscle protein synthesis rate calculated using muscle free pool 13C6-phe enrichment to reflect precursor pool enrichment
aAverage plasma TTR measured during 11–12 h of the tracer infusion
bAverage of 3- and 12-h muscle free pool TTR measurements
cMixed muscle protein TTR increment from 3–12 h of the tracer infusion
Baseline mixed muscle protein FSR was low in these 60- to 75-year-old men (0.048 ± 0.009% per hour, range 0.036–0.073% per hour). A 100-mg testosterone injection increased muscle FSR above baseline on day 7 (p = 0.041). However, relative to placebo, average muscle FSR tended to increase 8% after 100 mg testosterone (p = 0.206) and 25% after 300 mg testosterone (p = 0.271; Fig. 2d). These statistical comparisons are limited by the variable response to testosterone (CV% = 11–44%) and the small number of participants in each group.
Muscle comparative proteomics
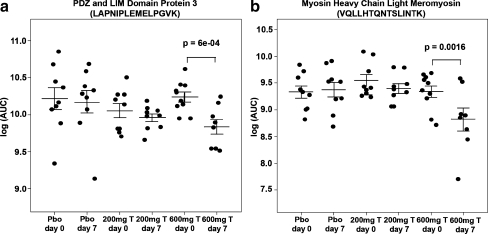
A primary objective for study 2 was the discovery of protein biomarkers for muscle anabolism using proteomics to detect peptides with statistically significant changes in expression levels from baseline to day 7 among the different testosterone/saline doses. A total of 83,420 features, each identified by a unique m/z and retention time, were detected and quantified using FTMS analysis. A subset of 65,152 features with liquid chromatography retention times between 10 and 45 min were selected, and platform variance was estimated to be 55% based on the mean coefficient of variance for these features. Visual inspection of the mass spectra for all of the samples identified one participant’s sample with a distinctly different base peak chromatogram. Reanalysis of the sample did not result in mass spectra that were comparable to other samples, so this sample was excluded from further analyses. Analysis of the 65,152 peptide features selected 692 peptide features with statistically significant difference (p < 0.01) between baseline and day 7 for the 600-mg testosterone treatment group. Scatter plots were created for 200 features with the smallest mixed model p values. Figure 3a is the scatter plot for a feature characterized by reduced expression in the 600-mg testosterone group compared with the 200-mg testosterone and saline groups (p < 6 × 10−4, −1.4-fold change). It corresponds to a peptide matching a region of the PDZ and LIM domain protein 3. Figure 3b depicts another feature characterized by reduced expression in the 600-mg testosterone group compared to the 200-mg testosterone and saline groups (p < 0.0016, −1.7-fold change). This peptide corresponds to skeletal myosin heavy chain–light meromyosin.
Fig. 3.
Skeletal muscle protein expression changes after 600-mg testosterone treatment. A plot of estimates of difference of relative intensity for the placebo, 200-mg, and 600-mg testosterone treatment groups at baseline (day 0) and after 1-week treatment (day 7) are shown. Two tryptic peptides of sequence LAPNIPLEMELPGVK and VQLLHTQNTSLINTK that correspond to the PDZ and LIM domain protein 3 and skeletal myosin heavy chain–light meromyosin region are shown. Y-axis is the area under the curve (AUC) on a natural log scale for the mass spectrometry feature. Values of p determined by a linear mixed model are shown
Muscle mRNA differential expression
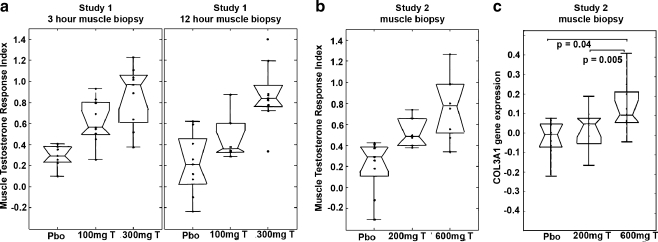
mRNA microarray analysis was used to discover gene expression changes that might represent candidate biomarkers associated with testosterone treatment. An unbiased analysis of the three sets of gene expression data (two from study 1 and one from study 2) yielded an overlap of 12 mRNA gene expression sets we call the muscle testosterone response (MTR) gene signature (Table 3). These 12 mRNA expression probes correspond to nine different genes with altered expression levels after testosterone in comparison to baseline. There were four genes with decreased expression (CAMTA1, CMBL, FEZ2, and IFIT1) and five genes with increased expression (GINS3, PSMD8, SORT1, NINJ2, and ANKDR2). Note that expression levels for two genes, CAMTA1 and CMBL, were decreased after testosterone treatment on two distinct microarray probes. One microarray RNA probe does not encode a known gene. The expression of these genes was converted into a composite MTR score using partial least squares discriminant analysis (Fig. 4). There was a dose-dependent increase in the MTR index with increasing testosterone dose. The relative MTR index for the 3- and 12-h muscle samples also demonstrated the reproducibility of this biomarker (Fig. 4a). (The RNA Microarray data will be available in the Gene Expression Omnibus (GEO) http://www.ncbi.nlm.nih.gov/geo/).
