Abstract
Although family history is a well-established risk factor for Parkinson's disease (PD), fewer than 5% of PD cases can be attributed to known genetic mutations. The etiology for the remainder of PD cases is unclear; however, neuronal accumulation of the protein α-synuclein is common to nearly all patients, implicating pathways that influence α-synuclein in PD pathogenesis. We report a genome-wide significant association (P = 3.97 × 10−8) between a polymorphism, rs1564282, in the cyclin-G-associated kinase (GAK) gene and increased PD risk, with a meta-analysis odds ratio of 1.48. This association result is based on the meta-analysis of three publicly available PD case–control genome-wide association study and genotyping from a new, independent Italian cohort. Microarray expression analysis of post-mortem frontal cortex from PD and control brains demonstrates a significant association between rs1564282 and higher α-synuclein expression, a known cause of early onset PD. Functional knockdown of GAK in cell culture causes a significant increase in toxicity when α-synuclein is over-expressed. Furthermore, knockdown of GAK in rat primary neurons expressing the A53T mutation of α-synuclein, a well-established model for PD, decreases cell viability. These observations provide evidence that GAK is associated with PD risk and suggest that GAK and α-synuclein interact in a pathway involved in PD pathogenesis. The GAK protein, a serine/threonine kinase, belongs to a family of proteins commonly targeted for drug development. This, combined with GAK's observed relationship to the levels of α-synuclein expression and toxicity, suggests that the protein is an attractive therapeutic target for the treatment of PD.
INTRODUCTION
Parkinson's disease (PD) is the second most common neurodegenerative disorder, trailing only Alzheimer's disease in prevalence. PD presents clinically with resting tremor, muscular rigidity and bradykinesia (1). Pathologically, the disease is characterized by the loss of dopaminergic neurons within the substantia nigra pars compacta and the presence of Lewy bodies (2). These hallmark intraneuronal inclusions are primarily composed of the small, lipid binding protein α-synuclein (3). Although a critical role for α-synuclein in PD pathogenesis is universally accepted, <5% of all PD cases can be attributed to genetic mutations in α-synuclein and the other known PD genes.
While >95% of PD cases are considered idiopathic, it is well established that between 16 and 27.5% have a positive family history and first-degree relatives of PD patients have a substantially increased risk for PD compared with first-degree relatives of controls (4–6). These observations led us to undertake the GenePD Study, an evaluation of familial PD cases aimed at identifying genes that influence PD susceptibility. The GenePD Study, in collaboration with the PROGENI Study, performed a genome-wide association study (GWAS) (dbGaP Accession: phs000126.v1.p1) focusing on families with multiple affected relatives (7). This was the first GWAS to implicate non-coding single nucleotide polymorphisms (SNPs) within the genes encoding α-synuclein (SNCA) and the micro-tubule-associated protein tau (MAPT) in PD risk (7). Both findings were confirmed in the subsequent GWAS of idiopathic PD (8,9).
We hypothesized that our familial PD cohort (7) was positioned to identify genes that GWAS of primarily idiopathic PD failed to detect (8–12). The region producing the strongest P-value for familial PD risk included the cyclin-G-associated kinase (GAK) gene with a minor allele conferring a 1.7 odds ratio (OR) for PD, which, although suggestive, failed to reach the conservative level for genome-wide significance at P< 5.0 × 10−8 (13). Recently, Hamza et al. (14) performed a PD GWAS in 2000 cases and 1986 controls, and in a meta-analysis with our study (7), the GAK region reached genome-wide significance (P= 3.2× 10−9). GAK was shown previously to be among 137 genes differentially expressed in the substantia nigra pars compacta of PD patients when compared with controls, with a 1.56-fold change (15). The combination of these independent findings prompted our further investigation of the involvement of GAK in PD pathogenesis.
GAK is a ubiquitously expressed protein (16), containing highly conserved serine/threonine kinase (17,18), PTEN (19,20) and J-domains (17,21). Interestingly, the clathrin-binding C-terminal domain of GAK has been shown to bind pre-cathepsin D (CTSD) and to directly sort the zymogen into clathrin-coated vesicles destined for the lysosome (22). Recently, CTSD was implicated as the main lysosomal enzyme involved in α-synuclein degradation (23–25). Mutations in CTSD have been shown to induce the pathological accumulation of α-synuclein in mice, sheep and human infants afflicted with fatal CTSD-deficient forms of neuronal ceroid lipofuscinosis (OMIM #610127) (24,25). Together, these findings led us to investigate the interrelationships between GAK, SNCA and CTSD and their potential role in a previously undescribed pathway for PD pathogenesis. Here we report compelling evidence for interactions between GAK, SNCA and CTSD as well as biological evidence that reduced GAK function enhances α-synuclein-mediated toxicity.
RESULTS
To substantiate the association between the GAK SNP rs1564282 and increased PD risk, we performed a meta-analysis of three publicly available PD case–control GWAS, combined with our genotyping of a new, independent sample of 862 Italian PD cases and 517 controls. We used P ≤ 5 × 10−8 as the threshold for genome-wide significance, a widely accepted threshold based on correction for 1 million independent variants in the genome (13). Our results confirm that the minor allele of the GAK SNP rs1564282 is associated with a 1.48 increased odds of PD at a genome-wide level of significance (P= 3.97 × 10−8; Table 1). While all four studies independently show increased risk of PD associated with this SNP, the largest effect size is observed in the familial PD cohort. Interestingly, restricting analysis of the new Italian cases to those with reported family history (n = 165) shows a stronger effect size (OR = 1.9) and a stronger P-value (P= 0.0026) than the idiopathic cases alone (OR = 1.2, P= 0.18), emphasizing the advantages of focusing on familial cases in the search for genetic contributors to PD.
Table 1.
Meta-analysis of three publicly available GWAS and replication genotyping
| SNP | Familial PD GWAS7 | 1st NIA PD-GWAS10 | 2nd NIA PD-GWAS9 | Milan, Italy cohort | Meta-analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | OR | P-value | n | OR | P-value | n | OR | P-value | n | OR | P-value | OR | P-value | |
| rs1564282 | 1728 | 1.70 | 5.9 × 10−6 | 505 | 1.55 | 0.047 | 1166 | 1.34 | 0.033 | 1379 | 1.32 | 0.0497 | 1.48 | 3.97 × 10−8a |
Each OR reflects the effect of the minor (T) allele, which has a frequency of ∼11% in the overall sample.
aAchieves genome-wide significance.
