Abstract
AIM: To investigate the utility of the cytomegalovirus (CMV) antigenemia assay for the diagnosis of CMV gastrointestinal disease (GID).
METHODS: One hundred and thirty immunocompromised patients were enrolled in this study. Patients with a history of anti-CMV treatment and who had not undergone examination using the antigenemia assay were excluded. CMV-GID was defined as the detection of large cells with intranuclear inclusions alone or associated with granular cytoplasmic inclusions by biopsy. Biopsy sections were stained with hematoxylin and eosin and immunohistochemically stained with anti-CMV. We evaluated the association between CMV-GID and patient characteristics (symptoms, underlying disease, medication, leukocyte counts, and antigenemia assay). All patients were checked with an human immunodeficiency virus (HIV) antibody test before endoscopic examination. White blood cell (WBC) counts were obtained from medical records within 1 wk of endoscopy. Leukopenia was defined as a total WBC count < 5000 cells/mm3. For HIV patients, we also checked CD4+ counts from medical records.
RESULTS: A total of 99 patients were retrospectively selected for analysis. Of the immunocompromised patients, 19 had malignant disease, 18 had autoimmune disease, 19 had disorders of biochemical homeostasis, three had undergone transplantation, and 45 had HIV infection. A total of 50 patients had received immunosuppressive therapy. No patients had inflammatory bowel disease. Fifty-five patients were diagnosed as having CMV-GID. Univariate analysis indicated an association between HIV infection, leukopenia, and positive antigenemia and CMV-GID (P < 0.05). Multivariate analysis using logistic regression revealed that HIV infection and positive antigenemia were the only independent factors related to CMV-GID (P < 0.01). The sensitivity, specificity, positive predictive value, and negative predictive value of antigenemia for CMV-GID were 65.4%, 93.6%, 91.9%, and 71.0%, respectively. In a subgroup analysis, patients with leukopenia displayed low sensitivity and high specificity. Minimal differences in accuracy were seen among patients with or without leukopenia. HIV-infected patients displayed low sensitivity and high specificity. Accuracy barely differed between HIV-positive and -negative patients. In HIV-infected patients, CD4 count < 50 cells/μL resulted in low sensitivity and high specificity. Differences in accuracy among patients were minor, regardless of CD4 count. In patients who had undergone both quantitative real-time polymerase chain reaction (PCR) and antigenemia assay, real-time PCR was slightly more accurate in terms of sensitivity than the antigenemia assay; however, this difference was not statistically significant (P = 0.312).
CONCLUSION: If the antigenemia test is positive, endoscopic lesions are acceptable for the diagnosis of CMV-GID without biopsy. The accuracy is not affected by HIV infection and leukopenia. Either PCR or the antigenemia assay are valid.
Keywords: Cytomegalovirus, Gastrointestinal disease, Antigenemia assay, Real-time polymerase chain reaction, Human immunodeficiency virus infection
INTRODUCTION
As the number of patients with immune deficiency has been increasing dramatically in recent years, the number of patients with cytomegalovirus (CMV) disease has also been increasing. CMV gastrointestinal disease (CMV-GID) frequently occurs in immunocompromised patients, particularly among those with human immunodeficiency virus (HIV) infection, transplantation, autoimmune diseases, or secondary immunodeficiency[1-8]. CMV-GID has also been described following the use of steroids, immunosuppressants, or cancer chemotherapy[1,2]. In immunocompromised patients, CMV-GID in the absence of therapy is a major cause of morbidity and mortality due to events such as massive bleeding or perforation. Therefore, diagnosis at an early stage is essential[1,2,9-12]. However, diagnosis of this infection is difficult because of wide variations in symptoms and endoscopic features depending on the infected organs[1,2].
Although the utility of various diagnostic tests for CMV-GID has been reported, the best approach is to conform the presence of CMV by histological analysis, including immunological staining by endoscopy[1-3,5,13,14]. Endoscopic examination is generally tolerated, but tissue biopsy can possibly lead to hemorrhage or perforation after endoscopic examination[10,11,15]. Endoscopists therefore hesitate to perform biopsy when deep, large, and bleeding ulcerous lesions are encountered. Patients receiving antithrombotic drugs or with thrombocytopenia also require careful consideration before biopsy.
