Abstract
AIM: To investigate the association between TP53 Arg72Pro polymorphism and esophageal cancer (EC) risk using meta-analysis.
METHODS: All eligible studies published before March 1, 2010 were selected by searching PubMed using keywords “p53” or “TP53”, “polymorphism” or “variation”, “esophageal” and “cancer” or “carcinoma”. Crude odds ratios (ORs) with 95% confidence intervals (CIs) were assessed for EC risk associated with TP53 Arg72Pro polymorphism using fixed- and random-effects models.
RESULTS: Nine case-control studies involving 5545 subjects were included in this meta-analysis. Significantly reduced risk of EC was associated with TP53 genotypes for Arg/Arg + Arg/Pro vs Pro/Pro (OR = 0.73, 95% CI: 0.57-0.94, P = 0.014). Subgroup analyses according to the source of controls and the specimens used for determining TP53 Arg72Pro genotypes or sample size showed that significantly reduced risk was observed only in studies which have population-based controls (Arg/Arg vs Pro/Pro: OR = 0.56, 95% CI: 0.47-0.66, P < 0.001), and use white blood cells or normal tissue to assess TP53 genotypes of cases (Arg/Arg vs Pro/Pro: OR = 0.56, 95% CI: 0.47-0.65, P < 0.001) or include at least 200 subjects (Arg/Arg vs Pro/Pro: OR = 0.56, 95% CI: 0.47-0.65, P < 0.001). Analysis restricted to well-designed studies also supported the significantly decreased risk of EC (Arg/Arg vs Pro/Pro: OR = 0.54, 95% CI: 0.46-0.64, P < 0.001).
CONCLUSION: TP53 Arg72 carriers are significantly associated with decreased EC risk. Nevertheless, more well-designed studies are needed to confirm our findings.
Keywords: TP53, Codon 72, Polymorphism, Esophageal cancer, Meta-analysis
INTRODUCTION
Esophageal cancer (EC) is the eighth most common cancer and sixth most deadly cancer worldwide. China and southern and eastern Africa are the relatively high risk areas[1,2]. There are two main forms of EC histologically: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EA). ESCC constitutes the majority (over 90%) of EC, but the incidence rates of EA have sharply increased in many Western countries recently[3-5]. The development of EC is a multifactorial process associated with a variety of risk factors. The two major risk factors of EC are tobacco smoking and alcohol drinking[6-8]. Inherited predisposition may also explain the high rates of EC[9].
TP53 is a major regulator of the cell response to stress and serves as a tumor suppressor by inducing cell cycle arrest or apoptosis[10]. Inactivation of the TP53 signaling pathway has been seen in most human cancers[11]. Previously, polymorphisms of TP53 have been reported to be the possible risk factors for some kinds of tumors[12]. The most common polymorphism of TP53 is at the 72nd amino acid residue, with an arginine (Arg) to proline (Pro) change because of a G→C transverse[13]. Differences in the biochemical or biological characteristics of these wild-type TP53 variants have been reported[14]. The Arg72 variant can better induce apoptosis than the Pro72 variant, indicating that the two polymorphic variants of TP53 are functionally distinct, which may influence the cancer risk or treatment[15].
A number of studies have reported the role of TP53 Arg72Pro polymorphism in cancers such as cervical cancer[16], lung cancer[17], breast cancer[18], and gastric cancer[19], but little is known about the association of TP53 polymorphism with EC. In recent years, several studies focused on the association between TP53 Arg72Pro polymorphism and EC susceptibility, with inconsistent results[20-29]. Hence, we performed a meta-analysis of all eligible studies to estimate the association between TP53 polymorphism and the risk of EC.
MATERIALS AND METHODS
Publication search
We searched the articles using the terms “p53” or “TP53”, “polymorphism” or “variation”, “esophageal” and “cancer” or “carcinoma” in Medline database utilizing the PubMed engine, and all eligible studies were published before March 1, 2010. We evaluated all associated publications to retrieve the most eligible literatures. Their reference lists were hand-searched to find other relevant publications. Articles were limited to English language papers.
Inclusion and exclusion criteria
The following inclusion criteria were used to select literatures for the meta-analysis: (1) published in peer-reviewed journals; (2) articles about TP53 Arg72Pro polymorphism and risk of EC; and (3) containing useful genotype frequencies. The exclusion criteria were: (1) none-case-control studies; (2) control population including malignant tumor patients; (3) the genotype frequencies of control group departing from Hardy-Weinberg equilibrium (HWE); and (4) duplicated publications.
Data extraction
Two investigators (Jiang and Yao) reviewed and extracted information from all eligible publications independently, according to the inclusion and exclusion criteria listed above. An agreement was reached by discussion between the two reviewers whenever there was a conflict. The following items were collected from each study: first author’s surname, year of publication, country of origin, ethnicity, source of controls, specimens used for assessment of TP53 Arg72Pro genotypes, total number of cases and controls as well as numbers of cases and controls with Arg/Arg, Arg/Pro and Pro/Pro genotypes, respectively.