As reviewed above, SNCA, GAK and CTSD appear to interact either directly or indirectly. To evaluate whether there is a correlation between the expression of these genes, as reflected by messenger ribonucleic acid (mRNA) levels, in control or PD brains, we analyzed four probes (one each for exons 26 and 28 of GAK, one for the 3′-UTR of SNCA, and one for exon 6 of CTSD) present on a commercially available microarray (One-Color Agilent 60-mer Whole Human Genome Microarray). We screened the microarray with total RNA purified from frontal cortex of 33 PD cases and 29 controls (see Table 2 for characteristics of samples). Despite the expression levels of the two C-terminal GAK probes showing marked correlation with one another in both PD (r2 = 0.8) and controls (r2 = 0.6), the expression of GAK exon 26, but not exon 28, was observed to have a positive correlation with expression of CTSD exon 6 in both PD and control brains (r2 = 0.5) (Table 3). Neither GAK nor CTSD expression was correlated with total SNCA expression in either PD or control brains.
Table 2.
Characteristics of RNA samples for microarray analysis
| Sample type (n) | Age at death, years (range) | PMI, hours (range) | RIN (range) | Tissue pHa (range) |
|---|---|---|---|---|
| Control (29) | 75.21 (58–97) | 13.70 (1.50–39.67) | 7.36 (4.8–8.5) | 6.641 (6.26–7.32) |
| PD (33) | 77.67 (64–94) | 7.43 (1.16–30.75) | 7.182 (5.2–8.4) | 6.652 (6.27–7.13) |
aThe pH was measured following a previously established protocol (46).
Table 3.
Correlation between mRNA expression levels
| SNCA (probe 29 939) | CTSD (probe 52 556) | GAK exon 26 (probe 155 700) | GAK exon 28 (probe 397 150) | |
|---|---|---|---|---|
| SNCA | 0.10 (0.58) | 0.15 (0.42) | −0.13 (0.47) | |
| CTSD | 0.28 (0.14) | 0.45 (0.009) | 0.18 (0.31) | |
| GAK exon 26 (probe 155 700) | 0.18 (0.34) | 0.50 (0.006) | 0.79 (<0.0001) | |
| GAK exon 28 (probe 397 150) | −0.27 (0.16) | 0.02 (0.93) | 0.58 (0.001) |
Correlations were computed separately for PD cases (above diagonal) and controls (below diagonal). P-values are indicated in parentheses.
We also used the microarray data from these four probes to analyze the relationship between several SNPs and expression levels of GAK, CTSD and SNCA, adjusting for PD/control status, RNA integrity number (RIN), post-mortem interval (PMI) and age at death (Table 4). In addition to the GAK SNP rs1564282, we evaluated three SNPs located within 100 000bp of SNCA, which were also implicated in PD risk in our familial GWAS (7). In order to account for the 16 different association tests (four SNPs and four expression probes) that were conducted, we used an adjusted alpha-level of significance of 0.007; this value was determined by applying a modified Bonferroni correction (26) that accounts for the high degree of correlation between the expression levels of the used probes (mean R2 = 0.3). We found that rs1564282 in GAK associated significantly with higher levels of total SNCA expression (P = 0.003). Additionally, an intronic SNCA SNP (rs356188) with a minor allele conferring a protective effect in the GWAS (7) was significantly associated with higher CTSD levels (P = 0.007). Some suggestive associations were also detected: rs1564282 GAK with lower levels of GAK exon 28 (P = 0.03), and rs356188 with higher levels of GAK exon 26 (P= 0.02). None of the SNPs within the SNCA gene region was significantly associated with SNCA expression levels (Table 4).
Table 4.
Association of SNPs with mRNA expression levelsa
| SNP | Position on chrom 4 | Gene | MAF | GWAS ORb | SNCA (probe 29 939) | CTSD (probe 52 556) | GAK exon 26 (probe 155 700) | GAK exon 28 (probe 397 150) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | P-value | Beta | P-value | Beta | P-value | Beta | P-value | |||||
| rs1564282 | 842,313 | GAK | 0.14 (T) | 1.7 | 0.35c | 0.003 | −0.06 | 0.49 | −0.12 | 0.16 | −0.15 | 0.03 |
| rs356229 | 90,825,620 | 3’ SNCA | 0.44 (C) | 1.35 | −0.03 | 0.70 | 0.02 | 0.73 | −0.01 | 0.87 | −0.01 | 0.83 |
| rs356188 | 90,910,560 | SNCA | 0.29 (C) | 0.7 | −0.08 | 0.27 | 0.12 | 0.007 | 0.11 | 0.02 | 0.07 | 0.1 |
| rs3775478 | 91,061,863 | MMRN1 | 0.12 (G) | 1.7 | 0.05 | 0.67 | 0.04 | 0.55 | −0.05 | 0.53 | −0.04 | 0.56 |
MAF, minor allele frequency; Beta, beta coefficient.
aAdjusted for PD/control, RIN, PMI and age at death.
bOR previously reported for SNPs association to PD in our Familial PD GWAS (7) with P-values ranging from 5.9 × 10−6 to 8.4 × 10−5.
ceSNP beta estimate in a subset of 32 brain samples with available ethnicity information (all Caucasians) was similar to the beta estimate value in the whole set of brains (Beta = 0.33).
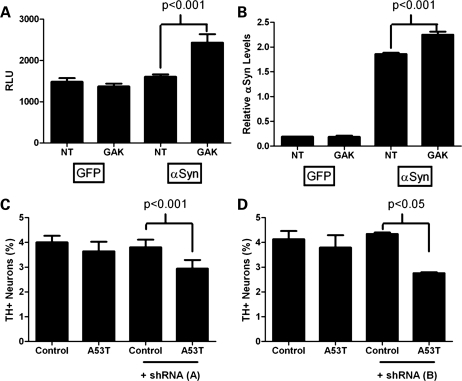
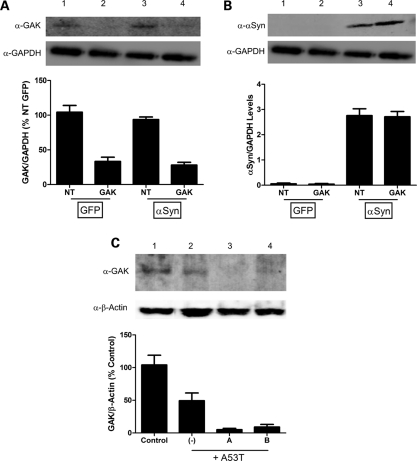
Previous reports have demonstrated that reduced expression of CTSD results in increased α-synuclein aggregation and toxicity (24,25), and that RNAi targeting of GAK in mammalian cell culture reduces CTSD protein levels in the lysosome (22). We decided to define the connection between GAK and α-synuclein by evaluating whether manipulating GAK expression affects α-synuclein-mediated toxicity and α-synuclein protein levels in a simple mammalian cell culture system. We performed siRNA knockdown of GAK in HEK 293 cells while overexpressing α-synuclein. Reducing GAK expression consistently increased toxicity in α-synuclein-overexpressing cells, as indicated by increased adenylate kinase release (Fig. 1A). Notably, knockdown of GAK had no effect on toxicity in the absence of α-synuclein overexpression, as demonstrated by the green fluorescent protein (GFP) overexpression control (Fig. 1A). Western blot analysis confirmed that GAK siRNA treatment dramatically suppressed GAK protein levels in these cells (Fig. 2A). ELISA revealed that this reduction in the GAK protein was accompanied by a significant increase in α-synuclein protein levels in α-synuclein-overexpressing cells (Fig. 1B), although the less-sensitive western blot analysis demonstrated only subtle changes in α-synuclein protein levels (Fig. 2B). Control siRNA had no effect on α-synuclein levels and GAK siRNA did not induce α-synuclein expression when GFP was overexpressed instead of α-synuclein (Fig. 1B). These cell culture results suggest a synergistic effect whereby an increase in α-synuclein combined with a decrease in GAK results in increased cytotoxicity.
