Abstract
High blood concentration of the N-terminal cleavage product of the B-type natriuretic peptide (NT-proBNP) is strongly associated with cardiac dysfunction and is increasingly used for heart failure diagnosis. To identify genetic variants associated with NT-proBNP level, we performed a genome-wide association analysis in 1325 individuals from South Tyrol, Italy, and followed up the most significant results in 1746 individuals from two German population-based studies. A genome-wide significant signal in the MTHFR-CLCN6-NPPA-NPPB gene cluster was replicated, after correction for multiple testing (replication one-sided P-value = 8.4 × 10−10). A conditional regression analysis of 128 single-nucleotide polymorphisms in the region of interest identified novel variants in the CLCN6 gene as independently associated with NT-proBNP. In this locus, four haplotypes were associated with increased NT-proBNP levels (haplotype-specific combined P-values from 8.3 × 10−03 to 9.3 × 10−11). The observed increase in the NT-proBNP level was proportional to the number of haplotype copies present (i.e. dosage effect), with an increase associated with two copies that varied between 20 and 100 pg/ml across populations. The identification of novel variants in the MTHFR-CLCN6-NPPA-NPPB cluster provides new insights into the biological mechanisms of cardiac dysfunction.
INTRODUCTION
NT-proBNP is the N-terminal signal peptide of pro-B-type natriuretic peptide (proBNP). ProBNP is synthesized in the heart as a reaction to increased wall distension, neurohormonal and immunological activation (1–4), and splits into BNP and NT-proBNP upon secretion from the cardiomyocyte. Although BNP and NT-proBNP, which have both emerged as important markers of cardiac function, have a common genetic precursor, blood concentrations of NT-proBNP differ markedly from and exceed those of BNP due to differential clearance mechanisms and a longer half-life (5–10).
Previously, an important role of genetic factors in the regulation of blood BNP level was suggested by a heritability estimate of 0.35, reported from a study of 1914 individuals of Caucasian origin (11). A similar estimate was obtained for NT-proBNP in 1325 individuals from our MICROS (Microisolates in South Tyrol) study in South Tyrol (data not published). A genome-wide association (GWA) analysis of BNP in the Framingham Heart Study (12) identified only one locus for BNP regulation, the nuclear factor I/A (NFIA) gene, expressed in the heart, which has not yet been replicated. No GWA analysis of NT-proBNP has been reported yet. Candidate gene studies revealed a significant association of NT-proBNP with its precursor gene NPPB, encoding the proBNP, in 164 normo-albuminuric type 1 diabetic patients (13) as well as with the natriuretic peptide precursor A (NPPA) gene, both in healthy individuals (14) and in patients with severe heart failure (15). These results were confirmed in a meta-analysis of 14 473 samples of European ancestry, which showed a strong association of variants in the NPPB and NPPA genes with BNP and NT-proBNP levels (16).
To improve our comprehension of genetic variants influencing the complex biology of the natriuretic peptide system, to identify potential genetic modifiers of cardiac hormonal response to increased wall stress and to better understand the genetic contribution to NT-proBNP serum concentration by identifying additional genetic variants involved in this system, we performed a GWA study of NT-proBNP blood level in 1325 individuals from the MICROS study, a population-based genetic survey of three semi-isolated Alpine villages in northern Italy (17). Results from this analysis were subsequently assessed for possible replication in two independent population-based studies from southern and northern Germany, namely KORA-F3 (from the MONICA/KORA study) and popgen.
RESULTS
GWA and replication analyses
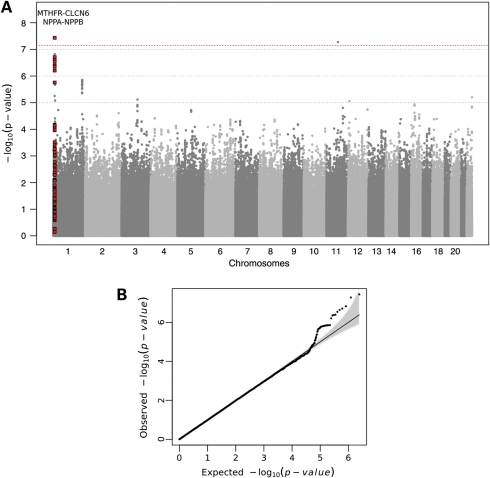
Characteristics of the study samples are reported in Table 1. The mean NT-proBNP level was 102.5 (standard deviation, SD = 365.0), 96.1 (SD = 171.2) and 144.3 (SD = 240.4)pg/ml in MICROS, popgen and KORA-F3, respectively. The Manhattan plot of the discovery GWA analysis in MICROS is given in Figure 1A together with the quantile–quantile (QQ) plot to illustrate the level of potential P-value inflation (Fig. 1B). The genomic control factor (λ) was 0.99, indicating that no cryptic relatedness or hidden population structure affected our GWA results. To account for multiple testing, the significance threshold in the GWA analysis was set to 7.2 × 10–8 (18). Two genome-wide significant hits were observed, namely for SNPs rs1023252, located within the chloride channel 6 (CLCN6) gene (P-value = 3.7 × 10−08), and rs1004565, located close to the odz, odd Oz/ten-m homolog 4 (Drosophila) (ODZ4) gene (P-value = 5.3 × 10−08) (Table 2).