Statistic analysis
Crude odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the association between TP53 Arg72Pro polymorphism and EC risk. The pooled ORs were performed for homozygote comparison (Arg/Arg vs Pro/Pro), dominant model (Arg/Arg + Arg/Pro vs Pro/Pro), and recessive model (Arg/Arg vs Arg/Pro + Pro/Pro), respectively. Stratified analyses were performed based on the source of controls, the specimens used for determining TP53 Arg72Pro genotypes and sample size (cases and controls in total). A Chi-square-based Q-test was performed to check the heterogeneity[30]. If P ≥ 0.1 was obtained in the heterogeneity test, ORs were pooled according to the fixed-effects model (the Mantel-Haenszel model)[31], otherwise the random-effects model (the DerSimonian and Laird model) was used[32]. One-way sensitivity analyses were performed to evaluate the stability of the meta-analysis results[33]. The potential publication bias was estimated using Egger’s linear regression test by visual inspection of the Funnel plot. P < 0.05 was considered statistically significant in publication bias[34]. If publication bias existed, the Duval and Tweedie non-parametric “trim and fill” method was used to adjust for it[35]. All statistical tests were performed with the software STATA version 10.0 (Stata Corporation, College station, TX).
RESULTS
Study characteristics
Eighteen studies were identified through literature search and selection based on the inclusion criteria. By the extraction of data, seven articles which are not case-control studies and one review article were excluded. Among the remaining 10 studies, one study[29] was excluded due to the genotype frequencies of controls deviated from HWE. In one study[24], two groups of controls (a high risk population and a low risk population) were used. However, the genotype frequencies of the low-risk population controls deviated from HWE. Therefore, only the high-risk population controls of this study were included in the final analysis.
The characteristics of nine eligible case-control studies are summarized in Table 1. The sample size of the 9 studies ranged from 89 to 2178. In total, 2114 EC cases and 3431 controls were included in the meta-analysis. Distribution of TP53 genotype frequencies among EC cases and controls of the nine studies are shown in Table 2. The frequencies of heterozygote genotype among the cases of the studies using the specimens of exfoliated esophageal cells[21] or tumor tissues[23] were obviously lower than those of other studies. In studies with at least 200 samples, there was not a wide variation of Arg72 and Pro72 allele frequencies among controls, with the Arg72 allele frequencies ranging from 53% to 60%[20,22,24,26-28]. But in studies with less than 200 samples, the control groups represented diverse frequencies of the Arg72 allele, which were 37%[21], 48%[23] and 59%[25], respectively.
Table 1.
Main characteristics of included studies in the meta-analysis
| First author (yr) | Country | Ethnicity | Source of controls | Specimens | Sample size (case/control) |
| Lee[20] (2000) | China (Taiwan) | Asian | Hospital | White blood cells | 90/254 |
| Peixoto[21] (2001) | China | Asian | Population | Exfoliated esophageal cells | 32/57 |
| Hamajima[22] (2002) | Japan | Asian | Hospital | White blood cells | 102/241 |
| Li[23] (2002) | China | Asian | Population/blood donors | Tumor tissue | 62/131 |
| Hu[24] (2003) | China | Asian | Population | White blood cells | 120/130 |
| Vos[25] (2003) | South Africa | African | Unknown | Tumor tissue, White blood cells | 73/115 |
| Hong[26] (2005) | China | Asian | Population | Normal Esophageal tissue | 758/1420 |
| Cai[27] (2006) | China | Asian | Population | White blood cells | 204/389 |
| Shao[28] (2008) | China | Asian | Population | White blood cells | 673/694 |
Table 2.
Distribution of TP53 Arg72Pro genotypes among esophageal cancer cases and controls included in the meta-analysis n (%)
| First author (yr) |
Cases |
Controls |
||||
| Arg/Arg | Arg/Pro | Pro/Pro | Arg/Arg | Arg/Pro | Pro/Pro | |
| Lee[20] (2000) | 20 (22.2) | 46 (51.1) | 24 (26.7) | 94 (37) | 116 (45.7) | 44 (17.3) |
| Peixoto[21] (2001) | 8 (25) | 13 (40.6) | 11 (34.4) | 9 (15.8) | 24 (42.1) | 24 (42.1) |
| Hamajima[22] (2002) | 37 (36.3) | 51 (50) | 14 (13.7) | 91 (37.8) | 107 (44.4) | 43 (17.8) |
| Li[23] (2002) | 27 (43.5) | 21 (33.9) | 14 (22.6) | 29 (22.1) | 67 (51.1) | 35 (26.7) |
| Hu[24] (2003) | 29 (24.2) | 60 (50) | 32 (26.7) | 38 (29.2) | 68 (52.3) | 24 (18.5) |
| Vos[25] (2003) | 26 (35.6) | 42 (57.5) | 5 (6.8) | 37 (32.2) | 62 (53.9) | 16 (13.9) |
| Hong[26] (2005) | 199 (26.3) | 340 (44.9) | 219 (28.9) | 425 (29.9) | 731 (51.5) | 264 (18.6) |
| Cai[27] (2006) | 41 (20.1) | 89 (43.6) | 74 (36.3) | 117 (30.1) | 178 (45.8) | 94 (24.2) |
| Shao[28] (2008) | 163 (24.2) | 306 (45.5) | 204 (30.3) | 195 (28.1) | 366 (52.7) | 133 (19.2) |
Arg: Arginine; Pro: Proline.