Figure 1.
Potentiation of α-synuclein-mediated toxicity in cell culture systems with reduced GAK expression. (A and B) HEK 293 cells were transiently transfected with CMV-GFP, CMV-α-synuclein and either non-targeted (NT) or GAK-targeted pools of siRNA. (A) Toxicity was assessed by release of adenylate kinase into the media 72 h after transfection. Enzymatic release was detected by luciferase reaction and quantified by relative luminescent units (RLU) compared with controls (Toxilight). (B) α-Synuclein protein levels in siRNA-treated cell lysates were quantified by ELISA. (C and D) Primary midbrain cultures were transduced with A53T adenovirus (MOI = 3) with or without lentivirus encoding either (C) GAK shRNA ‘A’ (MOI = 3) or (D) GAK shRNA ‘B’ (MOI = 1). Control cells were incubated in the absence of virus. Dopaminergic cell viability was determined by staining with antibodies specific for MAP2 and TH and is expressed as the percentage of MAP2-positive neurons that were also TH-positive.
Figure 2.
Western blot analyses confirming reduced GAK expression and enhanced α-synuclein accumulation. (A) GAK protein levels and (B) α-synuclein protein levels in HEK 293 cells transfected with either CMV-GFP (lanes 1 and 2) or CMV-α-synuclein (lanes 3 and 4) and non-targeted (lanes 1 and 3) or GAK-specific (lanes 2 and 4) pools of siRNA's. Western blot quantifications were performed via densitometric analysis using NIH ImageJ software. GAK band intensities were normalized first to the GAPDH signal and then to the values for non-targeted, GFP-transfected cells lane (set to 100%) (A), while α-synuclein band intensities were reported as relative expression levels of α-synuclein compared to GAPDH (B). Each experiment was performed in triplicate. (C) Representative western blot depicting shRNA-mediated GAK knockdown in the MES23.5 dopaminergic cell line (n ≥ 2 for each of the different treatment conditions). Lysates were prepared from control cells (lane 1), and cells transduced with A53T-α-synuclein adenovirus alone (lane 2, MOI = 30) or with A53T-α-synuclein adenovirus (MOI = 30) plus lentivirus encoding GAK-specific shRNA ‘A’ (lane 3, MOI = 30) or ‘B’ (lane 4, MOI = 15). The graph below the blot shows relative GAK protein levels determined via densitometric analysis using NIH ImageJ software. GAK band intensities were normalized first to the β-actin signal and then to the normalized GAK band intensity in the control lane (set to 100%). The data suggest that (i) A53T expression induces GAK downregulation in the MES23.5 dopaminergic cell line, and (ii) shRNAs A and B induce a further decrease in GAK levels in A53T-expressing cells.
We also investigated whether the interaction of SNCA and GAK was present for the A53T mutation in α-synuclein. In this experiment, we examined how the reduction in endogenous GAK influences α-synuclein-mediated toxicity in cultured primary rat midbrain neurons expressing the A53T form of α-synuclein, an in vitro model of PD (27–31). Primary rat midbrain neuron cultures consist of ∼4–5% tyrosine hydroxylase positive (TH+) cells, which have been described as being exquisitely sensitive to α-synuclein levels in PD (31). Primary rat midbrain cultures were treated with sub-lethal levels of an adenovirus containing the A53T-SNCA variant associated with early onset PD, with or without lentiviruses encoding one of two GAK-specific short-hairpin RNAs (shRNAs) (each targeting a different coding region of rat GAK). Ninety-six hours after transduction, we assessed relative dopaminergic cell viability by immunocytochemistry for TH, a marker of dopaminergic neurons and microtubule-associated protein 2 (MAP2), a general neuronal marker. Transduction with the A53T-SNCA virus or with either GAK shRNA virus alone did not affect dopaminergic cell viability compared with untransduced cultures. However, simultaneous exposure to the A53T-α-synuclein adenovirus and either of the GAK-specific shRNA lentiviruses dramatically reduced the number of TH+ neurons relative to MAP2+ neurons, suggesting that α-synuclein neurotoxicity is triggered or enhanced by the combination of overexpression of mutant SNCA and reduction in GAK (Fig. 1C and D). In other (control) experiments, we found that a non-specific lentivirus (lenti-LacZ) failed to exacerbate dopaminergic cell death elicited by the A53T adenovirus (Strathearn and Rochet, data not shown). To confirm that transduction with the GAK shRNA lentiviruses reduces GAK protein levels, we examined GAK expression in MES23.5 dopaminergic cells by immunoblot analysis and found that each of the shRNAs reduced GAK protein levels by ∼90–95% relative to control and 80–90% relative to cells expression A53T α-synuclein (Fig. 2C). These results are compatible with those obtained for the HEK 293 cells, indicating that GAK reduction plays an important role in cell toxicity not only when α-synuclein is over-expressed, but also for A53T mutant-α-synuclein.
DISCUSSION
Emerging evidence implicates endocytic pathways in the pathogenesis of the most common neurodegenerative disorders. Highlighting this concept are the results from a recent Alzheimer's disease GWAS implicating PICALM (32), a gene involved in clathrin-mediated endocytosis. In PD, the relationship between disrupted endocytic trafficking and α-synuclein accumulation has received increasing attention in a diverse number of models for the disorder (30,33,34). Notably, GAK (also known as auxilin-2) and PICALM have similar canonical interactions with clathrin (16,17,19,21,35) and both proteins are critical to clathrin-mediated endocytosis (36). Here we report that a SNP in the GAK gene (rs1564282), previously implicated as the top hit in a GWAS for familial PD (7), is significantly associated with PD risk at a genome-wide level of significance (P= 3.97 × 10−8). These results position GAK alongside SNCA and MAPT as a PD risk-associated gene, and also implicate clathrin-mediated endocytosis in the etiology of PD.