Table 1.
Characteristics of study samples [mean (SD) unless otherwise specified]
| Study name | MICROS (Discovery study) | popgen (Replication study) | KORA-F3 (Replication study) |
|---|---|---|---|
| Number of samples, n (%) | 1325 (100) | 1152 (100) | 594 (100) |
| Females, n (%) | 749 (57) | 512 (44) | 325 (55) |
| Age (years) | 45 (17) | 57 (14) | 62 (10) |
| NT-proBNP (pg/ml) | |||
| Median (range) | 45.6 (20.0–8116.0) | 48.4 (4.9–2304.0) | 77.6 (7.2–2504.0) |
| Distribution, n (%) | |||
| <20 | 241 (18.2) | 194 (16.8) | 28 (4.7) |
| 20–125 | 911 (68.7) | 754 (65.4) | 384 (64.7) |
| 125–300 | 124 (9.3) | 144 (12.5) | 127 (21.4) |
| >300 | 49 (3.7) | 60 (5.2) | 55 (9.3) |
| BMI | 24.7 (4.8) | 26.3 (4.4) | 27.6 (4.2) |
| DBP (mmHg) | 79.5 (11.4) | 80.9 (9.5) | 82.7 (17.4) |
| SBP (mmHg) | 132.5 (20.6) | 134.9 (17.3) | 131.9 (21.1) |
| Hypertensiona, n (%) | 452 (36.5) | 537 (46.9) | 364 (61.3) |
| Strokeb, n (%) | 19 (1.4) | 25 (2.2) | 14 (2.3) |
| Myocardial infarctionb, n (%) | 16 (1.2) | 33 (2.9) | 15 (2.5) |
| Atrial fibrillationb, n (%) | 6 (0.4) | 13 (1.2) | Not available |
SD, standard deviation; DBP, diastolic blood pressure; SBP, systolic blood pressure; BMI, body mass index.
aDefined as SBP ≥140 mmHg or DBP ≥90 mmHg or under treatment for hypertension.
bAssessed by questionnaire interview.
Figure 1.
(A) Manhattan plot of the results of the GWA analysis in the MICROS study. The locus marked in red satisfied the replication criterion. (B) The QQ plot in the upper-right corner compares expected versus observed –log10(P-value) for all 2.5 million SNPs included in the GWA analysis, with the dashed line corresponding to the null hypothesis of no association.
Table 2.
SNPs selected for replication by chromosome and position: results of the discovery, replication and combined analyses
| SNPs selected from the discovery analysis for replication |
Discovery analysis (MICROS) | Replication analysis (meta-analysis of data from popgen and KORA-F3) | Combined analysis (meta-analysis of data from MICROS, popgen and KORA-F3) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Chr | Position (NCBI build 36) | Gene annotation (distance to nearest gene) | Major/minor allelesa | MAFb | Effect (SE) | P-valuec | Effect (SE) | One-sided P-valued | Effect (SE) | P-value |
| rs1023252 | 1 | 11 821 620 | CLCN6 (intronic) | G/T | 0.26 | 0.21 (0.04) | 3.7 × 10−08 | 0.20 (0.03) | 8.4 × 10−10 | 0.20 (0.02) | 3.5 × 10−16 |
| rs10927698 | 1 | 15 376 990 | TMEM51 (intronic) | T/G | 0.39 | −0.16 (0.03) | 2.1 × 10−06 | −0.02 (0.03) | 0.275 | −0.08 (0.02) | 2.9 × 10−04 |
| rs7547072 | 1 | 152 905 879 | ADAR (38 kb) | C/T | 0.48 | 0.13 (0.03) | 8.7 × 10−05 | −0.01 (0.03) | 0.653 | 0.06 (0.02) | 0.014 |
| rs10864721 | 1 | 230 340 755 | DISC1 (97 kb) | G/C | 0.18 | 0.19 (0.04) | 1.4 × 10−06 | −0.05 (0.04) | 0.907 | 0.06 (0.03) | 0.019 |