Table 3.
Muscle testosterone response gene signature
| Gene name | Gene symbol | Transcript |
|---|---|---|
| GINS complex subunit 3 | GINS3 | NM_022770 |
| Calmodulin-binding transcriptional activator 1 | CAMTA1 | AB020640 |
| Carboxymethylenebutenolidase homolog | CMBL | ENST00000296658 |
| Nerve-induced injury protein 2 | NINJ2 | NM_016533 |
| Interferon-induced protein with tetricopeptide repeats 1 | IFIT1 | NM_001548 |
| none | none | ENST00000358677 |
| Fasiculation and elongation protein zeta 2 | FEZ2 | BC018032 |
| Proteasome 26S subunit, non-ATPase 8 | PSMD8 | NM_002812 |
| Sortilin 1 | SORT1 | NM_002959 |
| Ankryin repeat domain 2 | ANKRD2 | NM_020349 |
| Calmodulin binding transcription activator 1 | CAMTA1 | NM_015215 |
| Carboxymethylenebutenolidase homolog | CMBL | NM_138809 |
Fig. 4.
The MTR index gene signature demonstrates a dose-dependent increase with testosterone comparing change from baseline and 1-week treatment. a In study 1, the MTR was assessed twice at baseline and twice after 1-week treatment; muscle biopsy performed at 3 and 12 h after beginning the stable isotope infusion for measuring fractional synthetic rate. For comparison between groups, p < 3.9 × 10−5 (3 h) and p < 2.7 × 10−4 (12 h). b In study 2, MTR was assessed once at baseline and after 1-week treatment. For comparison between groups, p < 6.2 × 10−4. c Collagen 3 (COL3A1) RNA expression is increased with the testosterone 600-mg dose compared with testosterone 200-mg dose and placebo. The box has horizontal lines at the lower, median, and upper quartile values. Whiskers extend from each end of the box to the adjacent values in the data, the most extreme values within 1.5 times the interquartile range from the ends of the box. Outliers displayed are data with values beyond the ends of the whiskers
The RNA microarray analysis is also useful for the validation of other biomarkers associated with testosterone treatment and muscle anabolism. The increase in plasma P3NP after testosterone treatment was substantiated by the RNA microarray analysis of skeletal muscle. Expression of collagen III, alpha-1 (COL3A1) mRNA, which encodes P3NP, was increased with testosterone 600-mg treatment compared with testosterone 200 mg and saline (Fig. 4c). There were no statistically significant differences in COL3A1 expression when the lower (100, 200, 300 mg) testosterone doses were compared with saline placebo in studies 1 and 2 (data not shown and Fig. 4c).