Meta-analysis results
When all the eligible studies were pooled into the meta-analysis, evidence was found in an association between significantly decreased EC risk and the variant genotypes of TP53 in the dominant model (OR = 0.73, 95% CI: 0.57-0.94, P = 0.014, Table 3). However, significant inter-study heterogeneity existed in all genetic models (Table 3). In order to figure out the main reasons of the heterogeneity among studies and obtain exact consequence on the relationship between TP53 Arg72Pro polymorphism and EC susceptibility, stratified analyses were then performed.
Table 3.
Results of meta-analysis for TP53 Arg72Pro polymorphism and esophageal cancer risk
| Study groups | n1 | Sample size(case/control) |
Arg/Arg vs Pro/Pro |
Arg/Arg+Arg/Pro vs Pro/Pro |
Arg/Arg vs Arg/Pro+Pro/Pro |
||||||
| OR (95% CI) | P2 | P3 | OR (95% CI) | P2 | P3 | OR (95% CI) | P2 | P3 | |||
| Total | 9 | 2114/3431 | 0.76 (0.54-1.07)4 | 0.114 | 0.001 | 0.73 (0.57-0.94)4 | 0.014 | 0.009 | 0.89 (0.69-1.13)4 | 0.334 | 0.004 |
| Source of controls | |||||||||||
| Population | 5 | 1787/2690 | 0.56 (0.47-0.66) | < 0.001 | 0.273 | 0.57 (0.50-0.66) | < 0.001 | 0.404 | 0.80 (0.70-0.92) | 0.001 | 0.324 |
| Population/blood donors | 1 | 62/131 | 2.33 (1.03-5.24) | 0.041 | - | 1.25 (0.61-2.54) | 0.538 | - | 2.71 (1.42-5.20) | 0.003 | - |
| Hospital | 2 | 119/495 | 0.70 (0.22-2.18)4 | 0.533 | 0.022 | 0.80 (0.37-2.04)4 | 0.754 | 0.051 | 0.69 (0.36-1.31)4 | 0.252 | 0.08 |
| Unknown | 1 | 73/115 | 2.25 (0.73-6.91) | 0.157 | - | 2.20 (0.77-6.28) | 0.142 | - | 1.17 (0.63-2.16) | 0.626 | - |
| Specimen of cases | |||||||||||
| White blood cells or normal tissue | 6 | 1947/3128 | 0.56 (0.47-0.65) | < 0.001 | 0.233 | 0.58 (0.51-0.67) | < 0.001 | 0.219 | 0.78 (0.68-0.88) | < 0.001 | 0.321 |
| White blood cells/tumor tissue | 1 | 73/115 | 2.25 (0.73-6.91) | 0.157 | - | 2.20 (0.77-6.28) | 0.142 | - | 1.17 (0.63-2.16) | 0.626 | - |
| Tumor tissue | 1 | 62/131 | 2.33 (1.03-5.24) | 0.041 | - | 1.25 (0.61-2.54) | 0.538 | - | 2.71 (1.42-5.20) | 0.003 | - |
| Exfoliated esophageal cells | 1 | 32/57 | 1.94 (0.59-6.38) | 0.275 | - | 1.39 (0.56-3.41) | 0.474 | - | 1.78 (0.61-5.19) | 0.292 | - |
| Sample size | |||||||||||
| ≥ 200 subjects | 6 | 1947/3128 | 0.56 (0.47-0.65) | < 0.001 | 0.233 | 0.58 (0.51-0.67) | < 0.001 | 0.219 | 0.78 (0.68-0.88) | < 0.001 | 0.321 |
| < 200 subjects | 3 | 167/303 | 2.21 (1.24-3.93) | 0.007 | 0.969 | 1.47 (0.90-2.40) | 0.121 | 0.677 | 1.73 (1.15-2.61) | 0.009 | 0.182 |
1Number of comparisons; 2P value for the association; 3P value for the heterogeneity;
Random effects model was used when P value for heterogeneity test < 0.1, otherwise, fixed-effects model was used. Arg: Arginine; Pro: Proline; OR: Odds ratio.
In stratified analysis according to the source of controls, significant association between reduced EC risk and TP53 genotypes was found solely in subgroup of studies with population-based controls in all genetic models (homozygote comparison: OR = 0.56, 95% CI: 0.47-0.66, P < 0.001; dominant model: OR = 0.57, 95% CI: 0.50-0.66, P < 0.001; recessive model: OR = 0.80, 95% CI: 0.70-0.92, P = 0.001; Table 3). Significantly increased EC risk, however, was observed in the subgroup of a study with different source of controls selected from population and blood donors in homozygote comparison (OR = 2.33, 95% CI: 1.03-5.24, P = 0.041) and recessive model (OR = 2.71, 95% CI: 1.42-5.02, P = 0.003, Table 3). No evidence of association was observed in studies without clear presentation of hospital-based controls or the source of controls (Table 3).