On many occasions in recent years, noninvasive methods such as the CMV blood antigenemia assay have been applied instead of biopsy to avoid adverse effects[3,16-22]. However, few reports have examined the diagnostic value of the CMV antigenemia assay for CMV-GID, and the clinical utility of this method in immunodeficiency remains unclear[3,20-22]. Moreover, the CMV antigenemia assay requires sufficient granulocytes, and leukopenia and low CD4+ counts in patients with HIV infection could thus be expected to influence assay accuracy[3]. However, no reports have yet clarified this issue.
The aims of this study were to clarify the utility of the CMV antigenemia assay for diagnosing suspected CMV-GID, and to evaluate the accuracy of this assay under different clinical settings.
MATERIALS AND METHODS
Patient selection
One hundred and thirty immunocompromised patients with endoscopic findings who had undergone biopsy were enrolled in this study at the National Center for Global Health and Medicine (NCGM) from January 2002 to September 2009. Patients with a history of treatment with anti-CMV therapy were excluded, as were cases not examined using the CMV antigenemia assay test within 1 wk of endoscopy. Written informed consent was obtained from all patients prior to endoscopy and biopsy. All study protocols were approved by the ethics committee of NCGM.
Immunocompromised patients
Immunocompromised patients are associated with secondary immune deficiency, particularly HIV infection, hematopoietic stem cell transplantation, autoimmune diseases, malignancy, disorders of biochemical homeostasis, and use of steroids, immunosuppressants, or cancer chemotherapy.
Underlying autoimmune diseases included Rheumatoid arthritis, Systemic lupus erythematosis, Still’s disease, Behcet’s disease, Polymyositis, and Dermatomyositis. Diabetes mellitus, renal insufficiency/dialysis, and hepatic cirrhosis were included among the disorders of biochemical homeostasis. All patients were checked with an HIV antibody test before endoscopic examination.
Clinical manifestations
Gastrointestinal symptoms were collected from medical records written by the doctor who interviewed each person face-to-face before endoscopy. Those without records were treated as symptom free. Gastrointestinal symptoms included compromised odynophagia, epigastralgia, nausea, lower abdominal pain, diarrhea, and hematochezia. White blood cell (WBC) counts were obtained from medical records within 1 wk of endoscopy. Leukopenia was defined as a total WBC count < 5000 cells/mm3. For HIV patients, we also checked CD4+ counts from medical records.
Antigenemia assay and quantitative real-time polymerase chain reaction
Antigenemia assay using C10/C11 monoclonal antibodies (Mitsubishi Chemical Medience, Tokyo, Japan) was performed as previously reported[16,19,20]. A positive result for the CMV antigenemia assay was defined as ≥ 1 CMV-positive cell per 150 000 granulocytes applied.
A total of 47 patients underwent additional examination with real-time polymerase chain reaction (PCR), performed basically as previously reported[3,23,24]. The minimum detection level was 200 copies/mL of plasma. A positive result for real-time CMV PCR was defined as > 200 copies/mL.
Diagnosis of CMV-GID
CMV-GID was suspected based on endoscopic findings, such as patchy erythema, edematous mucosa, multiple erosions, and ulcers (Figure 1)[25,26]. Biopsy was therefore performed when such endoscopic findings were encountered. CMV-GID was defined as the detection of large cells with intranuclear inclusions alone or associated with granular cytoplasmic inclusions by histological testing of biopsy specimens[1]. Biopsy sections were stained with hematoxylin and eosin, and immunohistochemically stained with anti-CMV (Figure 2). The results were considered positive when the above-mentioned cells showed marked brown coloration in both nuclei and cytoplasm.
Figure 1.