Discussion
The objective of these studies was to use different but biochemically relevant analytical platforms (transcript profiling→protein profiling→targeted protein→protein synthesis) to identify muscle-specific candidate early biomarkers of testosterone-induced muscle anabolism that would be useful in the development of novel therapies for sarcopenia and cachexia. We confirmed that plasma P3NP concentration is a potentially useful early biomarker for high-dose testosterone-induced muscle anabolism. Skeletal muscle protein and RNA profiling analyses provided novel potential candidate biomarkers (gene signatures) for muscle anabolism, which also confirmed the value of plasma P3NP as an early biomarker for muscle anabolism.
During the conversion of procollagen to collagen, there is release of the P3NP propeptide of procollagen. Preclinical and clinical studies measuring serum or plasma P3NP and collagen III gene expression in muscle have demonstrated increased levels of P3NP or procollagen 3 mRNA following muscle anabolic interventions, including resistance exercise and growth hormone treatment [28–33]. As skeletal muscle protein remodels, the collagenous perimysium and endomysium surrounding muscle fibers must also remodel. Thus, P3NP release and appearance in the circulation should increase as muscle protein remodels, providing rationale that plasma P3NP could be an early biomarker for muscle anabolism. In the current studies, plasma P3NP increased from baseline within 1 week of testosterone exposure. These findings are consistent with those from the Hormonal Regulators of Muscle and Metabolism in Aging Study involving elderly men where P3NP increased over baseline within 16 weeks of testosterone and growth hormone administration [34]. Though P3NP may be elevated in diseased states such as acute respiratory distress syndrome [35], liver fibrosis [36], and myocardial infarction [37], this does not significantly limit the clinical science utility of P3NP because an increase in P3NP from baseline level can be utilized. Future study participants with these potentially confounding comorbid conditions can be excluded from randomized controlled trials. The possibility that serum P3NP increases are only sensitive and specific to testosterone or growth hormone exposure needs to be addressed.
The skeletal muscle origin of P3NP, as opposed to other tissues, is supported by the muscle RNA microarray data indicating that high-dose (but not lower doses) testosterone exposure increases muscle expression of COL3A; the transcript that encodes collagen and is the source of P3NP. A smaller change in serum P3NP level and muscle COL3A1 mRNA expression may occur with lower doses of testosterone, but this study may not have been adequately powered to detect this small change.
In comparison to placebo, mixed muscle protein FSR was not significantly increased 7 days after a testosterone injection. This is in contrast to prior reports that acute increases in mixed muscle protein FSR occur with androgen administration [38, 39]. A single dose of testosterone (200 mg, IM) to young men resulted in supraphysiologic levels of total testosterone (mean = 33 nmol/L) and a twofold increase in mixed muscle FSR after 5 days [38]. Oxandrolone administration to 72 ± 2-year-old men (10 mg twice daily for 5 days) increased mixed muscle FSR 66% and to 65 ± 2-year-old women (7.5 mg twice daily for 14 days) increased muscle protein FSR 57% over baseline [39]. However, neither study included an appropriate placebo control group. These findings suggest that higher total testosterone levels (>19.5 nmol/L) might be necessary to significantly and acutely increase mixed muscle protein FSR in 60- to 75-year-old men. As suggested by Ferrando et al. [40], perhaps testosterone reduced muscle protein breakdown rate; this was not measured in the current study. Mixed muscle protein synthesis was measured after an overnight fast, and this may have limited amino acid availability for muscle protein synthesis. Also, we may have waited too long after the testosterone injection (7 days) to detect an increase in mixed muscle FSR. We confirmed that resting mixed muscle protein FSR is low in 60- to 75-year-old men [17, 22, 24, 41–44] and lower than a previous report that used 2H5-phe to quantify FSR in older men and women [39]. For the purpose of identifying early biomarkers of muscle protein anabolism-promoting therapeutic interventions, serum P3NP levels, muscle protein, and transcript profiling may be easier and less cumbersome than quantifying in vivo mixed muscle protein FSR, but in vivo quantitation of muscle protein FSR is the gold standard for identifying protein anabolic interventions. However, in this study, it only tended to increase 7 days after testosterone injection, too few participants were enrolled, and their response varied (potential type II error). This limits the utility and applicability of the in vivo measurement of mixed muscle protein FSR in the clinical drug development setting, but it is suitable and warranted in well-controlled physiologic research studies.