We divided the included studies into four subgroups according to the specimens used. As a result, significantly reduced EC risk was found only in subgroups where white blood cells or normal tissue were used to determine TP53 genotypes in different genetic models (homozygote comparison: OR = 0.56, 95% CI: 0.47-0.65, P < 0.001; dominant model: OR = 0.58, 95% CI: 0.51-0.67, P < 0.001; recessive model: OR = 0.78, 95% CI: 0.68-0.88, P < 0.001; Table 3). Nevertheless, significantly excessive risk of EC was observed in the subgroup of a study using tumor tissue to extract genomic DNA for genotyping TP53 by homozygote comparison and recessive model (Table 3). This study also has different sources of controls. No significant association was observed in the studies using mixed specimens of white blood cells and tumor tissues or exfoliated esophageal cells to assess TP53 genotypes (Table 3).
We also stratified the included studies into two subgroups by sample size. One included studies with at least 200 participants, and the other included studies with less than 200 participants. Interestingly, studies in the former subgroup also used white blood cells or normal tissue as the specimens to assess TP53 genotypes, and significant association between reduced EC risk and TP53 genotypes was observed in all genetic models (Table 3). However, in the latter subgroup, significantly increased EC risk was found in homozygote comparison (OR = 2.21, 95% CI: 1.24-3.93, P = 0.007) and recessive model (OR = 1.73, 95% CI: 1.15-2.61, P = 0.009).
We performed the analysis only in well-designed studies with population-based controls with at least 200 participants using white blood cells or normal tissue to determine TP53 genotypes. Significantly decreased risk of EC was found in all genetic models (homozygote comparison: OR = 0.54, 95% CI: 0.46-0.64, P < 0.001; dominant model: OR = 0.56, 95% CI: 0.49-0.65, P < 0.001; recessive model: OR = 0.79, 95% CI: 0.69-0.91, P = 0.001; Figure 1).
Figure 1.
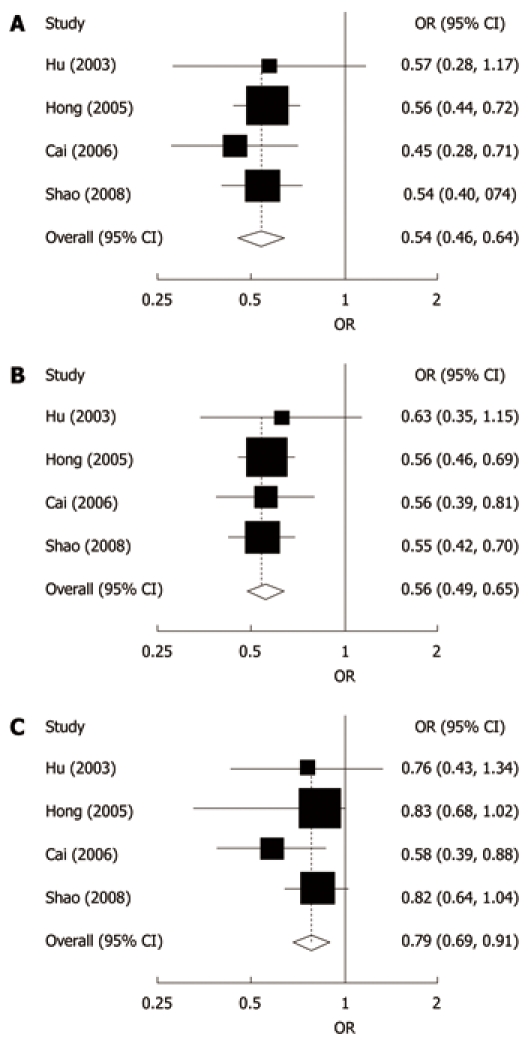
Forest plots for the relationship between TP53 Arg72Pro polymorphism and esophageal cancer risk in studies with population-based controls with at least 200 participants, using white blood cells or normal tissues to determine TP53 genotypes. A: Homozygote comparison; B: Dominant model; C: Recessive model. The first authors’ surname and year of publication are given in the left part of the figure. The size of the black square corresponding to each study is proportional to the sample size. The centre of each square represents the odds ratio (OR) and the horizontal line shows the corresponding 95% CI. The pooled OR was obtained using fixed-effects model and is represented by hollow diamond, where its centre indicates the OR and its ends correspond to the 95% CI. Arg: Arginine; Pro: Proline.
Sensitivity analysis
A single study involved in the meta-analysis was deleted each time to reflect the effects of individual data-set on the pooled ORs, and most of the corresponding pooled ORs were not materially altered (data not shown).
Publication bias
Both Begg’s funnel plot and Egger’s test were performed to assess the publication bias of literatures. Begg’s funnel plots did not reveal any evidence of obvious asymmetry except for heterozygote comparison and dominant model in the overall meta-analysis (figures not shown). The Egger’s test results suggested that publication bias was evident in heterozygote comparison (P = 0.003) and dominant model (P = 0.004), but not evident in homozygote comparison (P = 0.058) and recessive model (P = 0.389). The Duval and Tweedie non-parametric “trim and fill’ method was used to adjust for publication bias. Meta-analysis with and without using “trim and fill” method did not draw different conclusions (data not shown), indicating that our results were statistically robust.
