Abstract
The allele frequencies of two functional single-nucleotide polymorphisms (SNPs) in the p53 pathway, the MDM2 SNP309 and TP53 Arg72Pro, vary dramatically among populations. That the frequencies of the TP53 SNP follow a clinal distribution may suggest that selective pressure from environmental variables correlated with latitude contributed to these observed population differences. Recently, winter temperature and UV radiation were found to be significantly correlated with the TP53 and the MDM2 SNPs, respectively, in East Asians; whether these correlations are more extreme than expected based upon nonselective factors such as patterns of human migration remains unclear. Here, we genotyped these two SNPs in 971 unrelated individuals from 52 unique populations worldwide and tested for correlations with both latitude and a number of climate-related environmental variables on a global scale, controlling for these neutral processes. The TP53 SNP was associated with a significant selection signal for a few climate variables, such as short-wave radiation flux in the winter, but these signals were no longer significant after correction for multiple tests. The MDM2 SNP did not exhibit a significant signal with any climate variable. Therefore, these SNPs are unlikely to be under selective pressure driven by these variables. Thus, these data underscore the need to incorporate population history when assessing signatures of selection.
INTRODUCTION
The p53 tumor suppressor is central to the process by which cells sense and respond to a variety of stresses with carcinogenic potential. Underscoring its importance to the defense against cancer is the observation that TP53 acquires an inactivating mutation in half of all cases of cancer. In normal cells, upon induction, p53 initiates transcriptional response programs that regulate cell-cycle arrest, apoptosis, DNA repair and other functions. MDM2 is the key negative regulatory partner of p53. It is transcriptionally activated by p53, but serves to inactivate p53 and target it for destruction in the proteasome (1,2).
It was recently demonstrated that p53 regulates the UV-induced pigmentation response (suntanning) by acting as a sensor for UV irradiation and by directly regulating pro-opiomelanocortin (POMC) gene expression following UV exposure (3). POMC is a multicomponent precursor gene whose product is cleaved in melanocytes and keratinocytes to produce alpha-melanocyte-stimulating hormone, a propigmentation hormone, as well as β-endorphin and adrenocorticotropic hormone. Hence, it was proposed that p53 plays an important role in the suntan response by increasing melanin production, relieving local inflammation in UV-exposed skin and inducing sun-seeking behavior (4).
In humans, single-nucleotide polymorphisms (SNPs) in the p53-pathway have been reported that alter its function. One example is a C/G SNP in exon 4 of the TP53 gene (rs1042522) that results in the substitution of an arginine (Arg; G allele) for a proline (Pro; C allele) at codon 72. The Arg72 variant exists only in humans and is more efficient both at inducing apoptosis and at suppressing transformation than is the Pro72 allele (5). Another example is the MDM2 SNP309 (T/G) (rs2279744), which lies in a putative SP1 site near the p53-responsive promoter element in the first intron of MDM2. The G allele has been associated with increased levels of MDM2 mRNA and protein, a concomitant attenuation of the p53-mediated DNA damage response, and accelerated tumor development (6,7). The association between this SNP and cancer is most evident in premenopausal women (7).
The observation that individuals with Li–Fraumeni syndrome who carry the MDM2 SNP309 G allele develop cancer 7–10 years earlier than those homozygous for the T allele suggests that high levels of MDM2 may cooperate with a weakened p53 to compromise p53-mediated tumor suppression (6,8,9). Supporting this is evidence that the MDM2 SNP309 G allele and the TP53 Pro72 allele interact to increase risk for esophageal squamous cell carcinoma (10). An interactive effect was also observed in a study of risk for therapy-related acute myeloid leukemia (t-AML), in which it was found that whereas neither polymorphism alone influenced risk, individuals homozygous for the G allele in MDM2 and the TP53 Pro72 allele were at increased risk for t-AML (11).
Genetic variants responsible for adaptive traits are expected to carry detectable signatures of natural selection; if the selective pressures vary across different regions, the frequency of the advantageous allele may be correlated with the intensity of selection. Although the biological mechanism by which variation in skin pigmentation confers a selective advantage has not been completely elucidated, it is clear that variation in skin pigmentation reflects the action of geographically varying selective pressures (12,13). In turn, levels of UV radiation and latitude are strongly correlated with skin pigmentation (14).
