Abstract
AIM: To evaluate the association between p53 codon 72 polymorphism and liver cancer risk by means of meta-analysis.
METHODS: Two investigators independently searched the Medline, Embase and Chinese Biomedicine databases. Summary odds ratios and 95% CI for p53 codon 72 polymorphism and liver cancer were calculated in fixed-effects model (Mantel-Haenszel method) and random-effects model (DerSimonian and Laird method) when appropriate.
RESULTS: This meta-analysis included 1115 liver cancer cases and 1778 controls. The combined results based on all studies showed that there was a statistically significant link between Pro/Pro genotype and liver cancer, but not between Arg/Arg or Pro/Arg genotype and liver cancer. When stratifying for race, similar results were obtained, i.e. patients with liver cancer had a significantly higher frequency of Pro/Pro genotype than non-cancer patients among Asians. After stratifying the various studies by control source, gender, family history of liver cancer and chronic hepatitis virus infection, we found that (1) patients among hospital-based studies had a significantly higher frequency of Pro/Pro and a significantly lower frequency of Arg/Arg genotype than individuals without cancer; (2) female patients with liver cancer had a significantly lower frequency of Arg/Arg and a higher frequency of Pro/Arg+Pro/Pro genotypes than female individuals without cancer; (3) subgroup analyses for family history of liver cancer did not reveal any significant association between p53 codon 72 polymorphism and liver cancer development; and (4) patients with negative hepatitis virus infection had a significantly higher frequency of Pro/Pro and a significantly lower frequency of Arg/Arg genotype than individuals without cancer.
CONCLUSION: This meta-analysis suggests that the p53 codon 72 polymorphism may be associated with liver cancer among Asians.
Keywords: Liver cancer, p53 codon 72, Gene polymorphism, Meta-analysis
INTRODUCTION
Primary liver cancer is the sixth most common cancer in the world and the third most common cause of cancer mortality[1]. A 2005 analysis of the worldwide incidence of and mortality from cancer showed that 626 000 cases of liver cancer occurred in 2002, 82% of which are from developing countries and that 598 000 patients die annually of this disease[1].China alone accounts for 55% liver cancer death worldwide. Moreover, the 5-year survival rate was 8% in the United States during 1988-2001[2], 9% in Europe during 1995-1999[3], and 5% in developing countries in 2002[1]. The major etiologies of hepatocellular carcinoma (HCC) include infection with hepatitis B virus (HBV) and hepatitis C virus (HCV), cigarette smoking, alcohol drinking and aflatoxin B1 (AFB1) exposure[4-9]. However, not all individuals with exposure to the risk factors develop cancer even after a long-term follow-up. The pathogenesis of human HCC is a multistage process with the involvement of a series of genes, including oncogenes and tumor suppressor genes.
The p53 tumor suppressor gene, located on chromosome 17p13, is of critical importance for the regulation of cell cycle and maintenance of genomic integrity. Loss of p53 function has been suggested to be a critical step in multistage hepatocarcinogenesis[10]. A specific p53 mutation at codon 249 in exon 7 was associated with AFB1-induced HCC in certain areas of high AFB1 contamination[11]. The wild-type p53 gene exhibits a polymorphism at codon 72 in exon 4, with a single nucleotide change that causes a substitution of proline for arginine (Arg72Pro)[12]. The polymorphism occurs in the proline-rich domain of p53 protein, which is necessary for the protein to fully induce apoptosis. It is found that in cell lines containing inducible versions of alleles encoding the Pro and Arg variants, the Arg variant induces apoptosis more markedly than the Pro variant[13]. In other words, the two polymorphic variants of p53 are functionally distinct, and these differences may influence cancer risk. The polymorphism consists of a single base pair change of either arginine or proline which creates 3 distinct genotypes: homozygous for arginine (Arg/Arg), homozygous for proline (Pro/Pro) and a heterozygote (Pro/Arg)[14]. p53 codon 72 polymorphisms have been reported to be associated with cancers of the lung[15], esophagus[16], stomach[17], colorectum[18], breast[19], bladder[20] and cervix[21].
In recent years, a number of case-control studies were conducted to investigate the association between p53 codon 72 polymorphism and liver cancer susceptibility in humans. But these studies reported conflicting results. No quantitative summary of the evidence has ever been performed. The purpose of this meta-analysis was to quantitatively summarize the evidence for such a relationship.
MATERIALS AND METHODS
Literature search strategy
Search was applied to the following electronic databases: Medline (from 1966 to September 2010), Embase (from 1950 to September 2010) and Chinese Biomedicine databases (from 1979 to September 2010). The following key words were used: “p53” or “codon 72”, “liver” or “hepatocellular”, “carcinoma” or “cancer” or “tumor”. The search was without restriction in language, but with restriction in the studies conducted in human subjects. The reference lists of reviews and retrieved articles were hand searched at the same time. We did not include abstracts or unpublished reports. If more than one article was published by the same author using the same case series, we selected the study with the largest series.
Inclusion and exclusion criteria
We reviewed abstracts of all citations and retrieved studies. The following criteria were used to include published studies: (1) evaluating the association between p53 codon 72 polymorphism and liver cancer; (2) case-control study; and (3) sufficient genotype data were presented to calculate the odds ratio (OR) with confidence interval (CI). Major reasons for exclusion of studies were: (1) no control; (2) duplicate; and (3) no usable data reported.