DISCUSSION
Since the identification of TP53 Arg72Pro polymorphism[13], a number of studies[20-29] have investigated the genetic effect of this polymorphism on EC susceptibility, but the results are inconclusive. This led us to undertake the present meta-analysis, which could quantitify all the available data and might help us to distinguish the true from the false, to explore a more robust estimate of the effect of this polymorphism on EC risk. The main finding of our meta-analysis with 9 published studies including 2114 cases and 3431 controls is that TP53 Arg72 carriers are significantly associated with decreased EC risk, and the results of increased risk or no effect of this polymorphism on EC may be due to methodological errors such as selection bias, inappropriate specimens used for genotype assessment, or limited statistical power.
We found that the distribution of TP53 Arg72Pro genotypes in controls deviated from HWE in the study by Yang et al[29], although it has a relatively large sample size including 435 cases and 550 controls. Yang et al[29] reported that TP53 Arg/Arg genotype was associated with significantly increased EC risk (OR = 6.48, 95% CI: 4.65-9.03), which is contrary to our results of meta-analysis. It is well known that deviation from HWE may be due to genetic reasons including non-random mating, or the alleles reflecting recent mutations that have not reached equilibrium, as well as methodological reasons including biased selection of subjects from the population, or genotyping errors[36,37]. In despite of the reasons of disequilibrium, the results of genetic association studies might be spurious if the controls were not in HWE[38,39]. In order to guarantee the criteria for the eligible studies, only studies with controls in HWE were included in this meta-analysis.
In the present study, statistically significant inter-study heterogeneity of genotype effect was detected in different genetic models when all the eligible studies were pooled into the meta-analysis. Pooling despite the presence of heterogeneity may yield the mean of varying effect sizes, but the biological interpretation of such a mean and its clinical application would be very difficult[40,41]. Therefore, it is important to explore the source of heterogeneity rather than obtaining a potentially meaningless pooled summary measure[42]. In order to identify the source of heterogeneity and ascertain the exact genetic effect of TP53 Arg72Pro polymorphism on EC risk, we stratified the studies according to the source of controls, the specimens used for assessment of TP53 genotypes and sample size. We found that the heterogeneity was remarkably decreased when the studies were divided according to the specimens used and sample size, indicating that the two factors may contribute to the observed heterogeneity.
In two of the nine included studies where the specimens used for assessment of TP53 genotype were exfoliated esophageal cells[21] or tumor tissues[23], the frequencies of heterozygote genotype were obviously lower than those of other studies. This indicates that loss of heterozygosity (LOH) may exist and the distribution of TP53 genotypes in these cases may not be the same as that in normal tissue or cells. Generally, spurious results may be obtained from genetic association studies with inappropriate material for determining genotypes[16]. In our meta-analysis, significant association between TP53 Arg72Pro polymorphism and reduced EC risk was not observed in subgroups using inappropriate material to determine TP53 genotypes but only in the subgroups using white blood cells or normal tissues. Consequently, in genetic association studies, DNA from white blood cells or normal tissues should be used for determining genetic polymorphism, but not tumor tissue or exfoliated cells, in which LOH is a frequent event[43,44].
Lacking sufficient statistical power is an unnegligible problem in genetic association studies detecting the possible risk for the polymorphism[45]. It is likely that most genetic polymorphisms represent modest effects on disease susceptibility. An adequately powered study to detect single genetic associations would typically require a relatively large sample size, depending on the prevalence of the implicated polymorphism and the exact OR. Some of the eligible studies for our meta-analysis had a very small sample size and may have limited statistical power to detect a slight effect or may have generated a fluctuated risk estimate[46,47]. Carefully conducted meta-analysis of these data is essential to clarify whether these associations are true or not. Through stratified analysis, we found significantly increased EC risk associated with TP53 genotypes in subgroups of the studies with less than 200 participants, which was contrary to the results in subgroup of the studies with at least 200 participants. Given that all of the studies with a sample size of less than 200 used inappropriate specimens for determining TP53 genotypes, the results may be unreliable.
Some limitations of this meta-analysis should be addressed. Firstly, publication bias was detected for heterozygote comparison and dominant model in overall meta-analysis. The potential reason may be that results from small studies were more likely to be published if there was positive data reported. Therefore, well-designed studies with large sample size are required. Secondly, in the subgroup analyses by ethnicity, the included studies involved only Asians and Africans. Data concerning other ethnicities such as Caucasians were not found. For Africans, only one study was conducted, with a small sample size of 73 cases and 115 controls, which has not enough statistical power to find the real association. Thus, additional studies are warranted to evaluate the effect of this functional polymorphism on EC risk in different ethnicities, especially in Africans and Caucasians. Thirdly, lack of original data, including data of genotypes and environmental risk factors, of the included studies limited our further evaluation of potential gene-environment interaction, especially the interaction between human papillomavirus (HPV) infection and TP53 Arg72Pro polymorphism, which was investigated in several studies[23,48-50]. However, unlike HPV infection in cervical carcinoma, the role of HPV in the etiology of EC remains controversial[8]. A more precise analysis should be conducted if individual data are available.