Identification of the GAK gene in a GWAS of strictly familial PD supports the importance of studying PD cohorts enriched for family history of disease. The Italian sample studied for replication had detailed data on family history, allowing us to observe the enhanced effect and improved P-value among those cases with family history of PD. Hamza et al. (14) recently confirmed our association with GAK(P= 3.1 × 10−4) in a GWAS for 2000 PD cases and 1986 controls from the NeuroGenetics Research Consortium (NGRC). Thus, while the evidence for GAK SNPs associated with PD was first seen in a cohort of familial PD (7) and reported here in the subset of Italian families with family history of PD, the association of GAK in PD risk appears to be present in random PD samples as well. Nevertheless, our results support the power of the familial PD sub-population in identifying genetic contributors to PD. Our replication and functional studies further suggest that new pathways identified in this way will likely prove relevant to the pathogenesis of idiopathic PD.
We also found that rs1564282 was associated with higher levels of SNCA expression, as reflected by the mRNA level, in the total sample of brains, adjusting for case/control status. Using a stratified analysis, we observed this eSNP relationship to be stronger in PD (beta = 0.473, P = 0.013), than in control samples (beta = 0.163, P = 0.267), although the effect of the SNP on expression is consistent in the two groups. Altered mRNA levels could be an influence on stability or degradation of the transcripts, and may not reflect altered mRNA expression. However, higher SNCA levels are consistent with the previously reported increased PD risk observed in duplication and triplication of the SNCA gene, a rare, but recognized, cause of early-onset PD (37–39). We examined the expression levels of two C-terminal GAK exons, exons 26 and 28, in post-mortem brains. Exon 26 is known to encode part of the clathrin-binding domain of GAK, which is critically involved in the sorting of the CTSD pro-enzyme, while exon 28 encodes the J-domain of the protein (22). We found exon 26 to be correlated with CTSD exon 6 expression, which was not observed for GAK exon 28. This suggests the presence of alternative splicing of GAK brain transcripts, whereby transcripts that contain exon 26, but not exon 28, have distinct relationships to CTSD levels.
We evaluated the relationship between expression of GAK and α-synuclein toxicity in two biological models: (i) a HEK 293 cell line transiently overexpressing α-synuclein, and (ii) primary rat neuronal cells transduced with the virus expressing SNCA with the A53T mutation. In both models, knockdown of GAK expression resulted in dramatic increases in cell death. Specifically, in the HEK 293 cells, siRNA knockdown of GAK increased toxicity and overall α-synuclein protein levels. In the primary neuronal cultures, expression of shRNA for GAK reduced the survival of TH+ dopaminergic neurons relative to that of total (MAP2+) neurons. From these data we infer that down-regulation of GAK enhances the preferential toxicity of A53T α-synuclein to dopaminergic neurons (29–31,40). Interestingly, the substantia nigra expression study performed by Grunblatt et al. (15) showed a 1.56-fold increase in GAK expression in PD compared with control samples. One possible explanation is the presence of a compensatory effect in the PD-affected brain region, which would lead to increased GAK expression. The minor allele of the rs1564282 GAK SNP is nominally associated with decreased expression of GAK exon 28 in frontal cortex, and decreased expression of GAK might be a possible mechanism by which this SNP is linked to increased risk for PD.
The described studies implicate GAK, SNCA and CTSD in PD pathogenesis, suggesting a role for these genes within a clathrin-mediated endocytic pathway involved in promoting neuronal toxicity. Additionally, the combination of GAK knockdown and α-synuclein over-expression may provide new in vitro models for PD, a disease that has been historically difficult to model. Finally, while the specific mechanisms by which GAK depletion enhances α-synuclein toxicity are not known, further studies targeted at enhancing GAK's function may provide novel approaches for PD treatment.
MATERIALS AND METHODS
Brain samples and microarray expression data
Brain tissue from the frontal cortex Brodmann 9 area was obtained from three different brain banks: the Harvard Brain Tissue Resource Center McLean Hospital, Belmont, Massachusetts; the Human Brain and Spinal Fluid Resource Center VA West Los Angeles Healthcare Center, California; and the Sun Health Research Institute Sun City, Arizona. Thirty-four PD and 29 control samples were identified to be the best available group of samples for a microarray study from an available pool of 118 PD and 87 control brains. We selected samples for inclusion in the micro-array by the following four criteria: (i) lack of Alzheimer disease pathology (specified by available neuropathological reports), (ii) pH value >6.25, (iii) similar age for PD cases and controls, and (iv) male gender. Total RNA for the 34 PD and 29 control samples was extracted with TRIzol (Invitrogen, Carlsbad, CA, USA). RNA was purified using the RNeasy MinElute Cleanup columns (Qiagen Sciences Inc, Germantown, MD, USA) and its quality was assessed with an Agilent Bioanalyzer Nano Chip (Agilent, Foster City, CA, USA). Two micrograms of each RNA sample were labeled and hybridized to the One-Color Agilent 60-mer Whole Human Genome Microarray at the Microarray Facility of the Whitehead Institute for Biomedical Research (Cambridge, MA, USA). The dye-normalized and post-surrogate processed signal for the green channel, gProcessedSignal, obtained from Agilent's Feature Extraction Software was used for downstream analyses. The QC reports for each of the 63 samples were individually inspected, and no systematic errors were determined for any sample. All the microarray processing analyses were performed in R (http://www.R-project.org).
To ensure that only the high-quality probes were retained for analysis, standard exclusion criteria were used as follows: (i) control probes were removed; (ii) each probe was flagged for a specific sample if (values explained in the Agilent Feature Extraction Reference Guide): gIsWellAboveBG equal to 0, gIsFeatNonUnifOL equal to 1, gIsBGNonUnifOL equal to 1, gIsFeatPopnOL equal to 1, gIsBGPopnOL equal to 1, or gIsSaturated equal to 1. If more than 50% of the PD samples or more than 50% of the control samples had a specific probe flagged, the respective probe was removed; (iii) each probe was flagged for a specific sample if the expression value for the probe was outside the range for the spike-in controls. A probe was removed if it was flagged in at least 50% PD cases and 50% controls; in addition, (iv) each probe was flagged for a specific sample if its expression was below the median expression value of the sample. If a probe was flagged in more than 80% cases and more than 80% controls, the probe was removed. To make sure we were not removing an excessive number of probes, we compared the list of genes corresponding to the retained probes with the list of ubiquitously expressed genes published by Ramskold et al. (41). 86.98% of the genes determined as ubiquitously expressed by RNA-Seq in the Ramskold et al. study were present in our set; we considered this value to be acceptable. Out of the total 45 015 probes present on the microarray chips, 22 393 (corresponding to 16 567 genes) were analyzed.