Data extraction
All data were extracted independently by two reviewers (Chen X and Liu F) according to the prespecified selection criteria. Disagreement was resolved by discussion. The following data were extracted: the last name of the first author, study design, publication year, statistical methods, ethnicity of the population, genotyping methods, number of liver cancer cases and controls studied and results of studies.
Statistical analysis
The statistical analysis was conducted using STATA 8.2 (Stata Corp LP, College Station, TX, USA), P < 0.05 was considered statistically significant. Dichotomous data were presented as OR with 95% CI. Statistical heterogeneity was measured using the Q statistic test (P < 0.10 was considered statistically significant heterogeneity)[22]. Either a random- effects model (DerSimonian-Laird method[23]) or fixed-effects model (Mantel-Haenszel method[24]) was used to calculate pooled effect estimates in the presence or absence of heterogeneity, respectively. To establish the effect of clinical heterogeneity among the studies on the conclusions of this meta-analysis, subgroup analyses were conducted based on race, study design, gender, chronic hepatitis virus status and family history of liver cancer patients. Several methods were used to assess the potential for publication bias. Visual inspection of funnel plot asymmetry was conducted. The Begg’s rank correlation method[25] and the Egger’s weighted regression method[26] were used to statistically assess publication bias. P < 0.05 was considered statistically significant.
RESULTS
Study characteristics
There were 2248 papers relevant to the search words. Through screening the title and reading the abstract and the entire article, 12 cohort studies were identified[27-38]. Six of them were excluded (four studies reported duplicate data[31-34] and three are not related to liver cancer[27,28,37]). As a result, six studies[29-31,35,36,38] were selected, including 1115 liver cancer cases and 1778 controls. These studies were carried out in China, Spain, Italy, Morocco and Korea. Characteristics of the studies included in the meta-analysis are presented in Table 1.
Table 1.
Characteristics of studies included in the meta-analysis
| First author | Design | Yr | Country | Ethnicity |
Case/control |
Genotyping | HWE | |||
| n | Arg/Arg | Arg/Pro | Pro/Pro | |||||||
| Yu et al[38] | HCC | 1999 | China | Asian | 80/328 | 28/112 | 35/141 | 17/75 | PCR-RFLP | 0.02 |
| Anzola et al[36] | HCC | 2003 | Spain | Caucasian | 97/111 | 46/65 | 47/42 | 4/4 | PCR-SSCP | 0.38 |
| Leveri et al[35] | PCC | 2004 | Italy | Caucasian | 86/254 | 46/122 | 33/113 | 7/19 | PCR-RFLP | 0.30 |
| Zhu et al[31] | HCC | 2005 | China | Asian | 469/567 | 135/197 | 252/284 | 82/86 | PCR-RFLP | 0.32 |
| Ezzikouri et al[30] | PCC | 2007 | Morocco | Caucasian | 96/222 | 52/129 | 31/79 | 13/14 | PCR-RFLP | 0.69 |
| Yoon et al[29] | HCC | 2008 | Korea | Asian | 287/296 | 110/124 | 111/135 | 66/37 | PCR-RFLP | 0.98 |
HCC: Hospital-based case-control; PCC: Population-based case-control; PCR-RFLP: Polymerase chain reaction-restriction fragment length polymorphism; PCR-SSCP: Single strand conformation polymorphism analysis of polymerase chain reaction products; HWE: Hardy-Weinberg equilibrium of genotypes in control group. χ2-test is used, if P > 0.05, frequencies of genotypes in control group was in Hardy-Weinberg equilibrium.
Quantitative data synthesis
The combined results based on all studies showed that there was a statistically significant link between Pro/Pro genotype and liver cancer (OR = 1.38, 95% CI: 1.11-1.72, P = 0.004), but not between Arg/Arg or Pro/Arg and liver cancer (Arg/Arg, OR = 0.85, 95% CI: 0.72-1.00; Pro/Arg, OR = 0.99, 95% CI: 0.85-1.16). When stratifying for race, similar results were obtained, i.e. patients with liver cancer had a significantly higher frequency of Pro/Pro (OR = 1.35, 95% CI: 1.06-1.71, P = 0.014) genotype than non-cancer patients among Asians (Figure 1).
Figure 1.