Despite some limitations, the results of this meta-analysis still suggest that TP53 Arg72 allele is a protective factor for EC. The significantly reduced EC risk was found only in subgroup analyses of well-designed studies. Therefore, it is necessary to conduct large-sample studies using appropriate materials for assessment of genotypes, as well as homogeneous EC patients and unbiased selected controls. Such studies taking these factors into account may eventually lead to a better and comprehensive understanding of the association between TP53 Arg72pro polymorphism and EC risk.
COMMENTS
Background
Esophageal cancer (EC) is the eighth most common cancer and sixth most deadly cancer worldwide. A common polymorphism of TP53 at the 72nd amino acid residue, with an arginine (Arg) to proline (Pro) change because of a G→C transverse has been implicated as a risk factor for EC, but individual studies have been inconclusive or controversial.
Research frontiers
A number of studies have reported the role of TP53 Arg72Pro polymorphism in cancers such as cervical cancer, lung cancer, breast cancer, and gastric cancer, but the association of TP53 polymorphism with EC is not fully understood.
Innovations and breakthroughs
The present study demonstrated that TP53 Arg72 carriers are significantly associated with decreased EC risk, and suggested that increased risk or no effect of Arg72 variant on EC reported may be due to methodological errors such as selection bias, inappropriate specimens used for genotype assessment, or limited statistical power.
Applications
In this report, the association between TP53 Arg72Pro polymorphism and EC risk was observed, and the Arg72 allele decreased the EC risk, which is meaningful to early diagnosis, prevention and individual-based treatment of EC. Therefore, Arg72Pro polymorphism of the TP53 gene might be a potential therapeutic target for EC.
Terminology
TP53 is a major regulator of the cellular response to stress and serves as a tumor suppressor by inducing cell cycle arrest or apoptosis. Inactivation of the TP53 signaling pathway has been seen in most human cancers and polymorphisms of TP53 have also been reported to be the possible risk factors for some kinds of tumors.
Peer review
This study is an interesting meta-analysis on the association of TP53 ArgPro polymorphism with EC risk. Out of the 9 studies that survived the selection criteria, they found that TP53 Arg72 carriers were significantly associated with decreased EC risk. The authors concluded that previous reports of increased risk or no effect of this polymorphism on EC may be due to methodological errors such as selection bias, inappropriate specimen or limited statistical power and they give guidelines on how to avoid these pitfalls.
Footnotes
Supported by the National 973 Program of China (No. 2004CB518605), the National 863 Project of China (No.2006AA020501), the National Key Sci-Tech Special Project of China (No.2008ZX10002-020), the Project of the Shanghai Municipal Science and Technology Commission (03dz14086) and the National Natural Science Foundation of China (No.30024001and 30771188)
Peer reviewer: Yoshiharu Motoo, MD, PhD, FACP, FACG, Professor and Chairman, Department of Medical Oncology, Kanazawa Medical University, 1-1 Daigaku, Uchinada, Ishikawa 920-0293, Japan
S- Editor Sun H L- Editor Ma JY E- Editor Ma WH
References
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Botterweck AA, Schouten LJ, Volovics A, Dorant E, van Den Brandt PA. Trends in incidence of adenocarcinoma of the oesophagus and gastric cardia in ten European countries. Int J Epidemiol. 2000;29:645–654. doi: 10.1093/ije/29.4.645. [DOI] [PubMed] [Google Scholar]
- 4.Brown LM, Devesa SS, Chow WH. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J Natl Cancer Inst. 2008;100:1184–1187. doi: 10.1093/jnci/djn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vizcaino AP, Moreno V, Lambert R, Parkin DM. Time trends incidence of both major histologic types of esophageal carcinomas in selected countries, 1973-1995. Int J Cancer. 2002;99:860–868. doi: 10.1002/ijc.10427. [DOI] [PubMed] [Google Scholar]
- 6.Fan Y, Yuan JM, Wang R, Gao YT, Yu MC. Alcohol, tobacco, and diet in relation to esophageal cancer: the Shanghai Cohort Study. Nutr Cancer. 2008;60:354–363. doi: 10.1080/01635580701883011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamangar F, Chow WH, Abnet CC, Dawsey SM. Environmental causes of esophageal cancer. Gastroenterol Clin North Am. 2009;38:27–57, vii. doi: 10.1016/j.gtc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JM, Xu B, Rao JY, Shen HB, Xue HC, Jiang QW. Diet habits, alcohol drinking, tobacco smoking, green tea drinking, and the risk of esophageal squamous cell carcinoma in the Chinese population. Eur J Gastroenterol Hepatol. 2007;19:171–176. doi: 10.1097/MEG.0b013e32800ff77a. [DOI] [PubMed] [Google Scholar]