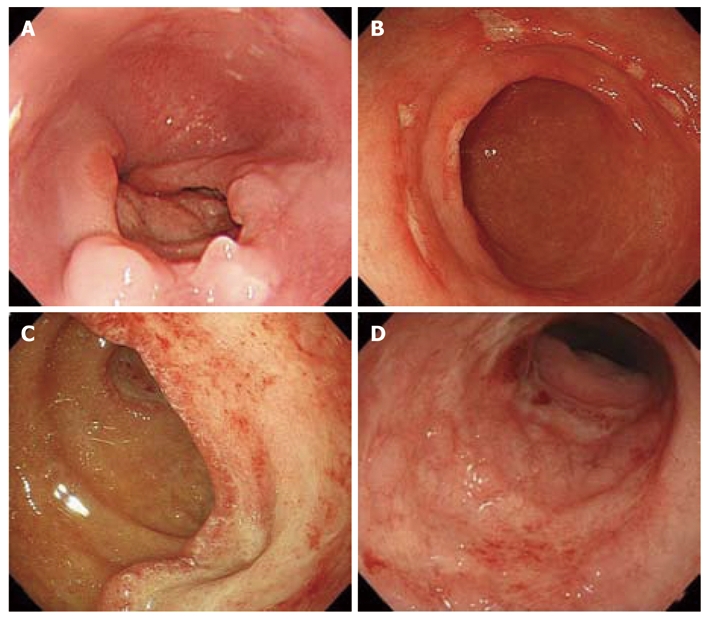
Endoscopic features in cytomegalovirus gastrointestinal disease. A: Deep, punched-out ulcer in the esophagus; B: Multiple, shallow ulcers in the gastric antrum; C: Large, deep ulcer in the duodenum; D: Multiple erosions and edematous mucosa with ulcer in the sigmoid colon.
Figure 2.
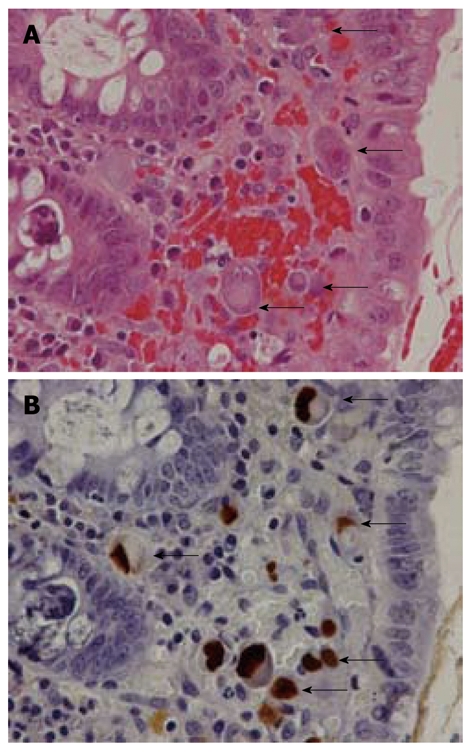
Pathological features in cytomegalovirus gastrointestinal disease. A: Large cells with intranuclear inclusions or associated with granular cytoplasmic inclusions (hematoxylin and eosin stain); B: Cytomegalovirus (CMV)-infected cells (arrows) show brown coloration in both nuclei and cytoplasm (immunohistochemical staining with anti-CMV).
Statistical analysis
We divided patients into two groups based on the presence or absence of CMV-GID. Patient characteristics and clinical findings were then compared between groups. Fisher’s exact test was used to compare frequencies for patient characteristics and clinical findings, and Mann-Whitney U test was used for comparing age and CD4 counts. To identify clinical factors independently associated with a diagnosis of CMV-GID, stepwise logistic regression modeling was used. Sensitivity, specificity, and positive and negative predictive values of CMV antigenemia for diagnosing CMV-GID were calculated. The difference in accuracy between CMV real-time PCR and CMV antigenemia assay was compared according to the area under the curve (AUC). Values of P < 0.05 were considered significant. All statistical analyses were performed using Stata software (version 10, Stata Co., USA).
RESULTS
Clinical features
We excluded 10 patients who had received anti-CMV treatment, along with 21 patients who had not been examined using the CMV antigenemia assay. Thus, a total of 99 patients were retrospectively selected for analysis (Figure 3). Of the immunocompromised patients, 19 (19.1%) had malignant disease, 18 (18.1%) had autoimmune disease, 19 (19.1%) had disorders of biochemical homeostasis, three (3%) had undergone transplantation, and 45 (45.5%) had HIV infection. A total of 50 patients (50.1%) had received immunosuppressive therapy. No patients had inflammatory bowel disease (IBD). Fifty-five patients were histologically diagnosed with CMV-GID. Univariate analysis (Table 1) identified HIV infection (P < 0.001), leukopenia (P = 0.023), and positive CMV antigenemia assay (P < 0.001) as being associated with CMV-GID. Multivariate analysis revealed HIV infection [odds ratio (OR), 6.57; 95% CI: 2.1-20.2, P = 0.001] and positive CMV antigenemia assay (OR, 33.3; 95% CI: 8.1-136.2, P < 0.001) as the only factors independently correlated with CMV-GID.
Figure 3.