The expression data for the retained probes of the 63 arrays were quantile normalized, and the obtained values were ln (natural logarithm) transformed. Two different probes for GAK (A_23_P155700, targeting expression of the gene's exon 26, sequence: TCCTCCAGGTCTGACAAGAAAGGGCCAAAGACCATTGCAGAGATGAGGAAGCAGGACCTG and A_24_P397150, targeting expression of the gene's exon 28, sequence: CCGTACGAGCAGCACGCCAAGATGATCTTCATGGAGCTGAATGACGCCTGGTCGGAGTTT) were available on the microarray chip. One probe was available for CTSD (A_23_P52556, targeting expression of the gene's exon 6, sequence: GTATTACAAGGGTTCTCTGTCCTACCTGAATGTCACCCGCAAGGCCTACTGGCAGGTCCA). The SNCA gene was targeted by a single probe (A_23_P29939, targeting the 3′-UTR of the gene, sequence: TGACAGATGTTCCATCCTGTACAAGTGCTCAGTTCCAATGTGCCCAGTCATGACATTTCT), repeated 10 times on the microarray. All these probes passed our strict filtering criteria. The expression data for SNCA used in this manuscript is the average value for the 10 repeated probes.
Sixty-two out of the 63 microarray samples had an RIN value above 4.8, and only these 62 samples were analyzed in the current study (42) (Table 2).
Genotyping
The genotyping in the Milan, Italy cohort and the available brain samples was performed using the TaqMan technology implemented on an ABI PRISM® 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) in the Neurogenetics Laboratory at Boston University School of Medicine. In brains, the GAK SNP rs1564282 and three SNPs around the SNCA gene on chromosome 4 (Table 4) were genotyped.
Statistical analysis of SNP data
The rs1564282 SNP genotyping was obtained from three publicly available GWAS data sets available from dbGaP: a sample of familial PD cases and NINDS controls (all self-reported white, non-Hispanic and screened for population outliers using the multidimensional scaling method; n = 1728, 865 cases, 863 controls) (7), an NIA sponsored case/control GWA (all self-reported white, non-Hispanic; n = 505, 256 cases, 249 controls) (10) and the second release of publicly available data from the NIA (all self-reported white, non-Hispanic; n = 1166, 644 cases, 522 controls) (9). Subjects in the three GWASs were cleaned with identical QC thresholds, requiring 98% or higher call rate and screening for sex inconsistency. The rs1564282 SNP met all quality control criteria including call rate above 98%, consistency with Hardy–Weinberg equilibrium (HWE) in controls, and did not appear differentially missing by genotype (the MISHAP test), by case/control status or by sex. Local genotyping in the Milan, Italy cohort [all white, non-Hispanic samples; case (n = 862), controls (n = 517)] was also consistent with HWE among controls and had a 96% call rate. SNP analysis was implemented in PLINK (43) separately for each of the four cohorts using logistic regression with adjustment for age (onset for cases/enrollment for controls) and sex. Meta-analysis was implemented with METAL using the inverse variance method.
The relationship of SNPs to expression levels (eSNP studies) was performed using linear regression in SAS v9.1. The normalized mRNA levels were modeled as the dependent variable and the association of SNPs was adjusted for PD/control status, RIN, PMI and age at death. The RIN and pH were the most highly correlated variables in our data (Spearman correlation coefficient = 0.403, P-value = 0.001) and we decided to include in the linear regression model only one of these two variables, to avoid the problem of over-adjustment. We chose the RIN variable, given its larger range of values compared with pH (Table 2). The results obtained with pH in the model instead of RIN are very similar (unreported). SNPs were modeled to estimate the effect of the minor allele with an additive genetic model except when fewer than five rare allele homozygotes were present in the data (corresponding to MAF ≤ 0.14), when a dominant model was implemented (rs1564282, rs3775478).
Knockdown of GAK in HEK 293 cells, cytotoxicity and ELISA assays
HEK 293 cells (ATCC) were maintained at 37°C, 5% CO2 in DMEM supplemented with 10% fetal bovine serum (Sigma), 2 mm glutamine, 10 μg/ml penicillin and 10 μg/ml streptomycin (GIBCO). GAK knockdown experiments were conducted in 96-well plates by siRNA treatment with either ON-TARGETplus SMART pool L-005005-00-0005, human GAK or ON-TARGETplus Non-targeting Pool D-001810-10-05 (Dharmacon/Thermo), as well as plasmids DNA encoding cytomegalovirus (CMV)-α-synuclein or CMV-GFP by Lipofectamine 2000. The protocol provided by the manufacturer was followed for co-transfection of siRNA and plasmid DNA in 96-well format. Each experiment had six separate wells per condition and the experiments were repeated three times. Toxicity was accessed by adenylate kinase release (ToxiLight, Amaxa) 72 h post-transfection and subsequent luminescent readings were acquired on a Perkin Elmer Envision 2103 Multilabel reader. The α-human α-synuclein ELISA kit was obtained from Invitrogen and the protocol was described by the manufacturer.
Antibodies
The following antibodies were used in this study: mouse anti-β-actin (Clone AC-74, Sigma-Aldrich, St Louis, MO, USA); chicken anti-MAP2 (EnCor Biotechnology); rabbit anti-TH (Chemicon); mouse anti-GAK (clone 1C2, Medical and Biological Laboratories); rabbit anti-GAPDH (Abcam); mouse anti-α-synuclein (BD Biosciences); anti-rabbit IgG-Alexa Fluor 488 and anti-chicken IgG-Alexa Fluor 594 (Invitrogen) and anti-mouse IgG-conjugated with alkaline phosphatase (Promega, Madison, WI, USA).
Western blotting
Protein expression levels in MES23.5 cultures were determined via western blot analysis. The cells were dislodged from the plate by pipetting, collected by centrifugation, washed with phosphate-buffered saline (PBS) (136 mm NaCl, 0.268 mm KCl, 10 mm Na2HPO4, 1.76 mm KH2PO4, pH 7.4), and lysed in RIPA buffer [25 mm Tris–HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% (v/v) Triton X-100, 0.1% (v/v) SDS, 1 sodium deoxycholate and protease inhibitor cocktail (Sigma)]. After centrifugation at 13 000g, the detergent-soluble (supernatant) fraction was recovered. The protein concentration in the soluble fraction was measured using the bicinchoninic acid Protein Assay Kit (Pierce Biotechnology, Rockford, IL, USA), and equal amounts of protein were separated via sodium dodecyl sulfate polyacrylamide gel electrophoresis on a 4–20% (w/v) polyacrylamide gel. The proteins were transferred to a polyvinylidenedifluoride membrane, which was then probed with a primary antibody specific for GAK (1:1000). The membrane was treated with a secondary anti-mouse alkaline phosphatase-conjugated antibody (1:6000). Chemifluorescence images were obtained and analyzed using a Typhoon imaging system (GE Health Sciences, Piscataway, NJ, USA). Densitometry analysis of the western blots was performed using NIH ImageJ software, where targeted proteins (GAK or α-synuclein) were compared with house keeping genes, GAPDH or β-actin.