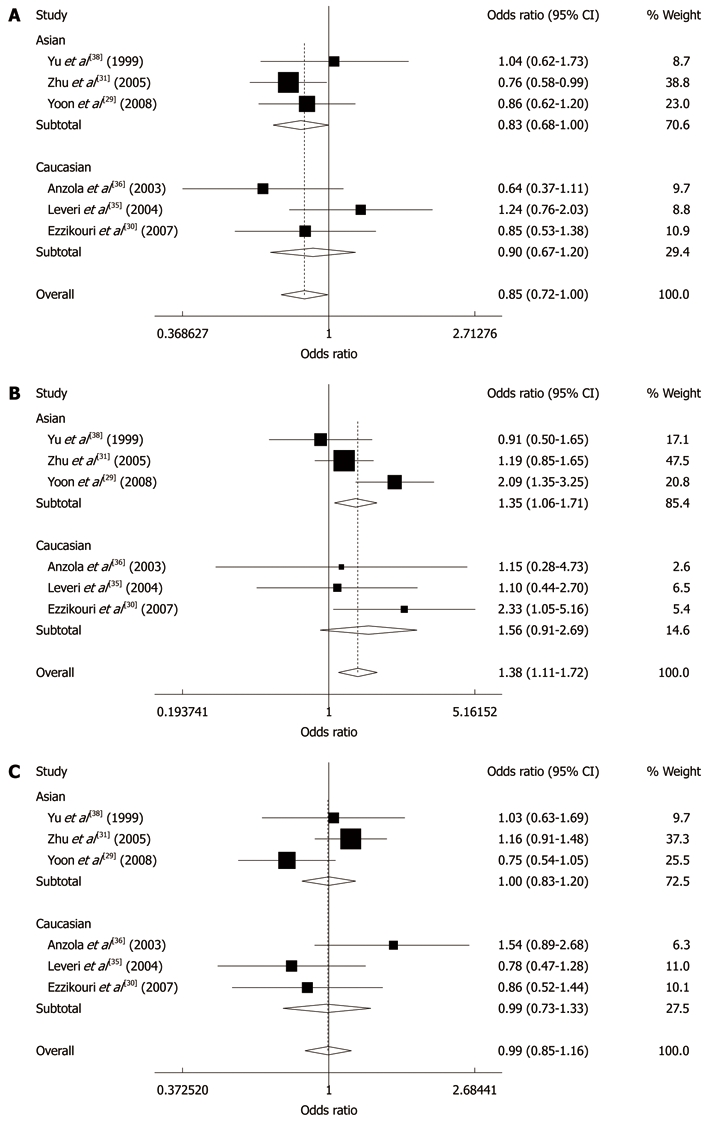
Meta-analysis of p53 codon 72 Arg/Arg (A), Pro/Pro (B) and Pro/Arg (C) and liver cancer risk.
When stratifying by control source, we found that patients among hospital-based studies had a significantly higher frequency of Pro/Pro (OR = 1.34, 95% CI: 1.06-1.70, P = 0.014) and a significantly lower frequency of Arg/Arg (OR = 0.80, 95% CI: 0.67-0.96, P = 0.018) genotype than patients without cancer, but not in population-based studies. When stratifying for gender, we found that female patients with liver cancer had a significantly lower frequency of Arg/Arg (OR = 0.49, 95% CI: 0.26-0.94, P = 0.031) and a higher frequency of Pro/Arg+Pro/Pro (OR = 2.03, 95% CI: 1.07-3.85, P = 0.031) genotypes than female individuals without cancer. When we stratified the various studies by family history of liver cancer, no statistically significant results were observed for all the analyses. When stratifying by chronic hepatitis virus status, we found that patients with negative hepatitis virus infection had a significantly higher frequency of Pro/Pro (OR = 2.07, 95% CI: 1.29-3.30, P = 0.002) and a significantly lower frequency of Arg/Arg (OR = 0.55, 95% CI: 0.32-0.94, P = 0.028) genotype than individuals without cancer, but not in patients with positive hepatitis virus infection (Table 2).
Table 2.
Meta-analysis of p53 codon 72 polymorphism and liver cancer, odds ratio (95% CI)
| Subgroups | Arg/Arg | P value | Pro/Pro | P value | Pro/Arg | P value | Pro/Arg + Pro/Pro | P value |
| Race | ||||||||
| Asian | 0.83 (0.68-1.00) | 0.543 | 1.35 (1.06-1.71) | 0.048 | 1.00 (0.83-1.20) | 0.120 | NA | NA |
| Caucasian | 0.90 (0.67-1.20) | 0.199 | 1.56 (0.91-2.69) | 0.420 | 0.99 (0.73-1.33) | 0.160 | NA | NA |
| Gender | ||||||||
| Male | 0.72 (0.47-1.09) | 0.023 | NA | NA | NA | NA | 1.39 (0.91-2.12) | 0.023 |
| Female | 0.49 (0.26-0.94) | 0.862 | NA | NA | NA | NA | 2.03 (1.07-3.85) | 0.862 |
| Control source | ||||||||
| HCC | 0.80 (0.67-0.96) | 0.575 | 1.34 (1.06-1.70) | 0.106 | 1.04 (0.88-1.24) | 0.094 | NA | NA |
| PCC | 1.03 (0.73-1.45) | 0.280 | 1.65 (0.92-2.79) | 0.220 | 0.82 (0.57-1.17) | 0.772 | NA | NA |
| Family history | ||||||||
| Yes | 0.32 (0.07-1.48) | 0.667 | NA | NA | NA | NA | 3.08 0.67-14.08) | 0.667 |
| No | 0.72 (0.28-1.81) | 0.013 | NA | NA | NA | NA | 1.39 (0.55-3.53) | 0.013 |
| Hepatitis virus infection | ||||||||
| Positive | 1.08 (0.75-1.56) | 0.980 | 0.90 (0.56-1.44) | 0.459 | 0.99 (0.70-1.40) | 0.550 | NA | NA |
| Negative | 0.55 (0.32-0.94) | 0.204 | 2.07 (1.29-3.30) | 0.373 | 1.19 (0.83-1.71) | 0.338 | NA | NA |
NA: Due to lack of data, meta-analyses cannot be performed. HCC: Hospital based case-control studies; PCC: Population based case-control studies. P value for heterogeneity. If P < 0.10, random effect model was used; otherwise, fixed effect model was used.