A primary objective of study 2 was the discovery of protein biomarkers of muscle anabolism. We have previously used this protein expression profiling approach to identify biomarkers of pancreatitis [45], muscle injury [46], and Alzheimer’s disease [19]. We found two candidate proteomic markers associated with high-dose testosterone treatment: PDZ LIM domain protein and myosin heavy chain–light meromyosin region. The lack of a change in PDZ LIM and myosin heavy chain–light meromyosin protein expression with lower dose testosterone (200 mg) may be due to the presence of a threshold effect that was reached with testosterone 600 mg, but not with testosterone 200 mg. Another explanation could be that the effect of testosterone 200 mg is small, and this was not detected given the variability of the subject response to testosterone and the variability of the proteomics assay method utilized. In either case, the lack of response at 200 mg is clearly an impediment for utility in clinical drug development since the 600-mg dose is supraphysiological.
Microarray mRNA expression profiling of skeletal muscle identified a set of genes (MTR gene signature) associated with acute testosterone exposure. The genes were selected without bias for their function in muscle. However, several identified genes are associated with proteins that regulate muscle anabolism. GINS complex subunit 3 is thought to regulate DNA replication [47]. The Calmodulin binding transcriptional activators (CAMTA) are a family of transcription factors [48], and CAMTA2 is a regulator of cardiac hypertrophy [49]. Nerve-induced injury protein 2 is expressed in glial cells and is increased with nerve injury [50]. The fasciculation and elongation protein zeta 2 is required for neural axon outgrowth [51]. Sortilin 1 stimulates myogenic differentiation and is involved in the glucose transport pathway in C2C12 myocytes [52]. The Ankyrin repeat domain 2 is a titin-binding protein that regulates skeletal muscle hypertrophy [53]. These genes are associated with musculoskeletal and neural function and could be used to develop direct mRNA or protein biomarker assays for assaying muscle anabolism. The MTR gene signature will require validation in future clinical studies as a muscle anabolism biomarker.
In summary, plasma P3NP levels increased in a dose-dependent manner 1 week after a single injection of testosterone. Thus, P3NP is a potentially useful early biomarker for muscle anabolic therapy. Skeletal muscle protein and RNA profiling are useful tools for the successful discovery of novel candidate muscle anabolic biomarkers. Future studies should validate these candidate early biomarkers (P3NP and MTR) in larger clinical studies of longer duration and determine the association between these early candidate biomarkers and sarcopenia-relevant clinical endpoints, such as lean body mass, strength, and functional improvement.
Acknowledgments
We acknowledge Carolyn Hustad for writing and editorial assistance and Jennifer Pawlowski for manuscript submission. Jennifer (Xianghong) Wang and Sam Smith conducted the MS-13C6-phe enrichment analyses. The authors of this manuscript certify that they comply with the ethical guidelines for authorship and publishing in the Journal of Cachexia, Sarcopenia and Muscle [54].
Funding
This study was funded by Merck & Co. Inc.