Both the MDM2 SNP309 and the TP53 Arg72Pro SNP alleles are differentially distributed among geographically distinct ethnic groups (15,16). In addition, a survey of seven geographically diverse populations showed that the allele frequency of the TP53 Arg72Pro SNP is strongly correlated with latitude; this correlation was statistically significant relative to the expectations of a random relationship between allele frequency and distance from the equator (15).
Given the link between p53 and tanning and the functional interactions between p53 and MDM2, we hypothesized that the geographic distribution of the TP53 Arg72Pro SNP and MDM2 SNP309 allele frequencies was coordinately shaped by selective pressures due to environmental variables such as UV exposure. Here, we examined the relationship between allele frequency and environmental variables for these SNPs worldwide to investigate the contribution of selective pressures to the geographic distribution of these variants. Recently, the correlation between these SNPs and environmental variables was examined in a large cohort of East Asian individuals (17); an assumption inherent in this analysis was that the detection of a correlation was evidence for selection. Hancock et al. (18), however, recently demonstrated that SNPs in unconstrained regions of the human genome are often strongly correlated with latitude and climate variables, suggesting that this assumption is not valid and that neutral processes such as population migrations and bottlenecks can obscure the search for signatures of selection (19).
Because population structure can be a confounding variable in these studies, we controlled for population history in order to distinguish neutral demographic processes from selection. By contrasting the correlation between these two SNPs and environmental variables against those of over 35 000 frequency-matched SNPs genotyped in the same populations, we were able to assess whether the associations were more extreme than would have been expected from population history alone.
RESULTS
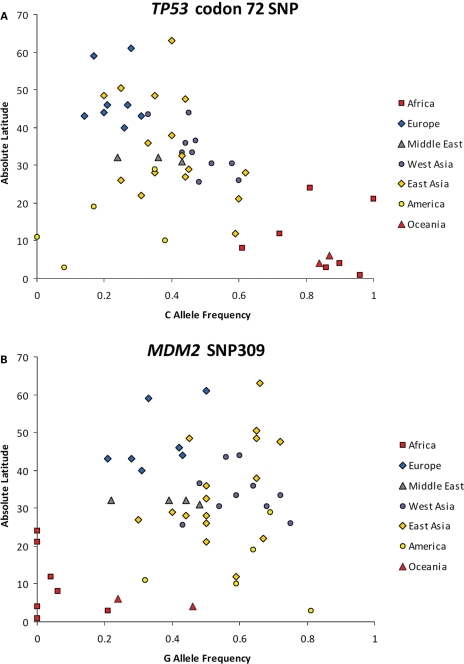
For the TP53 Arg72Pro SNP, we found that the frequency of the ancestral C allele was highest in Sub-Saharan Africa (0.96 in the Congo; 1.0 in Namibia), and lowest in Europe (0.21 in France; 0.20 in the Caucasus). The SNP was in Hardy–Weinberg equilibrium (HWE) in all 52 populations. Allele frequencies as well as the P-values for tests of HWE proportions for each geographic region are listed in Table 1, and for each of the 52 individual populations in Supplementary Material, Table S1. When plotted by absolute distance from the equator (Fig. 1A), there is a noticeable trend for the C allele frequency to decrease linearly with increasing distance (Spearman rank correlation score = −0.505). However, when this correlation score is compared in exactly the same populations to that of 36 953 control SNPs with global allele frequencies within 5% of that of the TP53 Arg72Pro SNP, this rank correlation did not reach significance (P = 0.065). Using the Bayesian method, again, the correlation between latitude and the distribution of allele frequencies was not significant (Bayes factor = 0.788, empirical P = 0.19). Thus, we cannot reject the null hypothesis that the geographic distribution of the TP53 Arg72Pro SNP was shaped by demography alone.
Table 1.