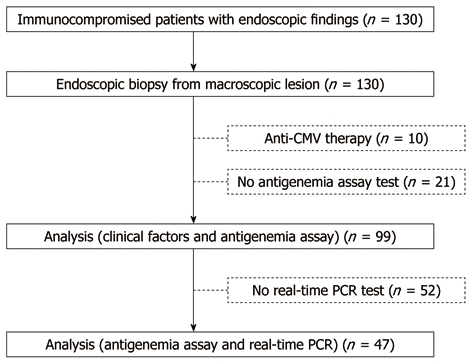
Study design. CMV: Cytomegalovirus; PCR: Polymerase chain reaction.
Table 1.
Clinical factors for cytomegalovirus gastrointestinal disease (univariate analysis)
| CMV-GID (n = 52) | Non-CMV-GID (n = 47) | P-value | |
| Age (yr, mean ± SD) | 46.8 ± 16.2 | 56.6 ± 17.8 | 0.050 |
| Male sex | 30 | 41 | 0.098 |
| Immunodeficiency disease | |||
| HIV infection | 33 | 12 | < 0.001 |
| Malignancy | 9 | 10 | 0.617 |
| Solid cancer | 1 | 3 | |
| Hematological cancer | 8 | 7 | |
| Autoimmune disease | 7 | 11 | 0.200 |
| Disorders of biochemical homeostasis | 8 | 11 | 0.312 |
| Chronic renal failure | 1 | 2 | |
| Liver cirrhosis | 0 | 2 | |
| Diabetes mellitus | 7 | 7 | |
| Transplantation | 1 | 2 | |
| Immunosuppressive therapy | 25 | 25 | 0.611 |
| Steroids | 22 | 19 | |
| Immunosuppressants | 8 | 4 | |
| Chemotherapy | 4 | 4 | |
| Positive CMV antigenemia | 34 | 3 | < 0.001 |
| Leukopenia | 35 | 21 | 0.023 |
| With gastrointestinal symptoms | 34 | 34 | 0.456 |
HIV: Human immunodeficiency virus; CMV: Cytomegalovirus; GID: Gastrointestinal disease.
HIV-infected patients included 44 men (97.8%) and their mean age was 42.1 years (range, 25-74 years). Median CD4 count was 57 (interquartile range, 17-111). Patients with CMV-GID showed significantly lower CD4 counts than those without CMV-GID (median CD4 count; CMV-GID vs non-CMV-GID: 24 vs 150, P < 0.001).
Accuracy of CMV antigenemia assay for diagnosing CMV-GID
A positive CMV antigenemia assay showed low sensitivity and high specificity (Table 2). In a subgroup analysis, patients with leukopenia displayed low sensitivity and high specificity. Minimal differences in accuracy were seen among patients with or without leukopenia. HIV-infected patients displayed low sensitivity and high specificity. Accuracy barely differed between HIV-positive and -negative patients. In HIV-infected patients, CD4 count < 50 cells/μL resulted in low sensitivity and high specificity. Differences in accuracy among patients were minor, regardless of CD4 count.
Table 2.
Diagnostic accuracy of cytomegalovirus antigenemia for detecting cytomegalovirus gastrointestinal disease
| Subgroups | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) |
| All patients (n = 99) | 65.40% (55.4-74.9) | 93.60% (87.3-97.7) | 91.90% (84.7-96.4) | 71.00% (60.7-79.4) |
| Patients with leukopenia (n = 56) | 68.60% (54.0-79.7) | 100% (93.6-100) | 100% (93.6-100) | 65.60% (52.2-78.2) |
| Patients without leukopenia (n = 43) | 58.80% (42.1-73.0) | 88.50% (74.9-96.1) | 76.90% (61.4-88.2) | 76.70% (61.4-88.2) |
| HIV-infected patients (n = 45) | 63.60% (48.8-78.1) | 100% (92.2-100) | 100% (92.2-100) | 50.00% (35.8-66.3) |
| Non-HIV-infected patients (n = 54) | 68.40% (54.5-80.5) | 91.40% (79.7-96.9) | 81.30% (68.6-90.7) | 84.20% (70.7-92.1) |
| HIV-infected patients with CD4 count < 50 (n = 22) | 61.90% (40.7-82.8) | 100% (84.6-100) | 100% (84.6-100) | 11.10% (1.12-29.2) |
| HIV-infected patients with CD4 count ≥ 50 (n = 23) | 66.70% (42.7-83.6) | 100% (85.2-100) | 100% (85.2-100) | 73.30% (51.6-89.8) |
HIV: Human immunodeficiency virus; PPV: Positive predictive value; NPV: Negative predictive value.