Heterogeneity and publication bias
No statistically significant heterogeneity was observed among trials for all the analyses with the Q statistic (Arg/Arg P = 0.458; Pro/Pro P = 0.152; Pro/Arg P = 0.161). In addition, L’Abbe plots did not show evidence of heterogeneity (Figure 2A). Review of funnel plots could not rule out the potential for publication bias for all the analyses. Publication bias was not evident when the Begg rank correlation method (Arg/Arg P = 1.00; Pro/Pro P = 1.00; Pro/Arg P = 0.707) and the Egger weighted regression method (Arg/Arg P = 0.440; Pro/Pro P = 0.995; Pro/Arg P = 0.818) were used (Figure 2B and C).
Figure 2.
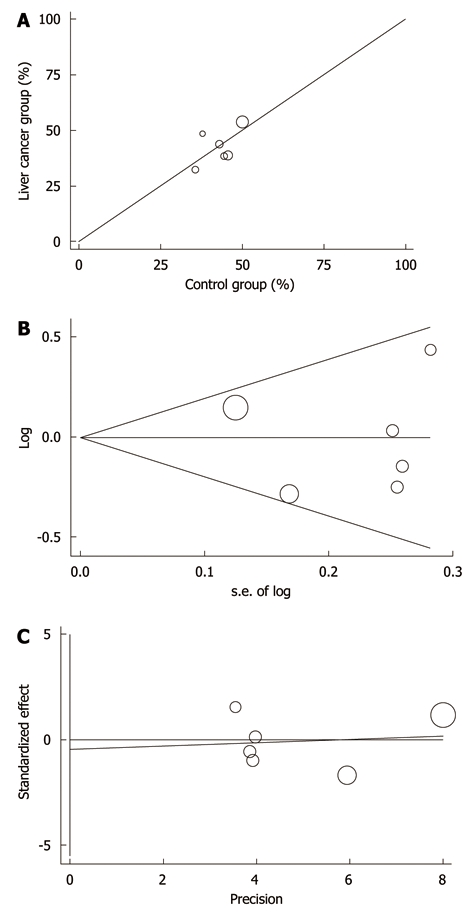
L’Abbe plots (A), Begg’s funnel plot (B) and Egger’s publication bias plot (C) of p53 codon 72 polymorphism and liver cancer risk.
DISCUSSION
Although many environmental factors are found to correlate with the tumorigenesis of liver cancer[4-9], the risk factors still need to be further elucidated. It has been recognized that the most important risk factor for the development of HCC is cirrhosis[39]. Chronic infections with HBV and HCV are the most frequent causes of cirrhosis worldwide. A large number of cohort and case-control studies have shown that alcohol consumption causes liver cirrhosis and is an independent risk factor for primary liver cancer[6,40,41]. Epidemiological studies reported elevated HCC risks associated with exposure to aflatoxins after adjustment for HBV exposure[42]. Cigarette smoking has been causally associated with the risk of HCC[6,43]. However, there are a portion of patients without known risk factors who eventually developed liver cancer[44]. Previous studies had shown an interaction of environmental factors and genetic predisposition in the development of liver cancer[31,38]. Therefore, genetic predisposition may contribute to the process of tumorigenesis.
A genetic predisposition to liver cancer has been suggested by many studies[45-47]. Recent studies suggest that single nucleotide polymorphism may be related to the tumorigenesis of liver cancer[48,49]. The p53 gene and its encoded protein controls cell cycle, cell growth and apoptosis, which has a common polymorphism at codon 72 of exon 4 that encodes either Pro or Arg. Until recently, a number of studies have been conducted to find the relationship between p53 codon 72 polymorphism and liver cancer risk. Most of these studies were based on small sample sizes. Moreover, there are still some conflicting results. As a powerful statistical method, meta-analysis can provide a quantitative approach for pooling the results of different researches on the same topic, and for estimating and explaining their diversity[50,51].
We found that Pro/Pro genotype had a 1.38-fold statistically significant increased risk of liver cancer in this meta-analysis. When stratifying for race, patients with liver cancer had a significantly higher frequency of Pro/Pro genotype than individuals without cancer among Asians. When stratifying the various studies by control source, gender, family history of liver cancer and chronic hepatitis virus infection, we found that (1) patients in hospital-based studies had a significantly higher frequency of Pro/Pro and a significantly lower frequency of Arg/Arg genotype than patients without cancer; (2) female patients with liver cancer had a significantly lower frequency of Arg/Arg and a higher frequency of Pro/Arg+Pro/Pro genotypes than female individuals without cancer; (3) subgroup analyses for family history of liver cancer did not reveal any significant association between p53 codon 72 polymorphism and liver cancer development; and (4) patients with negative hepatitis virus infection had a significantly higher frequency of Pro/Pro and a significantly lower frequency of Arg/Arg genotype than individuals without cancer.
A number of studies have shown significant differences in the biochemical properties of the p53 protein, depending on the particular polymorphic form. It has been shown that the Arg/Arg and Pro/Pro variants differ in binding activity, transcriptional activation, apoptosis induction and cell cycle arrest[13,52]. The p53 Arg variant induces apoptosis faster and more efficiently than the p53 Pro variant[13]. One explanation of such higher apoptotic potential is the greater ability of the Arg variant to localize to the mitochondria; this localization is accompanied by the proapoptotic release of cytochrome C into the cytosol[13]. In addition, p53 Arg72 is more active than p53 Pro72 in the induction of apoptosis through a transcription-dependant pathway. Pim et al[53] also found that the Arg72 form of p53 is significantly more efficient than the Pro72 form in inducing apoptosis. In contrast, the Pro72 form appears to induce a higher level of G1 arrest than the Arg72 form. These data indicate that the two polymorphic variants of p53 are functionally distinct, and these differences may influence cancer risk. From our meta-analyses, we found that patients with liver cancer had a significantly higher frequency of Pro/Pro than non-cancer patients (P = 0.004), which can be explained by the points of view mentioned above.