TP53 Arg72Pro and MDM2 SNP309 allele frequencies by major population group within the HGDP
| Geographic regiona | TP53 codon 72 SNP | MDM2 SNP309 | ||
|---|---|---|---|---|
| Allele frequency, G | HWE P-value | Allele frequency, G | HWE P-value | |
| 0 | 0.58 | 0 | 0.44 | 0 |
| 100 | 0.19 | 0.84 | 0.04 | 0.63 |
| 200 | 0.76 | 0.08 | 0.37 | 0.94 |
| 300 | 0.69 | 0.06 | 0.39 | 0.27 |
| 400 | 0.51 | 0.08 | 0.6 | 0.59 |
| 500 | 0.6 | 0.08 | 0.55 | 0.75 |
| 600 | 0.78 | 0.94 | 0.6 | 0.09 |
| 700 | 0.14 | 0.69 | 0.34 | 0.72 |
aCountries/areas included in geographic regions are as follows: 0—global; 100—Kenya, Central African Republic, Senegal, Nigeria, Democratic Republic of Congo, Namibia, Nigeria, South Africa; 200—Russia, Italy, France, Scotland; 300—Algeria, Israel; 400—Pakistan, China; 500—China, Cambodia, Japan; 600—Colombia, Brazil, Mexico; 700—Melanesia, Papua New Guinea.
Figure 1.
Relationship between latitude and the allele frequencies of the TP53 Arg72Pro SNP (A) and the MDM2 SNP309 (B). The minor allele frequency of each SNP in each of the 52 populations comprising the HGDP panel is plotted with respect to absolute latitude. For the TP53 Arg72Pro SNP, the Spearman rank correlation score = −0.505 (empirical P = 0.07) and the Bayes factor = 0.788 (empirical P = 0.23). For the MDM2 SNP309, the Spearman rank correlation score = 0.27 (empirical P = 0.38) and the Bayes factor = 0.23 (empirical P = 0.86).
When examining the global association of TP53 Arg72Pro SNP with winter and summer values of temperature (minimum, maximum and mean) and precipitation rate, again, neither statistical approach showed evidence for selection at this SNP after correction for multiple testing (Table 2). The Spearman rank correlation for winter short-wave radiation did exhibit a significant association (empirical P = 0.02), and both the Spearman rank correlation and Bayes factor of winter relative humidity with TP53 Arg72Pro were significant at the nominal empirical P < 0.05 level. However, when corrected for multiple testing, the association of the environmental score with allele frequency did not remain significant (maximum environmental P-value = 0.103). These data indicated that although the allele frequency distribution of the TP53 Arg72Pro across populations is unusual, we did not obtain statistical evidence that it was more extreme than could be explained by neutral processes alone.
Table 2.
Spearman rank correlation and Bayesian analysis rank scores of the TP53 Arg72Pro SNP and the MDM2 SNP309 and environmental variables
| Season | Environmental variable | Spearman correlation | Bayesian analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|
| TP53 (n = 36 953) | MDM2 (n = 35 686) | TP53 (n = 36 953) | MDM2 (n = 35 686) | ||||||
| Rank | P-value | Rank | P-value | Rank | P-value | Rank | P-value | ||
| — | Absolute latitude | 34 533 | 0.07 | 22 407 | 0.38 | 28 568 | 0.23 | 5121 | 0.86 |
| Summer | Minimum temperature | 30 529 | 0.17 | 22 935 | 0.36 | 2305 | 0.94 | 22 461 | 0.37 |
| Maximum temperature | 30 902 | 0.16 | 16 969 | 0.53 | 26 201 | 0.29 | 3620 | 0.90 | |
| Mean temperature | 31 085 | 0.16 | 15 831 | 0.56 | 20 568 | 0.44 | 317 | 0.99 | |
| Precipitation rate | 6824 | 0.82 | 2764 | 0.92 | 10 787 | 0.71 | 25 129 | 0.30 | |
| Short-wave radiation flux | 6689 | 0.82 | 11 444 | 0.68 | 18 455 | 0.50 | 30 950 | 0.14 | |
| Relative humidity | 7333 | 0.80 | 5364 | 0.85 | 35 694 | 0.03 | 24 769 | 0.31 | |
| Winter | Minimum temperature | 16 802 | 0.55 | 31 919 | 0.11 | 1414 | 0.96 | 24 795 | 0.31 |
| Maximum temperature | 30 228 | 0.18 | 28 983 | 0.19 | 12 805 | 0.65 | 22 594 | 0.37 | |
| Mean temperature | 25 234 | 0.32 | 29 580 | 0.18 | 3047 | 0.92 | 21 369 | 0.40 | |
| Precipitation rate | 19 334 | 0.48 | 15 800 | 0.56 | 9901 | 0.73 | 15 115 | 0.58 | |
| Short-wave radiation flux | 36 251 | 0.02 | 16 263 | 0.55 | 31 844 | 0.14 | 27 785 | 0.23 | |
| Relative humidity | 36 493 | 0.01 | 2571 | 0.93 | 35 133 | 0.05 | 1367 | 0.96 | |
Boldfaced P-values indicate nominal statistical significance at the 0.05 level; no correlation was significant after correction for multiple testing, as described in the text.