In patients who had undergone both quantitative real-time PCR and antigenemia assay (Table 3), real-time PCR was slightly more accurate in terms of sensitivity than the antigenemia assay; however, this difference was not statistically significant (P = 0.312).
Table 3.
Comparison of diagnostic accuracy for detecting cytomegalovirus gastrointestinal disease between antigenemia assay and quantitative real-time polymerase chain reaction (n = 47)
| Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) | |
| CMV real-time PCR | 73.00% (57.4-84.4) | 100% (92.5-100) | 100% (92.5-100) | 50.00% (36.1-65.9) |
| CMV antigenemia assay | 64.90% (50.7-79.1) | 100% (92.5-100) | 100% (92.5-100) | 43.50% (28.3-57.8) |
CMV: Cytomegalovirus; PPV: Positive predictive value; NPV: Negative predictive value; PCR: Polymerase chain reaction.
DISCUSSION
CMV-GID is a major cause of morbidity and mortality in immunocompromised patients; therefore, diagnosis at an early stage is essential[1,2,5,8,9]. However, clinical diagnosis of this disease can be difficult, as physicians need to consider various underlying diseases and clinical presentations. Patients at high risk of CMV-GID have been reported as those with HIV infection or undergoing steroid therapy or cancer therapy[1]. The present study identified HIV infection as one of the independent factors in secondary immunodeficiency diseases. This is because the number of eligible subjects was small and included immunocompromised patients while excluding immunocompetent patients.
Among the various clinical manifestations, a positive CMV antigenemia assay was found to be a useful factor for diagnosing CMV-GID. The CMV antigenemia assay is one of the most widely used methods for detecting reactivation of CMV infection, but few studies have examined the diagnostic value for CMV-GID[3,21,22]. Our findings demonstrated 65% sensitivity and 94% specificity of the CMV antigenemia assay for diagnosing CMV-GID. Mori et al[3] reported that only four of 19 patients (21%) developed a positive CMV antigenemia assay before developing CMV-GID; however, all 19 patients subsequently tested positive for CMV antigenemia after diagnosis of CMV-GID. There is a possibility that patients with CMV-GID will develop a positive CMV antigenemia assay at follow-up, but our study did not assess this process after diagnosis of CMV-GID. Fica et al[21] also reported that the CMV antigenemia assay result was positive for 18 of 31 patients (58%) with CMV end-organ disease, with CMV-GID (71%) as the most frequent cause. However, these studies were limited in that the number of subjects was small and the specificity of the CMV antigenemia assay was unknown. Jang et al[22] recently reported that the sensitivity and specificity of the CMV antigenemia assay for diagnosing CMV-GID were 54% and 88%, respectively, in patients with secondary immunodeficiency disease. The reports mentioned above showed that the CMV antigenemia assay has low sensitivity for the diagnosis of CMV-GID, which is consistent with our results.
It has been reported that sufficient granulocytes are essential in evaluating CMV using the antigenemia assay. Previous studies using the antigenemia assay to diagnose CMV-GID have reported that most of the patients were transplant recipients and were mostly HIV-negative[3,21,22]. No studies have compared the assay among groups of HIV-positive/-negative patients and among groups with or without leukopenia. In patients with HIV infection, most cases of CMV-GID have known to occur with CD4 counts < 50 cells/μL[2,4]. However, whether the accuracy of the antigenemia assay is affected by the immunosuppressed state has not been elucidated. We suspected that such different groups would show differences in the accuracy of CMV antigenemia assay, but found little difference. This suggests that our results are applicable to these different groups in clinical practice.
Besides the CMV antigenemia assay, quantitative real-time PCR is also used for detecting reactivation of CMV infection, and is considered more useful for predicting CMV disease than the CMV antigenemia assay[23,24]. In our study, quantitative real-time PCR and CMV antigenemia assay were performed simultaneously on 47 patients. The PCR method showed a tendency toward slightly higher sensitivity, but no significant differences were evident. In Japan, the CMV PCR method has not been widely used in clinical practice because of the higher costs compared to the antigenemia assay. We thus do not recommend use of PCR methods in the sub-diagnosis of CMV-GID, as the antigenemia assay is just as valid.