Another major finding of this study was the different associations of p53 codon 72 gene polymorphism with the risk of liver cancer based on race. In fact, race-specific variation in the distribution of genotypes in the p53 codon 72 polymorphism has been demonstrated[54]. Because race may be related to the disease, either through common risk factors or other genes in linkage disequilibrium with p53, confounding by race, or population stratification, may lead to result bias in studies conducted on ethnically diverse populations that did not account for possible confounding[55]. In this subgroup analysis, the frequency of Pro/Pro genotype showed distinct differences among Asians and Caucasians. The pooled OR associated with p53 codon 72 gene polymorphism was statistically significant among Asians, but not in Caucasians. The discrepancy might be due to genetic background and/or environmental exposure differences.
Results of meta-analyses often depend on control selection procedures[56]. Arg/Arg and Pro/Pro genotype frequency might be different between the two control sources (hospital-based and population-based) (Table 1). In subgroup analysis stratified by the different study designs, the hospital-based controls resulted in a significantly stronger association between p53 Arg72 Pro polymorphism and development of liver cancer than population-based controls.
It is widely accepted that family history of liver cancer and chronic hepatitis virus infection are obvious risk factors for development of liver cancer. By pooling the available data that evaluated associations and interaction between p53 Arg72 Pro genotype and family history of liver cancer risk, the p53 Arg72 Pro genotype was not found to be associated with increased risk of liver cancer in those either with or without family history of liver cancer. Interestingly, we found that patients with negative hepatitis virus infection were at higher risk for liver cancer than patients with positive hepatitis virus infection. One explanation for the preferentially increased liver cancer risk of the p53 Arg72Pro polymorphism among hepatitis virus-negative but not hepatitis virus-positive subjects, is that the effect of the Pro allele may be concealed by chronic HBV infection since the relative risk of HCC among chronic HBV carriers is 10-200-folds higher than among non-carriers[57,58]. Moreover, we demonstrated that there is an association between p53 Arg72Pro and enhanced risk of liver cancer in female patients. Such differences between men and women have already been reported in colorectal cancer, which were explained by exogenous hormones intake[18]. However, because of the limited study sample size, these results should be interpreted with caution.
However, there are still some limitations in this meta-analysis: (1) only published studies were included in the meta-analysis; therefore, publication bias may have occurred, even though the use of a statistical test did not show it; (2) these results should be interpreted with caution because the population from five countries and controls were not uniform; (3) the number of cases and controls in the included studies was low; and (4) meta-analysis is a retrospective research that is subject to the methodological limitations. In order to minimize the bias, we developed a detailed protocol before initiating the study, and performed a meticulous search for published studies using explicit methods for study selection, data extraction and data analysis. Nevertheless, our results still should be interpreted with caution.
In conclusion, this meta-analysis suggests that the p53 codon 72 polymorphism may be associated with liver cancer, and that difference in genotype distribution may be associated with race, gender and chronic hepatitis virus status of patients. Due to limited number of cases in this analysis, it is critical that larger and well-designed multi-center studies based on the same ethnic group are needed to confirm our results.
COMMENTS
Background
The wild-type p53 gene exhibits a polymorphism at codon 72 in exon 4, with a single nucleotide change that causes a substitution of proline for arginine (Arg72Pro). This change has been implicated as a risk factor for liver cancer, but individual studies have been inconclusive or controversial. The aim of this meta-analysis was to clarify the effect of p53 Arg72Pro polymorphism on the risk of liver cancer.
Research frontiers
There have been many studies on the association between p53 genetic polymorphism Arg72Pro and liver cancer risk, but no meta-analysis has been conducted.
Innovations and breakthroughs
To the best of our knowledge, this is the first systematic review that has investigated the association between p53 codon 72 polymorphisms and liver cancer. This meta-analysis revealed that the p53 codon 72 polymorphism may be associated with liver cancer among Asians.
Applications
It can be seen from this paper that p53 polymorphism Arg72Pro could alter the susceptibility to liver cancer. It suggests that, even a common variant in the functional region of a definitively meaningful gene had an effect on human diseases, such as cancer.
Terminology
Meta-analysis is a means of increasing the effective sample size under investigation through the pooling of data from individual association studies, thus enhancing the statistical power of the analysis.
Peer review
The authors analyzed the association between p53 codon 72 polymorphism and liver cancer susceptibility in humans through this meta-analysis, and quantitatively summarized the evidence for such a relationship. They found that patients with liver cancer had a significantly higher frequency of Pro/Pro than non-cancer patients among Asians. Female patients with liver cancer had a significantly lower frequency of Arg/Arg and a higher frequency of Pro/Arg+Pro/Pro than female individuals without cancer.