- 9.Hiyama T, Yoshihara M, Tanaka S, Chayama K. Genetic polymorphisms and esophageal cancer risk. Int J Cancer. 2007;121:1643–1658. doi: 10.1002/ijc.23044. [DOI] [PubMed] [Google Scholar]
- 10.Levine AJ, Finlay CA, Hinds PW. P53 is a tumor suppressor gene. Cell. 2004;116:S67–S69, 1 p following S69. doi: 10.1016/s0092-8674(04)00036-4. [DOI] [PubMed] [Google Scholar]
- 11.Hrstka R, Coates PJ, Vojtesek B. Polymorphisms in p53 and the p53 pathway: roles in cancer susceptibility and response to treatment. J Cell Mol Med. 2009;13:440–453. doi: 10.1111/j.1582-4934.2008.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 13.Matlashewski GJ, Tuck S, Pim D, Lamb P, Schneider J, Crawford LV. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7:961–963. doi: 10.1128/mcb.7.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19:1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumont P, Leu JI, Della Pietra AC 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 16.Klug SJ, Ressing M, Koenig J, Abba MC, Agorastos T, Brenna SM, Ciotti M, Das BR, Del Mistro A, Dybikowska A, et al. TP53 codon 72 polymorphism and cervical cancer: a pooled analysis of individual data from 49 studies. Lancet Oncol. 2009;10:772–784. doi: 10.1016/S1470-2045(09)70187-1. [DOI] [PubMed] [Google Scholar]
- 17.Dai S, Mao C, Jiang L, Wang G, Cheng H. P53 polymorphism and lung cancer susceptibility: a pooled analysis of 32 case-control studies. Hum Genet. 2009;125:633–638. doi: 10.1007/s00439-009-0664-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Wang M, Wu D, Wang M, Tong N, Tian Y, Zhang Z. P53 codon 72 polymorphism contributes to breast cancer risk: a meta-analysis based on 39 case-control studies. Breast Cancer Res Treat. 2010;120:509–517. doi: 10.1007/s10549-009-0480-4. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y, Li N, Zhuang W, Liu GJ, Wu TX, Yao X, Du L, Wei ML, Wu XT. P53 codon 72 polymorphism and gastric cancer: a meta-analysis of the literature. Int J Cancer. 2007;121:1481–1486. doi: 10.1002/ijc.22833. [DOI] [PubMed] [Google Scholar]
- 20.Lee JM, Lee YC, Yang SY, Shi WL, Lee CJ, Luh SP, Chen CJ, Hsieh CY, Wu MT. Genetic polymorphisms of p53 and GSTP1,but not NAT2,are associated with susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer. 2000;89:458–464. doi: 10.1002/1097-0215(20000920)89:5<458::aid-ijc10>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 21.Peixoto Guimaraes D, Hsin Lu S, Snijders P, Wilmotte R, Herrero R, Lenoir G, Montesano R, Meijer CJ, Walboomers J, Hainaut P. Absence of association between HPV DNA, TP53 codon 72 polymorphism, and risk of oesophageal cancer in a high-risk area of China. Cancer Lett. 2001;162:231–235. doi: 10.1016/s0304-3835(00)00643-1. [DOI] [PubMed] [Google Scholar]
- 22.Hamajima N, Matsuo K, Suzuki T, Nakamura T, Matsuura A, Hatooka S, Shinoda M, Kodera Y, Yamamura Y, Hirai T, et al. No associations of p73 G4C14-to-A4T14 at exon 2 and p53 Arg72Pro polymorphisms with the risk of digestive tract cancers in Japanese. Cancer Lett. 2002;181:81–85. doi: 10.1016/s0304-3835(02)00041-1. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Lu ZM, Guo M, Wu QJ, Chen KN, Xing HP, Mei Q, Ke Y. p53 codon 72 polymorphism (C/G) and the risk of human papillomavirus-associated carcinomas in China. Cancer. 2002;95:2571–2576. doi: 10.1002/cncr.11008. [DOI] [PubMed] [Google Scholar]
- 24.Hu N, Li WJ, Su H, Wang C, Goldstein AM, Albert PS, Emmert-Buck MR, Kong LH, Roth MJ, Dawsey SM, et al. Common genetic variants of TP53 and BRCA2 in esophageal cancer patients and healthy individuals from low and high risk areas of northern China. Cancer Detect Prev. 2003;27:132–138. doi: 10.1016/s0361-090x(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 25.Vos M, Adams CH, Victor TC, van Helden PD. Polymorphisms and mutations found in the regions flanking exons 5 to 8 of the TP53 gene in a population at high risk for esophageal cancer in South Africa. Cancer Genet Cytogenet. 2003;140:23–30. doi: 10.1016/s0165-4608(02)00638-6. [DOI] [PubMed] [Google Scholar]
- 26.Hong Y, Miao X, Zhang X, Ding F, Luo A, Guo Y, Tan W, Liu Z, Lin D. The role of P53 and MDM2 polymorphisms in the risk of esophageal squamous cell carcinoma. Cancer Res. 2005;65:9582–9587. doi: 10.1158/0008-5472.CAN-05-1460. [DOI] [PubMed] [Google Scholar]