For the MDM2 SNP309, the minor allele (G) frequency worldwide was 0.44, but as with the TP53 Arg72Pro SNP, there was substantial variation among populations. The frequency of the G allele was highest among West Asian (0.72 in Hazara, and 0.75 in Makrani) and Native American populations (0.81 in Piapoco/Curripaco), and almost as high in East Asian populations (0.72 in Hezhen). In contrast, it was extremely uncommon in Sub-Saharan Africa (0 in Biaka Pygmy, 0.06 in Yoruba). The SNP was in HWE in all populations. Overall allele frequencies as well as HWE P-values are shown in Table 1, with population frequencies listed in Supplementary Material, Table S2. When plotted by absolute distance from the equator, no significant correlation with latitude was observed (rank correlation score = 0.268, P = 0.38; Fig. 1B). Likewise, no significant association between the MDM2 SNP309 and environmental variables was noted by either Spearman's correlation or Bayesian geographic analysis (Table 2).
Based on power calculations using the Bayesian method to detect a signal with latitude, it was estimated for each variant that there is 80% power to detect a linear effect size ≥0.08 and close to 100% power to detect an effect size ≥0.10 (20). Therefore, a lack of significant signal with the Bayesian method is consistent with no more than moderately low effect sizes. More generally, to estimate the power to detect a signal of spatially varying selection in a worldwide set of human populations would require a model of both population structure including the 52 populations tested and natural selection including a large number of unknown parameters that would have to be specified. Owing to the complexity of this model, sensible power estimates are virtually impossible to obtain. In a genome-wide analysis, however, it was shown that there is a significant excess of genic and non-synonymous SNPs with signals of correlation with environmental variables relative to non-genic SNPs, thus providing evidence that spatially varying selection at the genome level can be detected (21). Therefore, we took an empirical approach in which we chose a set of candidate selected variants that were genotyped in the same population samples and subjected them to the same Bayesian analysis as the TP53 and MDM2 SNPs (22). As candidate selected variants, we chose all the SNPs in the genome-wide association study (GWAS) catalog (23) that had been associated with a pigmentation phenotype in a GWAS (24). Because variation in pigmentation is likely to confer a selective advantage as a result of different levels of UV radiation (12), these SNPs are expected to be targets of spatially varying selection and to exhibit strong correlations between allele frequency and short-wave radiation. As shown in Supplementary Material, Table S3, 6 out of 22 pigmentation SNPs have a significant correlation signal with short-wave radiation flux, and 10 out of 22 have a significant signal with at least one of the variables tested. This suggests that we have reasonable power to detect a significant signal of selection using the Bayesian method in this set of population samples. It should also be noted that, even though these SNPs are associated with pigmentation phenotypes, not all of them are necessarily targets of selection; therefore, our power is likely to be higher than the proportion of significant SNPs shown in Supplementary Material, Table S3.