Disclosure summary
All authors are employees of Merck & Co. Inc., except A.S., who is an employee of MDS Pharma, and K. E. Y., who was partially supported by NIH grants (P41 RR000954, P60 DK020579, P30 DK056341, R01 DK049393, and R01 DK059531) and who received support from Merck & Co., Inc. for a portion of the analytical work presented here.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- ANCOVA
Analysis of covariance
- ANOVA
Analysis of variance
- FSR
Fractional synthesis rate
- FTMS
Fourier transform mass spectrometry
- GH
Growth hormone
- IM
Intramuscular
- MS
Mass spectrometry
- MTR
Muscle testosterone response gene expression signature
- P3NP
Procollagen type III N-terminal propeptide
Footnotes
ClinicalTrials.gov identifier: NCT00812396 NCT00816712
References
- 1.Baumgartner RN, Koehler KM, Gallagher D, et al. Epidemiology of sarcopenia among the elderly in New Mexico. Am J Epidemiol. 1998;147:755–763. doi: 10.1093/oxfordjournals.aje.a009520. [DOI] [PubMed] [Google Scholar]
- 2.Von Haehling S, Morley JE, Anker SD. An overview of sarcopenia: facts and numbers on prevalence and clinical impact. J Cachexia Sarcopenia Muscle. 2010;1:129–133. doi: 10.1007/s13539-010-0014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenk K, Schuler G, Adams V. Skeletal muscle wasting in cachexia and sarcopenia: molecular pathophysiology and impact of exercise training. J Cachexia Sarcopenia Muscle. 2010;1:9–21. doi: 10.1007/s13539-010-0007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tellado JM, Garcia-Sabrido JL, Hanley JA, Shizgal HM, Christou NV. Predicting mortality based on body composition analysis. Ann Surg. 1989;209:81–87. doi: 10.1097/00000658-198901000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhasin S, Woodhouse L, Casaburi R, et al. Testosterone dose–response relationships in healthy young men. Am J Physiol Endocrinol Metab. 2001;281:E1172–E1181. doi: 10.1152/ajpendo.2001.281.6.E1172. [DOI] [PubMed] [Google Scholar]
- 6.Bhasin S, Woodhouse L, Casaburi R, et al. Older men are as responsive as young men to the anabolic effects of graded doses of testosterone on the skeletal muscle. J Clin Endocrinol Metab. 2005;90:678–688. doi: 10.1210/jc.2004-1184. [DOI] [PubMed] [Google Scholar]
- 7.Ferrando AA, Sheffield-Moore M, Yeckel CW, et al. Testosterone administration to older men improves muscle function: molecular and physiological mechanisms. Am J Physiol Endocrinol Metab. 2002;282:E601–E607. doi: 10.1152/ajpendo.00362.2001. [DOI] [PubMed] [Google Scholar]
- 8.Prockop DJ, Kivirikko KI, Tuderman L, Guzman NA. The biosynthesis of collagen and its disorders (first of two parts) N Engl J Med. 1979;301:13–23. doi: 10.1056/NEJM197907053010104. [DOI] [PubMed] [Google Scholar]
- 9.Risteli J, Niemi S, Trivedi P, Maentausta O, Mowat AP, Risteli L. Rapid equilibrium radioimmunoassay for the amino-terminal propeptide of human type III procollagen. Clin Chem. 1988;34:715–718. [PubMed] [Google Scholar]
- 10.Biolo G, Fleming RY, Maggi SP, Wolfe RR. Transmembrane transport and intracellular kinetics of amino acids in human skeletal muscle. Am J Physiol. 1995;268:E75–E84. doi: 10.1152/ajpendo.1995.268.1.E75. [DOI] [PubMed] [Google Scholar]
- 11.Biolo G, Declan Fleming RY, Wolfe RR. Physiologic hyperinsulinemia stimulates protein synthesis and enhances transport of selected amino acids in human skeletal muscle. J Clin Invest. 1995;95:811–819. doi: 10.1172/JCI117731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirkman MA. Comparative determination of protein amino acids in plant materials by automated cation exchange and gas–liquid chromatography of the amino acid N-heptafluorobutyryl, n-propyl esters. J Chromatogr. 1974;97:175–191. doi: 10.1016/S0021-9673(00)95596-1. [DOI] [PubMed] [Google Scholar]