Footnotes
Peer reviewer: Lisa J Herrinton, PhD, Division of Research, Kaiser Permanente, Kaiser Permanente, 2000 Broadway Avenue, Oakland, CA 94612, United States
S- Editor Tian L L- Editor Ma JY E- Editor Zheng XM
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Surveillance, Epidemiology, and End Results (SEER) Program (www. seer.cancer. gov) SEER*Stat Database: Incidence - SEER 17 Regs Limited-Use, Nov; 2006. pp. Sub (1973–2004 varying), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2007, based on the November 2006 submission. [Google Scholar]
- 3.Berrino F, De Angelis R, Sant M, Rosso S, Bielska-Lasota M, Coebergh JW, Santaquilani M. Survival for eight major cancers and all cancers combined for European adults diagnosed in 1995-99: results of the EUROCARE-4 study. Lancet Oncol. 2007;8:773–783. doi: 10.1016/S1470-2045(07)70245-0. [DOI] [PubMed] [Google Scholar]
- 4.Tang ZY. Hepatocellular carcinoma--cause, treatment and metastasis. World J Gastroenterol. 2001;7:445–454. doi: 10.3748/wjg.v7.i4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu ZZ, Cong WM. [Roles of hepatitis B virus and hepatitis C virus in hepato-carcinogenesis] Zhonghua Ganzangbing Zazhi. 2003;11:574–576. [PubMed] [Google Scholar]
- 6.Kuper H, Tzonou A, Kaklamani E, Hsieh CC, Lagiou P, Adami HO, Trichopoulos D, Stuver SO. Tobacco smoking, alcohol consumption and their interaction in the causation of hepatocellular carcinoma. Int J Cancer. 2000;85:498–502. [PubMed] [Google Scholar]
- 7.Chen CJ, Wang LY, Lu SN, Wu MH, You SL, Zhang YJ, Wang LW, Santella RM. Elevated aflatoxin exposure and increased risk of hepatocellular carcinoma. Hepatology. 1996;24:38–42. doi: 10.1002/hep.510240108. [DOI] [PubMed] [Google Scholar]
- 8.Ross RK, Yuan JM, Yu MC, Wogan GN, Qian GS, Tu JT, Groopman JD, Gao YT, Henderson BE. Urinary aflatoxin biomarkers and risk of hepatocellular carcinoma. Lancet. 1992;339:943–946. doi: 10.1016/0140-6736(92)91528-g. [DOI] [PubMed] [Google Scholar]
- 9.Qian GS, Ross RK, Yu MC, Yuan JM, Gao YT, Henderson BE, Wogan GN, Groopman JD. A follow-up study of urinary markers of aflatoxin exposure and liver cancer risk in Shanghai, People's Republic of China. Cancer Epidemiol Biomarkers Prev. 1994;3:3–10. [PubMed] [Google Scholar]
- 10.Staib F, Hussain SP, Hofseth LJ, Wang XW, Harris CC. TP53 and liver carcinogenesis. Hum Mutat. 2003;21:201–216. doi: 10.1002/humu.10176. [DOI] [PubMed] [Google Scholar]
- 11.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 12.Buchman VL, Chumakov PM, Ninkina NN, Samarina OP, Georgiev GP. A variation in the structure of the protein-coding region of the human p53 gene. Gene. 1988;70:245–252. doi: 10.1016/0378-1119(88)90196-5. [DOI] [PubMed] [Google Scholar]
- 13.Dumont P, Leu JI, Della Pietra AC 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 14.Shepherd T, Tolbert D, Benedetti J, Macdonald J, Stemmermann G, Wiest J, DeVoe G, Miller MA, Wang J, Noffsinger A, et al. Alterations in exon 4 of the p53 gene in gastric carcinoma. Gastroenterology. 2000;118:1039–1044. doi: 10.1016/s0016-5085(00)70356-8. [DOI] [PubMed] [Google Scholar]
- 15.Matakidou A, Eisen T, Houlston RS. TP53 polymorphisms and lung cancer risk: a systematic review and meta-analysis. Mutagenesis. 2003;18:377–385. doi: 10.1093/mutage/geg008. [DOI] [PubMed] [Google Scholar]
- 16.Lee JM, Shun CT, Wu MT, Chen YY, Yang SY, Hung HI, Chen JS, Hsu HH, Huang PM, Kuo SW, et al. The associations of p53 overexpression with p53 codon 72 genetic polymorphism in esophageal cancer. Mutat Res. 2006;594:181–188. doi: 10.1016/j.mrfmmm.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Hiyama T, Tanaka S, Kitadai Y, Ito M, Sumii M, Yoshihara M, Shimamoto F, Haruma K, Chayama K. p53 Codon 72 polymorphism in gastric cancer susceptibility in patients with Helicobacter pylori-associated chronic gastritis. Int J Cancer. 2002;100:304–308. doi: 10.1002/ijc.10483. [DOI] [PubMed] [Google Scholar]
- 18.Koushik A, Tranah GJ, Ma J, Stampfer MJ, Sesso HD, Fuchs CS, Giovannucci EL, Hunter DJ. p53 Arg72Pro polymorphism and risk of colorectal adenoma and cancer. Int J Cancer. 2006;119:1863–1868. doi: 10.1002/ijc.22057. [DOI] [PubMed] [Google Scholar]
- 19.Tommiska J, Eerola H, Heinonen M, Salonen L, Kaare M, Tallila J, Ristimäki A, von Smitten K, Aittomäki K, Heikkilä P, et al. Breast cancer patients with p53 Pro72 homozygous genotype have a poorer survival. Clin Cancer Res. 2005;11:5098–5103. doi: 10.1158/1078-0432.CCR-05-0173. [DOI] [PubMed] [Google Scholar]