One limitation of this study was the single-center, retrospective nature of the investigation. A significant difference might not have been confirmed among independent factors due to the small number of patients. Further studies of more patients are needed. Another limitation is the verification bias, which is dependent on the physician’s decision to perform the antigenemia assay.
The diagnosis of CMV-GID is considered as the gold standard for identifying CMV cells in tissue samples from endoscopic biopsy[1,2,13]. Various endoscopic findings are present in CMV-GID, such as ulcer and mucosal inflammation[25,26]; however, physicians may not perform a biopsy in cases only showing mucosal inflammation without ulcer. Even in cases of severe ulceration that is deep or bleeding, physicians may hesitate to perform a biopsy. In such cases, a diagnosis of CMV-GID may not be reached. Our results suggest that the CMV antigenemia assay is useful for the sub-diagnosis of CMV-GID in immunocompromised patients with endoscopic findings. Considering the high specificity of the test, the use of this method before endoscopy could potentially avoid complications due to biopsy. Positive antigenemia is also useful for evaluating improvements in CMV-GID after anti-CMV treatment. However, the low sensitivity means that if the antigenemia assay yields negative results, biopsy and immunohistochemical staining of specimens with anti-CMV will be required for diagnosis. Negative antigenemia assay results may require a repeat examination at a different time[3]. Moreover, the use of different non-invasive methods such as quantitative PCR should be considered.
In conclusion, the CMV antigenemia assay is highly useful for diagnosing CMV-GID. If the antigenemia assay provides positive results, the presence of endoscopic lesions should allow diagnosis of CMV-GID without biopsy. The accuracy of the test is unaffected by the presence of HIV infection or leukopenia.
COMMENTS
Background
Cytomegalovirus (CMV) gastrointestinal disease (GID) is a major cause of morbidity and mortality in immunocompromised patients; therefore, diagnosis at an early stage is essential. However, clinical diagnosis of this disease can be difficult, as physicians need to consider various underlying diseases and clinical presentations.
Research frontiers
The diagnosis of CMV-GID requires an endoscopic biopsy, which is invasive and may lead to complications. While the CMV antigenemia assay is one of the most widely used methods for detecting reactivation of CMV infection, few studies have examined its diagnostic value for CMV-GID. In this study, the authors demonstrate that the CMV antigenemia assay was highly useful for diagnosing CMV-GID.
Innovations and breakthroughs
There were no studies of diagnosis on CMV-GID related factors using multivariate analysis. In this study, among the various clinical manifestations, human immunodeficiency virus (HIV) infection and positive CMV antigenemia assay were found to be a useful factors for diagnosing CMV-GID by multivariate analysis. As for accuracy of CMV antigenemia for diagnosing CMV-GID, recent reports have highlighted that the sensitivity and specificity were 54% and 88%, respectively, in patients with secondary immunodeficiency disease. However, no studies have compared the assay among groups of HIV-positive/-negative patients and among groups with or without leukopenia. In this study, the sensitivity, specificity, positive predictive value, and negative predictive value of antigenemia for CMV-GID were 65.4%, 93.6%, 91.9%, and 71.0%, respectively. In addition, its accuracy was not affected by the presence of HIV infection and leukopenia. These results are very useful for diagnosing CMV-GID by clinical physicians.
Applications
Considering the high specificity of the test, use of this method before endoscopy could potentially avoid complications due to biopsy. However, the low sensitivity means that if the antigenemia assay yields negative results, biopsy and immunohistochemical staining of specimens with anti-CMV will be required for diagnosis. Negative antigenemia assay results may require repeat examination at a different time. Moreover, the use of different non-invasive methods such as quantitative polymerase chain reaction should be considered.
.Peer review
This paper is interesting and it could be valuable for other researchers.
Acknowledgments
We acknowledge Mr. Takashi Kurihara (Department, Mitsubishi Chemical Medience Corporation) for advice to this study on the antigenemia assay evaluation. We also acknowledge Shizuka Tanaka and Toshio Kitazawa for help with the pathological evaluation.