Preparation of lentiviral constructs
To ensure that changes in dopaminergic cell viability were due specifically to GAK knockdown and did not result from off-target effects, we conducted the shRNA experiments using three different shRNA constructs targeting different regions of the GAK gene. The use of multiple non-overlapping shRNAs (‘multiplicity control’) is an acceptable approach to ensure the validity (specificity) of an RNAi-mediated phenotype (44). All three shRNA constructs produced similar effects on dopaminergic cell viability (the data obtained using two of these constructs are included here).
The pLenti shRNA constructs targeting rat GAK were purchased from OpenBioSystems. Clone identification numbers for the two constructs presented in this study are TRCN0000027669 and TRCN0000027605. These constructs are referred to here as ‘shRNA A’ and ‘shRNA B’, respectively. The ViraPower Lentivirus Expression System (Invitrogen) was used to generate lentivirus encoding rat shRNA GAK (A or B). Lentiviral constructs were packaged into viruses as described (31,45). Lentiviral particles were titered using the Traditional p24 ELISA kit purchased from Cell Biolabs.
Preparation of adenoviral constructs
The ViraPower Adenovirus Expression System (Invitrogen) was used to generate adenovirus encoding human A53T-aSyn (downstream of the CMV promoter) as described (29,31). Adenoviral constructs were packaged into the virus via lipid-mediated transient transfection of the 293A packaging cell line. Adenoviral particles were titered using the QuickTiter Adenovirus Titer ELISA Kit (Cell Biolabs).
Preparation of primary mesencephalic cultures
Primary midbrain cultures were prepared via dissection of day 17 embryos obtained from pregnant Sprague–Dawley rats (Harlan, Indianapolis, IN, USA) as described previously (31). All of the procedures involving animal handling were approved by the Purdue Animal Care and Use Committee. The cells were plated on poly-l-lysine-treated 48-well plates at a density of 163 500 cells per well. Five days after plating, the cells were treated with cytosine arabinofuranoside (20 µm, 48 h) to inhibit the growth of glial cells. At this stage (i.e. 7 days in vitro), the glial cells accounted for ∼50% of the total cell population, and the neurons appeared differentiated with extended processes.
Lentiviral and adenoviral transductions
Primary cultures (7 days in vitro) were transduced with lentivirus and/or adenovirus in the presence of polybrene (6 µg/ml). The transductions were carried out for 72 h at a multiplicity of infection (MOI) of 3 in the case of A53T adenovirus and GAK shRNA lentivirus ‘A’, or an MOI of 1 in the case of GAK shRNA lentivirus ‘B’. The cells were then treated with fresh media for an additional 24 h and analyzed by immunocytochemistry. Control samples consisted of untransduced primary rat midbrain cultures (Fig. 1C and D), as well as primary rat midbrain cultures treated with sub-lethal levels of the A53T adenovirus and with a non-specific lentivirus (lenti-LacZ) (data not shown). In addition, the use of multiple GAK shRNA lentiviruses (multiplicity control) ensured that the phenotype produced by each shRNA was due to GAK knockdown rather than non-targeting effects (44). MES23.5 cells were transduced with A53T adenovirus (MOI = 30) and GAK shRNA lentivirus (MOI = 15 or 30) for 24 h. After incubating the cells in fresh media for 48 h, lysates were prepared and analyzed via western blotting.
Immunocytochemistry
Primary cells were fixed, permeabilized and blocked as described (31,45). After washing with PBS, the cells were treated overnight at 4°C with the primary antibodies anti-MAP2 (1:1000) and anti-TH (1:500). After washing with PBS, the cells were treated with goat anti-rabbit Alexa Fluor 488 and goat anti-chicken Alexa Fluor 594 for 1 h at room temperature. Prolong gold antifade reagent with DAPI (Invitrogen) was then applied to each before adding a cover slip.
Measurement of primary neuron viability
MAP2- and TH-immunoreactive primary neurons were counted in 10 randomly chosen observation fields (∼500–1000 neurons total) for each experimental condition using a Nikon TE2000-U inverted fluorescence microscope (Nikon Instruments, Melville, NY, USA) with a 20X objective. The data were expressed as the percentage of MAP2+ neurons that were also TH+ (this ratiometric approach was used to correct for variations in cell density). Each experiment was repeated three to four times using embryonic neurons isolated from different pregnant rats. Statistical analyses consisted of one-way analysis of variance with the Newman–Keuls post hoc test and were carried out using the program GraphPad Prism, Version 4.0 (http://www.graphpad.com/prism/Prism.htm).
Conflict of Interest statement. None declared.
FUNDING
This project was supported by two separate awards by the Robert P. & Judith N. Goldberg Foundation (to J.B.W. and S.L. supporting C.D.P.), the Bumpus Foundation, a Howard Hughes Collaborative Innovation Award and R01-NS036711. DNA samples contributed by the Parkinson Institute—Istituti Clinici di Perfezionamento, Milan, Italy—were from the ‘Human genetic bank of patients affected by PD and parkinsonisms' (http://www.parkinson.it/dnabank.html), supported by Italian Telethon grant no. GTB07001 and by the ‘Fondazione Grigioni per il Morbo di Parkinson'. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. Brain samples used in this study were provided by the Sun Health Research Institute in Sun City, Arizona [supported by the National Institute on Aging (P30 AG19610 Arizona Alzheimer's Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer's Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011 and 05-901 to the Arizona Parkinson's Disease Consortium) and the Prescott Family Initiative of the Michael J. Fox Foundation for Parkinson's Research]; the Harvard Brain Tissue Resource Center (supported in part by PHS grant number R24 MH 068855); and the Human Brain and Spinal Fluid Resource Center VA West Los Angeles Healthcare Center, 11301 Wilshire Blvd. Los Angeles, CA 90073 (sponsored by NINDS/NIMH, National Multiple Sclerosis Society, Department of Veterans).