DISCUSSION
In light of the critical roles played by p53 and MDM2 in the response to both potentially carcinogenic stresses and chemotherapy, elucidating the selective forces that shaped functional variation in these genes across human populations may shed significant light on the mechanisms by which the p53 pathway defends against cancer, and may also suggest targets to manipulate the anticancer activities of p53 therapeutically. Consequently, we undertook an agnostic analysis to assess the contribution of environmental factors to the evolution of two well-characterized functional variants in the p53 pathway.
For the TP53 Arg72Pro SNP, we confirmed that there are significant differences in allele frequencies among different populations, with the ancestral C allele, encoding the Pro variant, predominating among Africans, but the G variant, encoding the Arg variant, more common among Europeans. When compared with 36 953 SNPs with similar global allele frequencies, we found no evidence to suggest that this SNP is more significantly correlated with latitude than expected by ancestry alone. The Spearman rank correlation analysis showed that 7% of the control SNPs had a stronger correlation with latitude than the Arg72Pro SNP, and the Bayesian analysis showed that 23% of the control SNPs were more strongly correlated with latitude than was this SNP.
When we assessed the correlation between the TP53 Arg72Pro variant and environmental variables related to climate, we observed significant empirical P-values for relative humidity in the winter by both the Bayesian and the Spearman rank correlation methods, as well as short-wave radiation flux in the winter (Spearman rank correlation) and relative humidity in the summer (Bayesian analysis). Although the significant association with short-wave radiation flux is clearly interpretable based on the known involvement of p53 in the suntan response, there are considerable data demonstrating that p53 is a mediator of the cellular response to many cellular stresses in addition to short-wave radiation, some of which may be related to different environmental variables. For this reason, we subjected all variables to the same unbiased analysis free from prior assumptions of biological significance. When we correct for the multiple tests performed, none of these results remained significant. Similarly, for the MDM2 SNP309, we observed significant differences in allele frequencies among populations, but we were not able to demonstrate a significant association between the SNP and either latitude or any of the environmental variables investigated.
Recently, Shi et al. (17) genotyped these same SNPs in 67 East Asian populations over a wide range of environmental conditions as defined by latitude (11° south of the equator to 65° north of the equator). They concluded that the TP53 Arg72Pro G allele, encoding the Arg allele, was positively correlated with low winter temperature, and the MDM2 SNP309 G allele was negatively correlated with UV radiation; they further proposed that the reported relationships between allele frequencies and environmental variables reflect the action of spatially varying selective pressures.
Our results contrast with those of Shi et al. and underscore the importance of assessing evidence for signatures of selection in the context of human population history. Analyses undertaken using the Human Genome Diversity Project (HGDP) panel suggest that clines due to neutral processes such as population migrations and interbreeding are common in human populations (19,25,26). Evidence for natural selection should be based on detecting a correlation between a putative selective force and the frequencies of a candidate advantageous allele across populations that is greater than expected by taking into account the role of human population history in shaping geographic patterns of allelic variation.
Recently, global approaches have been developed for detecting the impact of selective pressures. For example, it has been shown that variants influencing risk to hypertension follow latitudinal clines; these clines are hypothesized to reflect adaptive changes in sodium homeostasis to different climates (27,28). A more recent study measured correlations of climate variables with the frequency distribution of SNPs in candidate genes associated with metabolic disorders, comparing the correlations of candidate SNPs with correlations of control SNPs that would be expected under neutrality (18). As in the present analysis, these studies differ from that of Shi et al. in that they aimed to detect a correlation with either an environmental or geographic variable stronger than expected under neutrality based on the history of population structure and migrations.
It might be predicted that the confounding effect of neutral processes would be exaggerated in a regional analysis of dynamic populations. As a result, to minimize these confounding effects, we undertook our study on a global rather than a regional scale. We were not able to detect evidence for the action of environmental variables on the evolution of the p53 pathway. One caveat, however, is that a global analysis such as the one here may require more data in order to detect a selective effect on the p53 pathway; although our analysis of data from a pigmentation GWAS indicates that we have sufficient power to detect significant signals of selection in the HGDP panel, we have limited power to detect modest effects. Consequently, further investigations into the causes of the unusual clinal distribution of the TP53 Arg72Pro SNP are warranted. Nonetheless, future studies must incorporate an evaluation of neutral processes into their design.