Footnotes
Peer reviewer: Beata Jolanta Jablońska, MD, PhD, Department of Digestive Tract Surgery, University Hospital of Medical University of Silesia, Medyków 14 St. 40-752 Katowice, Poland
S- Editor Tian L L- Editor Stewart GJ E- Editor Zheng XM
References
- 1.Goodgame RW. Gastrointestinal cytomegalovirus disease. Ann Intern Med. 1993;119:924–935. doi: 10.7326/0003-4819-119-9-199311010-00010. [DOI] [PubMed] [Google Scholar]
- 2.Baroco AL, Oldfield EC. Gastrointestinal cytomegalovirus disease in the immunocompromised patient. Curr Gastroenterol Rep. 2008;10:409–416. doi: 10.1007/s11894-008-0077-9. [DOI] [PubMed] [Google Scholar]
- 3.Mori T, Mori S, Kanda Y, Yakushiji K, Mineishi S, Takaue Y, Gondo H, Harada M, Sakamaki H, Yajima T, et al. Clinical significance of cytomegalovirus (CMV) antigenemia in the prediction and diagnosis of CMV gastrointestinal disease after allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 2004;33:431–434. doi: 10.1038/sj.bmt.1704369. [DOI] [PubMed] [Google Scholar]
- 4.Whitley RJ, Jacobson MA, Friedberg DN, Holland GN, Jabs DA, Dieterich DT, Hardy WD, Polis MA, Deutsch TA, Feinberg J, et al. Guidelines for the treatment of cytomegalovirus diseases in patients with AIDS in the era of potent antiretroviral therapy: recommendations of an international panel. International AIDS Society-USA. Arch Intern Med. 1998;158:957–969. doi: 10.1001/archinte.158.9.957. [DOI] [PubMed] [Google Scholar]
- 5.Fujita M, Hatachi S, Yagita M. Immunohistochemically proven cytomegalovirus gastrointestinal diseases in three patients with autoimmune diseases. Clin Rheumatol. 2008;27:1057–1059. doi: 10.1007/s10067-008-0846-8. [DOI] [PubMed] [Google Scholar]
- 6.Sultan SM, Ioannou Y, Isenberg DA. A review of gastrointestinal manifestations of systemic lupus erythematosus. Rheumatology (Oxford) 1999;38:917–932. doi: 10.1093/rheumatology/38.10.917. [DOI] [PubMed] [Google Scholar]
- 7.Falagas ME, Griffiths J, Prekezes J, Worthington M. Cytomegalovirus colitis mimicking colon carcinoma in an HIV-negative patient with chronic renal failure. Am J Gastroenterol. 1996;91:168–169. [PubMed] [Google Scholar]
- 8.Galiatsatos P, Shrier I, Lamoureux E, Szilagyi A. Meta-analysis of outcome of cytomegalovirus colitis in immunocompetent hosts. Dig Dis Sci. 2005;50:609–616. doi: 10.1007/s10620-005-2544-6. [DOI] [PubMed] [Google Scholar]
- 9.Toogood GJ, Gillespie PH, Gujral S, Warren BF, Roake JA, Gray DW, Morris PJ. Cytomegalovirus infection and colonic perforation in renal transplant patients. Transpl Int. 1996;9:248–251. doi: 10.1007/BF00335394. [DOI] [PubMed] [Google Scholar]
- 10.Almeida N, Romãozinho JM, Amaro P, Ferreira M, Cipriano MA, Leitão MC. Fatal mid-gastrointestinal bleeding by cytomegalovirus enteritis in an immunocompetent patient. Acta Gastroenterol Belg. 2009;72:245–248. [PubMed] [Google Scholar]
- 11.Frank D, Raicht RF. Intestinal perforation associated with cytomegalovirus infection in patients with acquired immune deficiency syndrome. Am J Gastroenterol. 1984;79:201–205. [PubMed] [Google Scholar]
- 12.Korkmaz M, Kunefeci G, Selcuk H, Unal H, Gur G, Yilmaz U, Arslan H, Demirhan B, Boyacioglu S, Haberal M. The role of early colonoscopy in CMV colitis of transplant recipients. Transplant Proc. 2005;37:3059–3060. doi: 10.1016/j.transproceed.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Drew WL. Diagnosis of cytomegalovirus infection. Rev Infect Dis. 1988;10 Suppl 3:S468–S476. doi: 10.1093/clinids/10.supplement_3.s468. [DOI] [PubMed] [Google Scholar]