REFERENCES
- 1.Goedert M. Alpha-synuclein and neurodegenerative diseases. Nat. Rev. Neurosci. 2001;2:492–501. doi: 10.1038/35081564. doi:10.1038/35081564. [DOI] [PubMed] [Google Scholar]
- 2.Fearnley J.M., Lees A.J. Ageing and Parkinson's disease: substantia nigra regional selectivity. Brain. 1991;114:2283–2301. doi: 10.1093/brain/114.5.2283. doi:10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- 3.Spillantini M.G., Schmidt M.L., Lee V.M.-Y., Trojanowski J.Q., Jakes R., Goedert M. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. doi:10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 4.Lazzarini A.M., Myers R.H., Zimmerman T.R., Jr, Mark M.H., Golbe L.I., Sage J.I., Johnson W.G., Duvoisin R.C. A clinical genetic study of Parkinson's disease: evidence for dominant transmission. Neurology. 1994;44:499–506. doi: 10.1212/wnl.44.3_part_1.499. [DOI] [PubMed] [Google Scholar]
- 5.Maher N.E., Currie L.J., Lazzarini A.M., Wilk J.B., Taylor C.A., Saint-Hilaire M.H., Feldman R.G., Golbe L.I., Wooten G.F., Myers R.H. Segregation analysis of Parkinson disease revealing evidence for a major causative gene. Am. J. Med. Genet. 2002;109:191–197. doi: 10.1002/ajmg.10335. doi:10.1002/ajmg.10335. [DOI] [PubMed] [Google Scholar]
- 6.Payami H., Larsen K., Bernard S., Nutt J. Increased risk of Parkinson's disease in parents and siblings of patients. Ann. Neurol. 1994;36:659–661. doi: 10.1002/ana.410360417. doi:10.1002/ana.410360417. [DOI] [PubMed] [Google Scholar]
- 7.Pankratz N., Wilk J.B., Latourelle J.C., DeStefano A.L., Halter C., Pugh E.W., Doheny K.F., Gusella J.F., Nichols W.C., Foroud T., et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum. Genet. 2009;124:593–605. doi: 10.1007/s00439-008-0582-9. doi:10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards T.L., Scott W.K., Almonte C., Burt A., Powell E.H., Beecham G.W., Wang L., Zuchner S., Konidari I., Wang G., et al. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann. Hum. Genet. 2010;74:97–109. doi: 10.1111/j.1469-1809.2009.00560.x. doi:10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon-Sanchez J., Schulte C., Bras J.M., Sharma M., Gibbs J.R., Berg D., Paisan-Ruiz C., Lichtner P., Scholz S.W., Hernandez D.G., et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat. Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. doi:10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung H.C., Scholz S., Matarin M., Simon-Sanchez J., Hernandez D., Britton A., Gibbs J.R., Langefeld C., Stiegert M.L., Schymick J., et al. Genome-wide genotyping in Parkinson's disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2006;5:911–916. doi: 10.1016/S1474-4422(06)70578-6. doi:10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- 11.Maraganore D.M., de Andrade M., Lesnick T.G., Strain K.J., Farrer M.J., Rocca W.A., Pant P.V., Frazer K.A., Cox D.R., Ballinger D.G. High-resolution whole-genome association study of Parkinson disease. Am. J. Hum. Genet. 2005;77:685–693. doi: 10.1086/496902. doi:10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satake W., Nakabayashi Y., Mizuta I., Hirota Y., Ito C., Kubo M., Kawaguchi T., Tsunoda T., Watanabe M., Takeda A., et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat. Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. doi:10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 13.Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. doi:10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 14.Hamza T.H., Zabetian C.P., Tenesa A., Laederach A., Montimurro J., Yearout D., Kay D.M., Doheny K.F., Paschall J., Pugh E., et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson's disease. Nat. Genet. 2010;42:781–785. doi: 10.1038/ng.642. doi:10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grunblatt E., Mandel S., Jacob-Hirsch J., Zeligson S., Amariglo N., Rechavi G., Li J., Ravid R., Roggendorf W., Riederer P., et al. Gene expression profiling of parkinsonian substantia nigra pars compacta; alterations in ubiquitin-proteasome, heat shock protein, iron and oxidative stress regulated proteins, cell adhesion/cellular matrix and vesicle trafficking genes. J. Neural. Transm. 2004;111:1543–1573. doi: 10.1007/s00702-004-0212-1. doi:10.1007/s00702-004-0212-1. [DOI] [PubMed] [Google Scholar]
- 16.Kanaoka Y., Kimura S.H., Okazaki I., Ikeda M., Nojima H. GAK: a cyclin G associated kinase contains a tensin/auxilin-like domain. FEBS Lett. 1997;402:73–80. doi: 10.1016/s0014-5793(96)01484-6. doi:10.1016/S0014-5793(96)01484-6. [DOI] [PubMed] [Google Scholar]
- 17.Umeda A., Meyerholz A., Ungewickell E. Identification of the universal cofactor (auxilin 2) in clathrin coat dissociation. Eur. J. Cell Biol. 2000;79:336–342. doi: 10.1078/S0171-9335(04)70037-0. doi:10.1078/S0171-9335(04)70037-0. [DOI] [PubMed] [Google Scholar]
- 18.Korolchuk V.I., Banting G. CK2 and GAK/auxilin2 are major protein kinases in clathrin-coated vesicles. Traffic. 2002;3:428–439. doi: 10.1034/j.1600-0854.2002.30606.x. doi:10.1034/j.1600-0854.2002.30606.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee D.W., Wu X., Eisenberg E., Greene L.E. Recruitment dynamics of GAK and auxilin to clathrin-coated pits during endocytosis. J. Cell Sci. 2006;119:3502–3512. doi: 10.1242/jcs.03092. doi:10.1242/jcs.03092. [DOI] [PubMed] [Google Scholar]
- 20.Massol R.H., Boll W., Griffin A.M., Kirchhausen T. A burst of auxilin recruitment determines the onset of clathrin-coated vesicle uncoating. Proc. Natl Acad. Sci. USA. 2006;103:10265–10270. doi: 10.1073/pnas.0603369103. doi:10.1073/pnas.0603369103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greener T., Zhao X., Nojima H., Eisenberg E., Greene L.E. Role of cyclin G-associated kinase in uncoating clathrin-coated vesicles from non-neuronal cells. J. Biol. Chem. 2000;275:1365–1370. doi: 10.1074/jbc.275.2.1365. doi:10.1074/jbc.275.2.1365. [DOI] [PubMed] [Google Scholar]