The p53 pathway is critical to healthy aging, the defense against cancer and other disorders and the response to anticancer therapy. Thus, insight into the forces that shaped its evolution could shed light on its normal function in health and mechanisms by which it is undermined in disease. The elucidation of these pressures and the investigation of their consequences will be of considerable interest as the interplay between genetics and the environment becomes increasingly integrated into routine medical care.
MATERIALS AND METHODS
SNP genotyping was performed on the entire HGDP–Centre d'Etude du Polymorphisme Humain panel, a collection of DNA samples from 1064 healthy individuals from 52 discrete populations from five continents. For details on these samples, see: http://www.cephb.fr/HGDP-CEPH-Panel/. The GenomiPhi kit (Amersham) was used for whole-genome amplification of the genomic DNA. Allele frequencies were determined by allele-specific PCR using the 5′ nuclease assay (TaqMan). Primer and probe sequences were designed using Primer Express v.2 software (ABI PRISM). They were manufactured as Assays-by-Design (ABI), and performed according to the manufacturer's specifications. In brief, 10 ml reactions were set up in 96-well plates with 2 ml amplified template genomic DNA, and cycled under standard conditions (50°C for 2 min), then a denaturation step at 95°C for 10 min, followed by 60 cycles of 92°C for 30 s, and 60°C for 1 min. Endpoint reads were conducted on the ABI 7300 sequence detection system. Cluster analysis was conducted on the scatter plot of Allele A Rn versus Allele B Rn. Genotype discrimination was determined and displayed in the allelic plot with four clusters: no calls, Allele A, Allele B and heterozygous. The data were then exported for further analysis.
For the TP53 Arg72Pro SNP, the amplification primers used were 5′-ATGAAGCTCCCAGAATGC and 5′-GCCGGTGTAGGAGCT. The G allele-specific probe was 5′-FAMCTGCTCCCCCCGTGGCCC-TAM. The C allele-specific probe was 5′-VIC-CTGCTCCCCGCGTGGCCC-TAM. For the MDM2 SNP309, the amplification primers used were 5′-CGGGAGTTCAGGGTAAAGGT and 5′-GCGCAGCGTTCACACTAG. The T allele-specific probe was 5′-VIC-CTCCCGCGCCGAAG-TAM. The G allele-specific probe was 5′-FAMTCCCGCGCCGCAG-TAM.
The allele frequency of each SNP was calculated for each population within the HGDP panel, using data only from the 971 unrelated individuals in our analysis. Average winter and summer values of minimum temperature, maximum temperature, mean surface temperature, precipitation rate, relative humidity and short-wave radiation flux for approximate locations of HGDP populations were taken from the IRI/LDEO Climate Data Library (http://ingrid.ldeo.columbia.edu/). Absolute latitude, as a measure of distance from the equator, was included as an environmental variable because it might capture the long-term climate of human populations better than the climate variables measured over the past 50 years.
Two complementary test statistics were computed to evaluate the relationship between SNP allele frequencies and each environmental variable. The first statistic is the Spearman rank correlation coefficient, which is non-parametric and does not assume a linear relationship. The second statistic is a Bayes factor that derives from a Bayesian geographical analysis method that does assume linearity (20). The Bayes factor measures the degree of support for a linear relationship between environmental variable and allele frequency relative to a null model that controls for the covariance structure of allele frequencies across populations. This method has the advantage over a correlation coefficient in that it explicitly takes into account both the covariance of allele frequencies due to population structure and the uncertainty in allele frequency estimates due to sampling.
To assess the significance of the results, first, we compared the test statistics generated by each method for the correlation between each SNP and variable to an empirical distribution of test statistics generated from a large set of frequency-matched control SNPs; an empirical P-value of association with each variable was determined for each SNP by calculating the proportion of control SNPs with more extreme test statistic values. Then, because the environmental variables analyzed are all highly correlated with each other and with absolute latitude, to control for the family-wise error rate associated with undertaking multiple highly correlated tests, we combined the Spearman rank correlation and Bayes factor for each environmental variable into a single test statistic, referred to as an environmental score, by summing the negative logarithm of their empirical P-values. We compared the maximum environmental score for each test SNP with the distribution of the maximum environmental scores for the set of frequency-matched control SNPs to generate an environmental P-value of association for each SNP.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the National Institutes of Health (DK56670 to A.D. and HD0433871 to K.O.).