- 20.Soulitzis N, Sourvinos G, Dokianakis DN, Spandidos DA. p53 codon 72 polymorphism and its association with bladder cancer. Cancer Lett. 2002;179:175–183. doi: 10.1016/s0304-3835(01)00867-9. [DOI] [PubMed] [Google Scholar]
- 21.Koushik A, Platt RW, Franco EL. p53 codon 72 polymorphism and cervical neoplasia: a meta-analysis review. Cancer Epidemiol Biomarkers Prev. 2004;13:11–22. doi: 10.1158/1055-9965.epi-083-3. [DOI] [PubMed] [Google Scholar]
- 22.Higgins JPT, Green S. Cochrane handbook for systematic reviews of interventions 4.2.6. (updated September 2006) Chichester, UK: John Wiley & Sons, Ltd; 2006. [Google Scholar]
- 23.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 24.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 25.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 26.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu ZZ, Liu B, Wang AZ, Jia HR, Jin XX, He XL, Hou LF, Zhu GS. Association of p53 codon 72 polymorphism with liver metastases of colorectal cancers positive for p53 overexpression. J Zhejiang Univ Sci B. 2008;9:847–852. doi: 10.1631/jzus.B0820100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Etienne-Grimaldi MC, Formento JL, Francoual M, François E, Formento P, Renée N, Laurent-Puig P, Chazal M, Benchimol D, Delpero JR, et al. K-Ras mutations and treatment outcome in colorectal cancer patients receiving exclusive fluoropyrimidine therapy. Clin Cancer Res. 2008;14:4830–4835. doi: 10.1158/1078-0432.CCR-07-4906. [DOI] [PubMed] [Google Scholar]
- 29.Yoon YJ, Chang HY, Ahn SH, Kim JK, Park YK, Kang DR, Park JY, Myoung SM, Kim do Y, Chon CY, et al. MDM2 and p53 polymorphisms are associated with the development of hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Carcinogenesis. 2008;29:1192–1196. doi: 10.1093/carcin/bgn090. [DOI] [PubMed] [Google Scholar]
- 30.Ezzikouri S, El Feydi AE, Chafik A, Benazzouz M, El Kihal L, Afifi R, Hassar M, Pineau P, Benjelloun S. The Pro variant of the p53 codon 72 polymorphism is associated with hepatocellular carcinoma in Moroccan population. Hepatol Res. 2007;37:748–754. doi: 10.1111/j.1872-034X.2007.00126.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhu ZZ, Cong WM, Zhu GS, Liu SF, Xian ZH, Wu WQ, Zhang XZ, Wang YH, Wu MC. [Association of p53 codon 72 polymorphism with genetic susceptibility to hepatocellular carcinoma in Chinese population] Zhonghua Yixue Yichuanxue Zazhi. 2005;22:632–635. [PubMed] [Google Scholar]
- 32.Zhu ZZ, Cong WM, Liu SF, Xian ZH, Wu WQ, Wu MC, Gao B, Hou LF, Zhu GS. A p53 polymorphism modifies the risk of hepatocellular carcinoma among non-carriers but not carriers of chronic hepatitis B virus infection. Cancer Lett. 2005;229:77–83. doi: 10.1016/j.canlet.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 33.Zhu ZZ, Cong WM, Liu SF, Dong H, Zhu GS, Wu MC. [p53 gene codon 72 polymorphism and genetic susceptibility to hepatocellular carcinoma in Chinese population] Zhonghua Yixue Zazhi. 2005;85:76–79. [PubMed] [Google Scholar]
- 34.Zhu ZZ, Cong WM, Liu SF, Dong H, Zhu GS, Wu MC. Homozygosity for Pro of p53 Arg72Pro as a potential risk factor for hepatocellular carcinoma in Chinese population. World J Gastroenterol. 2005;11:289–292. doi: 10.3748/wjg.v11.i2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leveri M, Gritti C, Rossi L, Zavaglia C, Civardi E, Mondelli MU, De Silvestri A, Silini EM. Codon 72 polymorphism of P53 gene does not affect the risk of cirrhosis and hepatocarcinoma in HCV-infected patients. Cancer Lett. 2004;208:75–79. doi: 10.1016/j.canlet.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 36.Anzola M, Cuevas N, López-Martínez M, Saiz A, Burgos JJ, de Pancorbo MM. Frequent loss of p53 codon 72 Pro variant in hepatitis C virus-positive carriers with hepatocellular carcinoma. Cancer Lett. 2003;193:199–205. doi: 10.1016/s0304-3835(03)00046-6. [DOI] [PubMed] [Google Scholar]
- 37.Chen WC, Tsai FJ, Wu JY, Wu HC, Lu HF, Li CW. Distributions of p53 codon 72 polymorphism in bladder cancer--proline form is prominent in invasive tumor. Urol Res. 2000;28:293–296. doi: 10.1007/s002400000117. [DOI] [PubMed] [Google Scholar]
- 38.Yu MW, Yang SY, Chiu YH, Chiang YC, Liaw YF, Chen CJ. A p53 genetic polymorphism as a modulator of hepatocellular carcinoma risk in relation to chronic liver disease, familial tendency, and cigarette smoking in hepatitis B carriers. Hepatology. 1999;29:697–702. doi: 10.1002/hep.510290330. [DOI] [PubMed] [Google Scholar]