- 13.CD WRR. Isotope tracers in metabolic research: principles and practice of kinetic analysis. 2. Hoboken: Wiley-Liss; 2005. [Google Scholar]
- 14.Bateman RJ, Munsell LY, Chen X, Holtzman DM, Yarasheski KE. Stable isotope labeling tandem mass spectrometry (SILT) to quantify protein production and clearance rates. J Am Soc Mass Spectrom. 2007;18:997–1006. doi: 10.1016/j.jasms.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yarasheski KE, Smith SR, Powderly WG. Reducing plasma HIV RNA improves muscle amino acid metabolism. Am J Physiol Endocrinol Metab. 2005;288:E278–E284. doi: 10.1152/ajpendo.00359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yarasheski KE, Smith K, Rennie MJ, Bier DM. Measurement of muscle protein fractional synthetic rate by capillary gas chromatography/combustion isotope ratio mass spectrometry. Biol Mass Spectrom. 1992;21:486–490. doi: 10.1002/bms.1200211004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasten DL, Pak-Loduca J, Obert KA, Yarasheski KE. Resistance exercise acutely increases MHC and mixed muscle protein synthesis rates in 78–84 and 23–32 yr olds. Am J Physiol Endocrinol Metab. 2000;278:E620–E626. doi: 10.1152/ajpendo.2000.278.4.E620. [DOI] [PubMed] [Google Scholar]
- 18.Meng F, Wiener MC, Sachs JR, et al. Quantitative analysis of complex peptide mixtures using FTMS and differential mass spectrometry. J Am Soc Mass Spectrom. 2007;18:226–233. doi: 10.1016/j.jasms.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Paweletz CP, Wiener MC, Bondarenko AY, et al. Application of an end-to-end biomarker discovery platform to identify target engagement markers in cerebrospinal fluid by high resolution differential mass spectrometry. J Proteome Res. 2010;9:1392–1401. doi: 10.1021/pr900925d. [DOI] [PubMed] [Google Scholar]
- 20.Jonckheere AR. A distribution-free k-sample test against ordered alternatives. Biometrika. 1954;41:133–145. [Google Scholar]
- 21.Terpstra TJ. The asymptomatic normality and consistency of Kendall's test against trend, when ties are present in one ranking. Inigationes Mathmaticae. 1952;14:327–333. [Google Scholar]
- 22.Nair KS, Halliday D, Griggs RC. Leucine incorporation into mixed skeletal muscle protein in humans. Am J Physiol. 1988;254:E208–E213. doi: 10.1152/ajpendo.1988.254.2.E208. [DOI] [PubMed] [Google Scholar]
- 23.Volpi E, Mittendorfer B, Rasmussen BB, Wolfe RR. The response of muscle protein anabolism to combined hyperaminoacidemia and glucose-induced hyperinsulinemia is impaired in the elderly. J Clin Endocrinol Metab. 2000;85:4481–4490. doi: 10.1210/jc.85.12.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welle S, Thornton C, Jozefowicz R, Statt M. Myofibrillar protein synthesis in young and old men. Am J Physiol. 1993;264:E693–E698. doi: 10.1152/ajpendo.1993.264.5.E693. [DOI] [PubMed] [Google Scholar]
- 25.Rennie MJ, Edwards RH, Halliday D, Matthews DE, Wolman SL, Millward DJ. Muscle protein synthesis measured by stable isotope techniques in man: the effects of feeding and fasting. Clin Sci (Lond) 1982;63:519–523. doi: 10.1042/cs0630519. [DOI] [PubMed] [Google Scholar]
- 26.Watt PW, Lindsay Y, Scrimgeour CM, et al. Isolation of aminoacyl-tRNA and its labeling with stable-isotope tracers: use in studies of human tissue protein synthesis. Proc Natl Acad Sci USA. 1991;88:5892–5896. doi: 10.1073/pnas.88.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ljungqvist OH, Persson M, Ford GC, Nair KS. Functional heterogeneity of leucine pools in human skeletal muscle. Am J Physiol. 1997;273:E564–E570. doi: 10.1152/ajpendo.1997.273.3.E564. [DOI] [PubMed] [Google Scholar]