ACKNOWLEDGEMENTS
We are grateful to Angela Hancock, Nathan Ellis and Andrew Skol for many productive discussions and thoughtful feedback that helped to shape this manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. doi:10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Levine A.J., Oren M. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. doi:10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui R., Widlund H.R., Feige E., Lin J.Y., Wilensky D.L., Igras V.E., D'Orazio J., Fung C.Y., Schanbacher C.F., Granter S.R., et al. Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell. 2007;128:853–864. doi: 10.1016/j.cell.2006.12.045. doi:10.1016/j.cell.2006.12.045. [DOI] [PubMed] [Google Scholar]
- 4.Oren M., Bartek J. The sunny side of p53. Cell. 2007;128:826–828. doi: 10.1016/j.cell.2007.02.027. doi:10.1016/j.cell.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 5.Dumont P., Leu J.I., Della Pietra A.C., III, George D.L., Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 2003;33:357–365. doi: 10.1038/ng1093. doi:10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 6.Bond G.L., Hu W., Bond E.E., Robins H., Lutzker S.G., Arva N.C., Bargonetti J., Bartel F., Taubert H., Wuerl P., et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. doi:10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 7.Bond G.L., Hirshfield K.M., Kirchhoff T., Alexe G., Bond E.E., Robins H., Bartel F., Taubert H., Wuerl P., Hait W., et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. doi:10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- 8.Bougeard G., Baert-Desurmont S., Tournier I., Vasseur S., Martin C., Brugieres L., Chompret A., Bressac-de Paillerets B., Stoppa-Lyonnet D., Bonaiti-Pellie C., et al. Impact of the MDM2 SNP309 and p53 Arg72Pro polymorphism on age of tumour onset in Li–Fraumeni syndrome. J. Med. Genet. 2006;43:531–533. doi: 10.1136/jmg.2005.037952. doi:10.1136/jmg.2005.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arva N.C., Gopen T.R., Talbott K.E., Campbell L.E., Chicas A., White D.E., Bond G.L., Levine A.J., Bargonetti J. A chromatin-associated and transcriptionally inactive p53-Mdm2 complex occurs in mdm2 SNP309 homozygous cells. J. Biol. Chem. 2005;280:26776–26787. doi: 10.1074/jbc.M505203200. doi:10.1074/jbc.M505203200. [DOI] [PubMed] [Google Scholar]
- 10.Hong Y., Miao X., Zhang X., Ding F., Luo A., Guo Y., Tan W., Liu Z., Lin D. The role of P53 and MDM2 polymorphisms in the risk of esophageal squamous cell carcinoma. Cancer Res. 2005;65:9582–9587. doi: 10.1158/0008-5472.CAN-05-1460. doi:10.1158/0008-5472.CAN-05-1460. [DOI] [PubMed] [Google Scholar]
- 11.Ellis N.A., Huo D., Yildiz O., Worrillow L.J., Banerjee M., Le Beau M.M., Larson R.A., Allan J.M., Onel K. MDM2 SNP309 and TP53 Arg72Pro interact to alter therapy-related acute myeloid leukemia susceptibility. Blood. 2008;112:741–749. doi: 10.1182/blood-2007-11-126508. doi:10.1182/blood-2007-11-126508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jablonski N.G., Chaplin G. The evolution of human skin coloration. J. Hum. Evol. 2000;39:57–106. doi: 10.1006/jhev.2000.0403. doi:10.1006/jhev.2000.0403. [DOI] [PubMed] [Google Scholar]
- 13.Roberts D.F. Body weight, race and climate. Am. J. Phys. Anthropol. 1953;11:533–558. doi: 10.1002/ajpa.1330110404. doi:10.1002/ajpa.1330110404. [DOI] [PubMed] [Google Scholar]
- 14.Jablonski D., Roy K., Valentine J.W. Out of the tropics: evolutionary dynamics of the latitudinal diversity gradient. Science. 2006;314:102–106. doi: 10.1126/science.1130880. doi:10.1126/science.1130880. [DOI] [PubMed] [Google Scholar]