- 27.Cai L, Mu LN, Lu H, Lu QY, You NC, Yu SZ, Le AD, Zhao J, Zhou XF, Marshall J, et al. Dietary selenium intake and genetic polymorphisms of the GSTP1 and p53 genes on the risk of esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:294–300. doi: 10.1158/1055-9965.EPI-05-0680. [DOI] [PubMed] [Google Scholar]
- 28.Shao Y, Tan W, Zhang S. P53 gene codon 72 polymorphism and risk of esophageal squamous cell carcinoma: a case/control study in a Chinese population. Dis Esophagus. 2008;21:139–143. doi: 10.1111/j.1442-2050.2007.00746.x. [DOI] [PubMed] [Google Scholar]
- 29.Yang W, Zhang Y, Tian X, Ning T, Ke Y. p53 Codon 72 polymorphism and the risk of esophageal squamous cell carcinoma. Mol Carcinog. 2008;47:100–104. doi: 10.1002/mc.20368. [DOI] [PubMed] [Google Scholar]
- 30.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:29. [Google Scholar]
- 31.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 32.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 33.Tobias A. Assessing the influence of a single study in the meta-analysis estimate. Stata Tech Bull. 1999;8:3. [Google Scholar]
- 34.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh IM, Matlashewski G, Banks L. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell AA, Cutler DJ, Chakravarti A. Undetected genotyping errors cause apparent overtransmission of common alleles in the transmission/disequilibrium test. Am J Hum Genet. 2003;72:598–610. doi: 10.1086/368203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosking L, Lumsden S, Lewis K, Yeo A, McCarthy L, Bansal A, Riley J, Purvis I, Xu CF. Detection of genotyping errors by Hardy-Weinberg equilibrium testing. Eur J Hum Genet. 2004;12:395–399. doi: 10.1038/sj.ejhg.5201164. [DOI] [PubMed] [Google Scholar]
- 38.Salanti G, Amountza G, Ntzani EE, Ioannidis JP. Hardy-Weinberg equilibrium in genetic association studies: an empirical evaluation of reporting, deviations, and power. Eur J Hum Genet. 2005;13:840–848. doi: 10.1038/sj.ejhg.5201410. [DOI] [PubMed] [Google Scholar]
- 39.Trikalinos TA, Salanti G, Khoury MJ, Ioannidis JP. Impact of violations and deviations in Hardy-Weinberg equilibrium on postulated gene-disease associations. Am J Epidemiol. 2006;163:300–309. doi: 10.1093/aje/kwj046. [DOI] [PubMed] [Google Scholar]
- 40.Thompson SG. Why sources of heterogeneity in meta-analysis should be investigated. BMJ. 1994;309:1351–1355. doi: 10.1136/bmj.309.6965.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson SG, Sharp SJ. Explaining heterogeneity in meta-analysis: a comparison of methods. Stat Med. 1999;18:2693–7208. doi: 10.1002/(sici)1097-0258(19991030)18:20<2693::aid-sim235>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 42.Thakkinstian A, McElduff P, D’Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Stat Med. 2005;24:1291–1306. doi: 10.1002/sim.2010. [DOI] [PubMed] [Google Scholar]
- 43.Lea IA, Jackson MA, Li X, Bailey S, Peddada SD, Dunnick JK. Genetic pathways and mutation profiles of human cancers: site- and exposure-specific patterns. Carcinogenesis. 2007;28:1851–1858. doi: 10.1093/carcin/bgm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spafford MF, Koch WM, Reed AL, Califano JA, Xu LH, Eisenberger CF, Yip L, Leong PL, Wu L, Liu SX, et al. Detection of head and neck squamous cell carcinoma among exfoliated oral mucosal cells by microsatellite analysis. Clin Cancer Res. 2001;7:607–612. [PubMed] [Google Scholar]
- 45.Ioannidis JP. Genetic associations: false or true? Trends Mol Med. 2003;9:135–138. doi: 10.1016/s1471-4914(03)00030-3. [DOI] [PubMed] [Google Scholar]
- 46.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 47.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawaguchi H, Ohno S, Araki K, Miyazaki M, Saeki H, Watanabe M, Tanaka S, Sugimachi K. p53 polymorphism in human papillomavirus-associated esophageal cancer. Cancer Res. 2000;60:2753–2755. [PubMed] [Google Scholar]
- 49.Lu XM, Zhang YM, Lin RY, Liang XH, Zhang YL, Wang X, Zhang Y, Wang Y, Wen H. p53 polymorphism in human papillomavirus-associated Kazakh’s esophageal cancer in Xinjiang, China. World J Gastroenterol. 2004;10:2775–2778. doi: 10.3748/wjg.v10.i19.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pantelis A, Pantelis D, Ruemmele P, Hartmann A, Hofstaedter F, Buettner R, Bootz F, Stoehr R. p53 Codon 72 polymorphism, loss of heterozygosity and high-risk human papillomavirus infection in a low-incidence German esophageal squamous cell carcinoma patient cohort. Oncol Rep. 2007;17:1243–1248. [PubMed] [Google Scholar]