| rs3919597 | 2 | 66 882 384 | MEIS1 (239 kb) | C/T | 0.47 | 0.13 (0.03) | 3.0 × 10−05 | 0.06 (0.03) | 0.026 | 0.09 (0.02) | 1.7 × 10−05 |
| rs1921649 | 2 | 223 977 434 | SCG2 (192 kb) | G/A | 0.47 | −0.15 (0.03) | 2.5 × 10−05 | −0.03 (0.03) | 0.171 | −0.08 (0.02) | 3.5 × 10−04 |
| rs1919008 | 3 | 108 312 022 | CCDC54 (266 kb) | C/A | 0.14 | 0.23 (0.05) | 1.2 × 10−05 | −0.08 (0.04) | 0.957 | 0.05 (0.03) | 0.139 |
| rs16842453 | 3 | 136 197 466 | EPHB1 (intronic) | G/C | 0.06 | −0.30 (0.07) | 5.1 × 10−05 | −0.06 (0.06) | 0.144 | −0.15 (0.04) | 8.8 × 10−04 |
| rs7669710 | 4 | 26 982 314 | STIM2 (346 kb) | G/A | 0.43 | 0.14 (0.03) | 3.9 × 10−05 | 0.05 (0.03) | 0.064 | 0.09 (0.02) | 8.7 × 10−05 |
| rs17071059 | 4 | 182 471 437 | ODZ3 (101 kb) | T/C | 0.15 | −0.18 (0.05) | 7.4 × 10−05 | 0.06 (0.04) | 0.944 | −0.04 (0.03) | 0.155 |
| rs4869419 | 5 | 92 593 295 | CCT7P2 (34 kb) | A/G | 0.08 | −0.25 (0.06) | 2.1 × 10−05 | −0.14 (0.06) | 0.014 | −0.20 (0.04) | 3.8 × 10−06 |
| rs9294233 | 6 | 82 327 882 | FAM46A (184 kb) | A/G | 0.31 | −0.14 (0.03) | 7.3 × 10−05 | 0.00 (0.03) | 0.522 | −0.06 (0.02) | 0.008 |
| rs12533743 | 7 | 24 435 064 | NPY (137 kb) | T/C | 0.38 | −0.14 (0.03) | 4.9 × 10−05 | −0.01 (0.03) | 0.399 | −0.07 (0.02) | 0.002 |
| rs7820107 | 8 | 93 765 031 | RUNX1T1 (588 kb) | T/C | 0.04 | 0.32 (0.08) | 4.4 × 10−05 | 0.07 (0.09) | 0.199 | 0.21 (0.06) | 3.2 × 10−04 |
| rs396250 | 9 | 105 127 157 | CYLC2 (306 kb) | G/A | 0.10 | 0.21 (0.05) | 6.5 × 10−05 | −0.12 (0.05) | 0.993 | −0.03 (0.04) | 0.340 |
| rs1004565 | 11 | 79 055 348 | ODZ4 (597 kb) | C/T | 0.02 | 0.82 (0.15) | 5.3 × 10−08 | −0.19 (0.11) | 0.955 | −0.18 (0.09) | 0.054 |
| rs11226914 | 11 | 105 446 714 | KBTBD3 (intronic) | T/G | 0.02 | 0.45 (0.11) | 3.2 × 10−05 | −0.06 (0.14) | 0.669 | 0.26 (0.09) | 0.002 |
| rs10845158 | 12 | 10 580 021 | KLRA1 (52 kb) | C/G | 0.33 | 0.16 (0.04) | 8.7 × 10−06 | 0.00 (0.03) | 0.496 | 0.08 (0.02) | 0.002 |
| rs10847578 | 12 | 127 272 840 | TMEM132C (393 kb) | T/C | 0.49 | 0.15 (0.03) | 1.8 × 10−05 | −0.03 (0.03) | 0.815 | 0.05 (0.02) | 0.031 |
| rs9510008 | 13 | 21 637 260 | FGF9 (463 kb) | C/T | 0.10 | −0.22 (0.06) | 8.4 × 10−05 | −0.08 (0.08) | 0.148 | −0.17 (0.04) | 1.5 × 10−04 |
| rs12920706 | 16 | 22 638 484 | HS3ST2 (94 kb) | T/C | 0.43 | 0.19 (0.04) | 1.2 × 10−05 | 0.01 (0.03) | 0.401 | 0.07 (0.03) | 0.005 |
| rs9940021 | 16 | 26 204 097 | HS3ST4 (149 kb) | G/T | 0.48 | 0.13 (0.03) | 3.2 × 10−05 | −0.01 (0.03) | 0.577 | 0.06 (0.02) | 0.006 |
| rs1943064 | 16 | 73 547 505 | WDR59 (intronic) | C/T | 0.39 | 0.14 (0.03) | 2.8 × 10−05 | −0.09 (0.03) | 0.998 | −0.02 (0.02) | 0.458 |
| rs9620688 | 22 | 26 109 122 | MN1 (365 kb) | G/A | 0.32 | 0.13 (0.03) | 9.8 × 10−05 | 0.03 (0.04) | 0.234 | 0.09 (0.03) | 6.1 × 10−04 |
| rs6009824 | 22 | 48 472 377 | BRD1 (81 kb) | G/A | 0.15 | −0.18 (0.04) | 6.3 × 10−06 | −0.08 (0.04) | 0.032 | −0.13 (0.03) | 6.7 × 10−06 |
aThe reference allele for association analysis is always the minor allele.