- 28.Takala TE, Vuori J, Anttinen H, Vaananen K, Myllyla R. Prolonged exercise causes an increase in the activity of galactosylhydroxylysyl glucosyltransferase and in the concentration of type III procollagen aminopropeptide in human serum. Pflugers Arch. 1986;407:500–503. doi: 10.1007/BF00657507. [DOI] [PubMed] [Google Scholar]
- 29.Crameri RM, Langberg H, Teisner B, et al. Enhanced procollagen processing in skeletal muscle after a single bout of eccentric loading in humans. Matrix Biol. 2004;23:259–264. doi: 10.1016/j.matbio.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Nelson AE, Meinhardt U, Hansen JL, et al. Pharmacodynamics of growth hormone abuse biomarkers and the influence of gender and testosterone: a randomized double-blind placebo-controlled study in young recreational athletes. J Clin Endocrinol Metab. 2008;93:2213–2222. doi: 10.1210/jc.2008-0402. [DOI] [PubMed] [Google Scholar]
- 31.Wallace JD, Cuneo RC, Lundberg PA, et al. Responses of markers of bone and collagen turnover to exercise, growth hormone (GH) administration, and GH withdrawal in trained adult males. J Clin Endocrinol Metab. 2000;85:124–133. doi: 10.1210/jc.85.1.124. [DOI] [PubMed] [Google Scholar]
- 32.Garma T, Kobayashi C, Haddad F, Adams GR, Bodell PW, Baldwin KM. Similar acute molecular responses to equivalent volumes of isometric, lengthening, or shortening mode resistance exercise. J Appl Physiol. 2007;102:135–143. doi: 10.1152/japplphysiol.00776.2006. [DOI] [PubMed] [Google Scholar]
- 33.Longobardi S, Keay N, Ehrnborg C, et al. Growth hormone (GH) effects on bone and collagen turnover in healthy adults and its potential as a marker of GH abuse in sports: a double blind, placebo-controlled study. The GH-2000 Study Group. J Clin Endocrinol Metab. 2000;85:1505–1512. doi: 10.1210/jc.85.4.1505. [DOI] [PubMed] [Google Scholar]
- 34.Bhasin S, He EJ, Kawakubo M, et al. N-terminal propeptide of type III procollagen as a biomarker of anabolic response to recombinant human GH and testosterone. J Clin Endocrinol Metab. 2009;94:4224–4233. doi: 10.1210/jc.2009-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meduri GU, Tolley EA, Chinn A, Stentz F, Postlethwaite A. Procollagen types I and III aminoterminal propeptide levels during acute respiratory distress syndrome and in response to methylprednisolone treatment. Am J Respir Crit Care Med. 1998;158:1432–1441. doi: 10.1164/ajrccm.158.5.9801107. [DOI] [PubMed] [Google Scholar]
- 36.Xie SB, Yao JL, Zheng RQ, Peng XM, Gao ZL. Serum hyaluronic acid, procollagen type III and IV in histological diagnosis of liver fibrosis. Hepatobiliary Pancreat Dis Int. 2003;2:69–72. [PubMed] [Google Scholar]
- 37.Poulsen SH, Host NB, Jensen SE, Egstrup K. Relationship between serum amino-terminal propeptide of type III procollagen and changes of left ventricular function after acute myocardial infarction. Circulation. 2000;101:1527–1532. doi: 10.1161/01.cir.101.13.1527. [DOI] [PubMed] [Google Scholar]
- 38.Ferrando AA, Tipton KD, Doyle D, Phillips SM, Cortiella J, Wolfe RR. Testosterone injection stimulates net protein synthesis but not tissue amino acid transport. Am J Physiol. 1998;275:E864–E871. doi: 10.1152/ajpendo.1998.275.5.E864. [DOI] [PubMed] [Google Scholar]
- 39.Sheffield-Moore M, Paddon-Jones D, Casperson SL, et al. Androgen therapy induces muscle protein anabolism in older women. J Clin Endocrinol Metab. 2006;91:3844–3849. doi: 10.1210/jc.2006-0588. [DOI] [PubMed] [Google Scholar]