- 15.Beckman G., Birgander R., Sjalander A., Saha N., Holmberg P.A., Kivela A., Beckman L. Is p53 polymorphism maintained by natural selection? Hum. Hered. 1994;44:266–270. doi: 10.1159/000154228. doi:10.1159/000154228. [DOI] [PubMed] [Google Scholar]
- 16.Sjalander A., Birgander R., Saha N., Beckman L., Beckman G. p53 polymorphisms and haplotypes show distinct differences between major ethnic groups. Hum. Hered. 1996;46:41–48. doi: 10.1159/000154324. doi:10.1159/000154324. [DOI] [PubMed] [Google Scholar]
- 17.Shi H., Tan S.J., Zhong H., Hu W., Levine A., Xiao C.J., Peng Y., Qi X.B., Shou W.H., Ma R.L., et al. Winter temperature and UV are tightly linked to genetic changes in the p53 tumor suppressor pathway in Eastern Asia. Am. J. Hum. Genet. 2009;84:534–541. doi: 10.1016/j.ajhg.2009.03.009. doi:10.1016/j.ajhg.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock A.M., Witonsky D.B., Gordon A.S., Eshel G., Pritchard J.K., Coop G., Di Rienzo A. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 2008;4:e32. doi: 10.1371/journal.pgen.0040032. doi:10.1371/journal.pgen.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novembre J., Di Rienzo A. Spatial patterns of variation due to natural selection in humans. Nat. Rev. Genet. 2009;10:745–755. doi: 10.1038/nrg2632. doi:10.1038/nrg2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coop G., Witonsky D., Di Rienzo A., Pritchard J.K. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–1423. doi: 10.1534/genetics.110.114819. doi:10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock A.M., Witonsky D.B., Ehler E., Alkorta-Aranburu G., Beall C., Gebremedhin A., Sukernik R., Utermann G., Pritchard J., Coop G., et al. Colloquium paper: human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc. Natl Acad. Sci. USA. 2010;107(Suppl. 2)):8924–8930. doi: 10.1073/pnas.0914625107. doi:10.1073/pnas.0914625107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. doi:10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 23.Hindorff L.A., Junkins H.A., Hall P.N., Mehta J.P., Manolio T.A. A Catalog of Published Genome Wide Association Studies. 2010. Available at: www.genome.gov/gwastudies. accessed 12/11/10.
- 24.Sulem P., Gudbjartsson D.F., Stacey S.N., Helgason A., Rafnar T., Magnusson K.P., Manolescu A., Karason A., Palsson A., Thorleifsson G., et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. doi:10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 25.Coop G., Pickrell J.K., Novembre J., Kudaravalli S., Li J., Absher D., Myers R.M., Cavalli-Sforza L.L., Feldman M.W., Pritchard J.K. The role of geography in human adaptation. PLoS Genet. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. doi:10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg N.A., Pritchard J.K., Weber J.L., Cann H.M., Kidd K.K., Zhivotovsky L.A., Feldman M.W. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. doi:10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 27.Thompson E.E., Kuttab-Boulos H., Witonsky D., Yang L., Roe B.A., Di Rienzo A. CYP3A variation and the evolution of salt-sensitivity variants. Am. J. Hum. Genet. 2004;75:1059–1069. doi: 10.1086/426406. doi:10.1086/426406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young J.H., Chang Y.P., Kim J.D., Chretien J.P., Klag M.J., Levine M.A., Ruff C.B., Wang N.Y., Chakravarti A. Differential susceptibility to hypertension is due to selection during the out-of-Africa expansion. PLoS Genet. 2005;1:e82. doi: 10.1371/journal.pgen.0010082. doi:10.1371/journal.pgen.0010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.