- 39.Collier J, Sherman M. Screening for hepatocellular carcinoma. Hepatology. 1998;27:273–278. doi: 10.1002/hep.510270140. [DOI] [PubMed] [Google Scholar]
- 40.Ogimoto I, Shibata A, Kurozawa Y, Nose T, Yoshimura T, Suzuki H, Iwai N, Sakata R, Fujita Y, Ichikawa S, et al. Risk of death due to hepatocellular carcinoma among drinkers and ex-drinkers. Univariate analysis of JACC study data. Kurume Med J. 2004;51:59–70. doi: 10.2739/kurumemedj.51.59. [DOI] [PubMed] [Google Scholar]
- 41.Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Bouvard V, Altieri A, Cogliano V. Carcinogenicity of alcoholic beverages. Lancet Oncol. 2007;8:292–293. doi: 10.1016/s1470-2045(07)70099-2. [DOI] [PubMed] [Google Scholar]
- 42.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Some traditional herbal medicines, some mycotoxins, naphthalene and styrene. IARC Monogr Eval Carcinog Risks Hum. 2002;82:1–556. [PMC free article] [PubMed] [Google Scholar]
- 43.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Tobacco smoke and involuntary smoking. IARC Monogr Eval Carcinog Risks Hum. 2004;83:1–1438. [PMC free article] [PubMed] [Google Scholar]
- 44.El-Serag HB, Mason AC. Risk factors for the rising rates of primary liver cancer in the United States. Arch Intern Med. 2000;160:3227–3230. doi: 10.1001/archinte.160.21.3227. [DOI] [PubMed] [Google Scholar]
- 45.Feo F, Frau M, Tomasi ML, Brozzetti S, Pascale RM. Genetic and epigenetic control of molecular alterations in hepatocellular carcinoma. Exp Biol Med (Maywood) 2009;234:726–736. doi: 10.3181/0901-MR-40. [DOI] [PubMed] [Google Scholar]
- 46.Dragani TA. Risk of HCC: genetic heterogeneity and complex genetics. J Hepatol. 2010;52:252–257. doi: 10.1016/j.jhep.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 47.Shih WL, Yu MW, Chen PJ, Wu TW, Lin CL, Liu CJ, Lin SM, Tai DI, Lee SD, Liaw YF. Evidence for association with hepatocellular carcinoma at the PAPSS1 locus on chromosome 4q25 in a family-based study. Eur J Hum Genet. 2009;17:1250–1259. doi: 10.1038/ejhg.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ezzikouri S, El Feydi AE, Benazzouz M, Afifi R, El Kihal L, Hassar M, Akil A, Pineau P, Benjelloun S. Single nucleotide polymorphism in DNMT3B promoter and its association with hepatocellular carcinoma in a Moroccan population. Infect Genet Evol. 2009;9:877–881. doi: 10.1016/j.meegid.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 49.Kim YJ, Lee HS. [Genetic epidemiological study on single nucleotide polymorphisms associated with hepatocellular carcinoma in patients with chronic HBV infection] Korean J Hepatol. 2009;15:7–14. doi: 10.3350/kjhep.2009.15.1.7. [DOI] [PubMed] [Google Scholar]
- 50.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 51.Munafò M. Replication validity of genetic association studies of smoking behavior: what can meta-analytic techniques offer? Nicotine Tob Res. 2004;6:381–382. doi: 10.1080/14622200410001676369. [DOI] [PubMed] [Google Scholar]
- 52.Bergamaschi D, Samuels Y, Sullivan A, Zvelebil M, Breyssens H, Bisso A, Del Sal G, Syed N, Smith P, Gasco M, et al. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat Genet. 2006;38:1133–1141. doi: 10.1038/ng1879. [DOI] [PubMed] [Google Scholar]
- 53.Pim D, Banks L. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int J Cancer. 2004;108:196–199. doi: 10.1002/ijc.11548. [DOI] [PubMed] [Google Scholar]
- 54.Själander A, Birgander R, Kivelä A, Beckman G. p53 polymorphisms and haplotypes in different ethnic groups. Hum Hered. 1995;45:144–149. doi: 10.1159/000154275. [DOI] [PubMed] [Google Scholar]
- 55.Thomas DC, Witte JS. Point: population stratification: a problem for case-control studies of candidate-gene associations? Cancer Epidemiol Biomarkers Prev. 2002;11:505–512. [PubMed] [Google Scholar]
- 56.Benhamou S, Lee WJ, Alexandrie AK, Boffetta P, Bouchardy C, Butkiewicz D, Brockmöller J, Clapper ML, Daly A, Dolzan V, et al. Meta- and pooled analyses of the effects of glutathione S-transferase M1 polymorphisms and smoking on lung cancer risk. Carcinogenesis. 2002;23:1343–1350. doi: 10.1093/carcin/23.8.1343. [DOI] [PubMed] [Google Scholar]
- 57.Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 58.Yang HI, Lu SN, Liaw YF, You SL, Sun CA, Wang LY, Hsiao CK, Chen PJ, Chen DS, Chen CJ. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med. 2002;347:168–174. doi: 10.1056/NEJMoa013215. [DOI] [PubMed] [Google Scholar]