- 14.Kambham N, Vij R, Cartwright CA, Longacre T. Cytomegalovirus infection in steroid-refractory ulcerative colitis: a case-control study. Am J Surg Pathol. 2004;28:365–373. doi: 10.1097/00000478-200403000-00009. [DOI] [PubMed] [Google Scholar]
- 15.Parente F, Cernuschi M, Rizzardini G, Lazzarin A, Valsecchi L, Bianchi Porro G. Opportunistic infections of the esophagus not responding to oral systemic antifungals in patients with AIDS: their frequency and treatment. Am J Gastroenterol. 1991;86:1729–1734. [PubMed] [Google Scholar]
- 16.Gondo H, Minematsu T, Harada M, Akashi K, Hayashi S, Taniguchi S, Yamasaki K, Shibuya T, Takamatsu Y, Teshima T. Cytomegalovirus (CMV) antigenaemia for rapid diagnosis and monitoring of CMV-associated disease after bone marrow transplantation. Br J Haematol. 1994;86:130–137. doi: 10.1111/j.1365-2141.1994.tb03263.x. [DOI] [PubMed] [Google Scholar]
- 17.Kurihara T, Hayashi J, Matusoka T, Ito A. HCMV pp65 antigenemia assay using indirect alkaline phosphatase staining method. Biomed Res. 1995;16:125–129. [Google Scholar]
- 18.Boeckh M, Gooley TA, Myerson D, Cunningham T, Schoch G, Bowden RA. Cytomegalovirus pp65 antigenemia-guided early treatment with ganciclovir versus ganciclovir at engraftment after allogeneic marrow transplantation: a randomized double-blind study. Blood. 1996;88:4063–4071. [PubMed] [Google Scholar]
- 19.Kanda Y, Mineishi S, Saito T, Seo S, Saito A, Suenaga K, Ohnishi M, Niiya H, Nakai K, Takeuchi T, et al. Pre-emptive therapy against cytomegalovirus (CMV) disease guided by CMV antigenemia assay after allogeneic hematopoietic stem cell transplantation: a single-center experience in Japan. Bone Marrow Transplant. 2001;27:437–444. doi: 10.1038/sj.bmt.1702805. [DOI] [PubMed] [Google Scholar]
- 20.Mori T, Okamoto S, Matsuoka S, Yajima T, Wakui M, Watanabe R, Ishida A, Iwao Y, Mukai M, Hibi T, et al. Risk-adapted pre-emptive therapy for cytomegalovirus disease in patients undergoing allogeneic bone marrow transplantation. Bone Marrow Transplant. 2000;25:765–769. doi: 10.1038/sj.bmt.1702227. [DOI] [PubMed] [Google Scholar]
- 21.Fica A, Cervera C, Pérez N, Marcos MA, Ramírez J, Linares L, Soto G, Navasa M, Cofan F, Ricart MJ, et al. Immunohistochemically proven cytomegalovirus end-organ disease in solid organ transplant patients: clinical features and usefulness of conventional diagnostic tests. Transpl Infect Dis. 2007;9:203–210. doi: 10.1111/j.1399-3062.2007.00220.x. [DOI] [PubMed] [Google Scholar]
- 22.Jang EY, Park SY, Lee EJ, Song EH, Chong YP, Lee SO, Choi SH, Woo JH, Kim YS, Kim SH. Diagnostic performance of the cytomegalovirus (CMV) antigenemia assay in patients with CMV gastrointestinal disease. Clin Infect Dis. 2009;48:e121–e124. doi: 10.1086/599116. [DOI] [PubMed] [Google Scholar]
- 23.Boeckh M, Boivin G. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin Microbiol Rev. 1998;11:533–554. doi: 10.1128/cmr.11.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caliendo AM, Schuurman R, Yen-Lieberman B, Spector SA, Andersen J, Manjiry R, Crumpacker C, Lurain NS, Erice A. Comparison of quantitative and qualitative PCR assays for cytomegalovirus DNA in plasma. J Clin Microbiol. 2001;39:1334–1338. doi: 10.1128/JCM.39.4.1334-1338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rene E, Marche C, Chevalier T, Rouzioux C, Regnier B, Saimot AG, Negesse Y, Matheron S, Leport C, Wolff B. Cytomegalovirus colitis in patients with acquired immunodeficiency syndrome. Dig Dis Sci. 1988;33:741–750. doi: 10.1007/BF01540440. [DOI] [PubMed] [Google Scholar]
- 26.Ljungman P, Griffiths P, Paya C. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin Infect Dis. 2002;34:1094–1097. doi: 10.1086/339329. [DOI] [PubMed] [Google Scholar]