- 40.Ferrando AA, Sheffield-Moore M, Paddon-Jones D, Wolfe RR, Urban RJ. Differential anabolic effects of testosterone and amino acid feeding in older men. J Clin Endocrinol Metab. 2003;88:358–362. doi: 10.1210/jc.2002-021041. [DOI] [PubMed] [Google Scholar]
- 41.Balagopal P, Ford GC, Ebenstein DB, Nadeau DA, Nair KS. Mass spectrometric methods for determination of [13C]Leucine enrichment in human muscle protein. Anal Biochem. 1996;239:77–85. doi: 10.1006/abio.1996.0293. [DOI] [PubMed] [Google Scholar]
- 42.Giannoulis MG, Jackson N, Shojaee-Moradie F, et al. The effects of growth hormone and/or testosterone on whole body protein kinetics and skeletal muscle gene expression in healthy elderly men: a randomized controlled trial. J Clin Endocrinol Metab. 2008;93:3066–3074. doi: 10.1210/jc.2007-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henderson GC, Dhatariya K, Ford GC, et al. Higher muscle protein synthesis in women than men across the lifespan, and failure of androgen administration to amend age-related decrements. FASEB J. 2009;23:631–641. doi: 10.1096/fj.08-117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yarasheski KE, Zachwieja JJ, Bier DM. Acute effects of resistance exercise on muscle protein synthesis rate in young and elderly men and women. Am J Physiol. 1993;265:E210–E214. doi: 10.1152/ajpendo.1993.265.2.E210. [DOI] [PubMed] [Google Scholar]
- 45.Yates NA, Deyanova EG, Geissler W, Wiener MC, Sachs JR, Wong KW, et al. Identification of peptidase substrates in human plasma by FTMS based differential mass spectrometry. Int J Mass Spectrom. 2007;259:174–183. doi: 10.1016/j.ijms.2006.09.020. [DOI] [Google Scholar]
- 46.Sietsema KE, Meng F, Yates NA, et al. Potential biomarkers of muscle injury after eccentric exercise. Biomarkers. 2010;15:249–258. doi: 10.3109/13547500903502802. [DOI] [PubMed] [Google Scholar]
- 47.MacNeill SA. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem J. 2010;425:489–500. doi: 10.1042/BJ20091531. [DOI] [PubMed] [Google Scholar]
- 48.Bouche N, Scharlat A, Snedden W, Bouchez D, Fromm H. A novel family of calmodulin-binding transcription activators in multicellular organisms. J Biol Chem. 2002;277:21851–21861. doi: 10.1074/jbc.M200268200. [DOI] [PubMed] [Google Scholar]
- 49.Song K, Backs J, McAnally J, et al. The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell. 2006;125:453–466. doi: 10.1016/j.cell.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 50.Araki T, Milbrandt J. Ninjurin2, a novel homophilic adhesion molecule, is expressed in mature sensory and enteric neurons and promotes neurite outgrowth. J Neurosci. 2000;20:187–195. doi: 10.1523/JNEUROSCI.20-01-00187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Assmann EM, Alborghetti MR, Camargo ME, Kobarg J. FEZ1 dimerization and interaction with transcription regulatory proteins involves its coiled-coil region. J Biol Chem. 2006;281:9869–9881. doi: 10.1074/jbc.M513280200. [DOI] [PubMed] [Google Scholar]
- 52.Ariga M, Nedachi T, Katagiri H, Kanzaki M. Functional role of sortilin in myogenesis and development of insulin-responsive glucose transport system in C2C12 myocytes. J Biol Chem. 2008;283:10208–10220. doi: 10.1074/jbc.M710604200. [DOI] [PubMed] [Google Scholar]
- 53.McKoy G, Hou Y, Yang SY, et al. Expression of Ankrd2 in fast and slow muscles and its response to stretch are consistent with a role in slow muscle function. J Appl Physiol. 2005;98:2337–2343. doi: 10.1152/japplphysiol.01046.2004. [DOI] [PubMed] [Google Scholar]
- 54.Von Haehling S, Morley JE, Coats AJS, Anker SD. Ethical guidelines for authorship and publishing in the journal of cachexia, sarcopenia and muscle. J Cachexia Sarcopenia Muscle. 2010;1:7–8. doi: 10.1007/s13539-010-0003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]