- 22.Kametaka S., Moriyama K., Burgos P.V., Eisenberg E., Greene L.E., Mattera R., Bonifacino J.S. Canonical interaction of cyclin G associated kinase with adaptor protein 1 regulates lysosomal enzyme sorting. Mol. Biol. Cell. 2007;18:2991–3001. doi: 10.1091/mbc.E06-12-1162. doi:10.1091/mbc.E06-12-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sevlever D., Jiang P., Yen S.H. Cathepsin D is the main lysosomal enzyme involved in the degradation of alpha-synuclein and generation of its carboxy-terminally truncated species. Biochemistry. 2008;47:9678–9687. doi: 10.1021/bi800699v. doi:10.1021/bi800699v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cullen V., Lindfors M., Ng J., Paetau A., Swinton E., Kolodziej P., Boston H., Saftig P., Woulfe J., Feany M.B., et al. Cathepsin D expression level affects alpha-synuclein processing, aggregation, and toxicity in vivo. Mol. Brain. 2009;2:5. doi: 10.1186/1756-6606-2-5. doi:10.1186/1756-6606-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao L., Hamamichi S., Caldwell K.A., Caldwell G.A., Yacoubian T.A., Wilson S., Xie Z.L., Speake L.D., Parks R., Crabtree D., et al. Lysosomal enzyme cathepsin D protects against alpha-synuclein aggregation and toxicity. Mol. Brain. 2008;1:17. doi: 10.1186/1756-6606-1-17. doi:10.1186/1756-6606-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sankoh A.J., Huque M.F., Dubey S.D. Some comments on frequently used multiple endpoint adjustment methods in clinical trials. Stat. Med. 1997;16:2529–2542. doi: 10.1002/(sici)1097-0258(19971130)16:22<2529::aid-sim692>3.0.co;2-j. doi:10.1002/(SICI)1097-0258(19971130)16:22<2529::AID-SIM692>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura K., Bindokas V.P., Kowlessur D., Elas M., Milstien S., Marks J.D., Halpern H.J., Kang U.J. Tetrahydrobiopterin scavenges superoxide in dopaminergic neurons. J. Biol. Chem. 2001;276:34402–34407. doi: 10.1074/jbc.M103766200. doi:10.1074/jbc.M103766200. [DOI] [PubMed] [Google Scholar]
- 28.Outeiro T.F., Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. doi:10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu F., Nguyen J.L., Hulleman J.D., Li L., Rochet J.C. Mechanisms of DJ-1 neuroprotection in a cellular model of Parkinson's disease. J. Neurochem. 2008;20:10–15. doi: 10.1111/j.1471-4159.2008.05333.x. [DOI] [PubMed] [Google Scholar]
- 30.Gitler A.D., Chesi A., Geddie M.L., Strathearn K.E., Hamamichi S., Hill K.J., Caldwell K.A., Caldwell G.A., Cooper A.A., Rochet J.C., et al. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat. Genet. 2009;41:308–315. doi: 10.1038/ng.300. doi:10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F., Hindupur J., Nguyen J.L., Ruf K.J., Zhu J., Schieler J.L., Bonham C.C., Wood K.V., Davisson V.J., Rochet J.C. Methionine sulfoxide reductase A protects dopaminergic cells from Parkinson's disease-related insults. Free Radic. Biol. Med. 2008;45:242–255. doi: 10.1016/j.freeradbiomed.2008.03.022. doi:10.1016/j.freeradbiomed.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Williams A., et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. doi:10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey N., Strider J., Nolan W.C., Yan S.X., Galvin J.E. Curcumin inhibits aggregation of alpha-synuclein. Acta Neuropathol. 2008;115:479–489. doi: 10.1007/s00401-007-0332-4. doi:10.1007/s00401-007-0332-4. [DOI] [PubMed] [Google Scholar]
- 34.Gitler A.D., Bevis B.J., Shorter J., Strathearn K.E., Hamamichi S., Su L.J., Caldwell K.A., Caldwell G.A., Rochet J.C., McCaffery J.M., et al. The Parkinson's disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc. Natl Acad. Sci. USA. 2008;105:145–150. doi: 10.1073/pnas.0710685105. doi:10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tebar F., Bohlander S.K., Sorkin A. Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol. Biol. Cell. 1999;10:2687–2702. doi: 10.1091/mbc.10.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parpura V., Doyle R.T., Basarsky T.A., Henderson E., Haydon P.G. Dynamic imaging of purified individual synaptic vesicles. Neuroimage. 1995;2:3–7. doi: 10.1006/nimg.1995.1003. doi:10.1006/nimg.1995.1003. [DOI] [PubMed] [Google Scholar]
- 37.Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., et al. Alpha-synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. doi:10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 38.Ibanez P., Bonnet A.M., Debarges B., Lohmann E., Tison F., Pollak P., Agid Y., Durr A., Brice A. Causal relation between alpha-synuclein gene duplication and familial Parkinson's disease. Lancet. 2004;364:1169–1171. doi: 10.1016/S0140-6736(04)17104-3. doi:10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 39.Chartier-Harlin M.C., Kachergus J., Roumier C., Mouroux V., Douay X., Lincoln S., Levecque C., Larvor L., Andrieux J., Hulihan M., et al. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. doi:10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 40.Su L.J., Auluck P.K., Outeiro T.F., Yeger-Lotem E., Kritzer J.A., Tardiff D.F., Strathearn K.E., Liu F., Cao S., Hamamichi S., et al. Compounds from an unbiased chemical screen reverse both ER-to-Golgi trafficking defects and mitochondrial dysfunction in Parkinson disease models. Dis. Model. Mech. 2010;3:194–208. doi: 10.1242/dmm.004267. doi:10.1242/dmm.004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramskold D., Wang E.T., Burge C.B., Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. doi:10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weis S., Llenos I.C., Dulay J.R., Elashoff M., Martinez-Murillo F., Miller C.L. Quality control for microarray analysis of human brain samples: the impact of postmortem factors, RNA characteristics, and histopathology. J. Neurosci. Methods. 2007;165:198–209. doi: 10.1016/j.jneumeth.2007.06.001. doi:10.1016/j.jneumeth.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whither RNAi? Nat. Cell Biol. 2003;5:489–490. doi: 10.1038/ncb0603-490. doi:10.1038/ncb0603-490. [DOI] [PubMed] [Google Scholar]
- 45.Cooper A.A., Gitler A.D., Cashikar A., Haynes C.M., Hill K.J., Bhullar B., Liu K., Xu K., Strathearn K.E., Liu F., et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. doi:10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrison P.J., Heath P.R., Eastwood S.L., Burnet P.W., McDonald B., Pearson R.C. The relative importance of premortem acidosis and postmortem interval for human brain gene expression studies: selective mRNA vulnerability and comparison with their encoded proteins. Neurosci. Lett. 1995;200:151–154. doi: 10.1016/0304-3940(95)12102-a. doi:10.1016/0304-3940(95)12102-A. [DOI] [PubMed] [Google Scholar]