bMAF: minor allele frequency as estimated by the sample-size-weighted mean of individual study MAFs.
cIn bold, genome-wide significant P-values.
dIn bold, P-values ≤threshold for replication; threshold for replication was set to 0.002, i.e. the nominal level of 0.05 divided by 25 independent tests (Bonferroni correction for multiple testing); in italics, P-values ≤0.05.
To assess the possible impact of pharmacological treatment upon our GWA results, sensitivity analyses were performed where individuals taking drugs potentially affecting the NT-proBNP level were removed from the analysis (details are given in Materials and Methods). Results from these sensitivity analyses did not differ from those of the main GWA analysis.
Twenty-five SNPs with P-values <10−05 were selected for replication, provided that at least another SNP with P-value <10−04 was located within 100 kb and that the power to replicate, when accounting for multiple testing, was ≥80%. The SNPs were chosen to represent independent loci, with independency defined as pair-wise r2 value <0.2 (see Materials and Methods). Among these, the power to replicate, after accounting for multiple testing, ranged from 82 to 99%. The replication analysis was performed separately in popgen and KORA-F3, and the results were pooled using an inverse-variance-weighted fixed-effect meta-analysis. We considered a locus to be replicated when the effect direction was the same in the discovery and replication analyses, and the corresponding one-sided replication P-value was ≤0.002 (i.e. Bonferroni correction). Results of both the discovery and the replication analyses are reported in Table 2. Also included are the results of a joint analysis of all discovery and replication samples, using an inverse-variance-weighted fixed-effect meta-analysis as well.
The association between rs1023252 and loge(NT-proBNP) observed in the GWA analysis was replicated in the German samples (one-sided P-value = 8.4 × 10–10), with an effect in the replication analysis that was remarkably similar to that of the discovery analysis (Table 2). In the combined analysis, the effect of the SNP was 0.20 loge pg/ml per allele (P-value = 3.5 × 10–16). This SNP identifies a locus that, in addition to the CLCN6 gene, also includes the methylenetetrahydrofolate reductase (MTHFR) gene, the NPPA gene and the NPPB gene. Given the high level of linkage disequilibrium (LD) within the MTHFR-CLCN6-NPPA-NPPB gene cluster, we investigated the whole locus using conditional single-marker and haplotype analyses (see what follows) to assess whether the signal in the CLCN6 gene was independent from that of the NPPB gene, the obvious biological candidate for regulating the NT-proBNP level.
Three additional SNPs, although not surviving the Bonferroni correction in the replication analysis, were found to be associated with NT-proBNP in the replication analysis at a nominal significance level of 0.05 and with effect in the same direction as in the discovery analysis, namely rs3919597 (one-sided P-value = 0.026), rs4869419 (one-sided P-value = 0.014) and rs6009824 (one-sided P-value = 0.032). SNP rs3919597 is located 239 kb downstream of the Meis homeobox 1 (MEIS1) gene, rs4869419 is located close to the chaperonin-containing TCP1, subunit 7 (eta) pseudogene 2 (CCT7P2) and 362 kb upstream of the nuclear receptor subfamily 2, group F, member 1 (NR2F1) gene, and rs6009824 is located nearby the bromodomain-containing protein 1 (BRD1) gene. Finally, despite an estimated replication power of 99% and a genome-wide significant P-value in the discovery phase, rs1004565 close to the ODZ4 gene did not replicate.
These results remained unchanged in a sample-size-weighted meta-analysis based on the Z-score method, which pools study-specific P-values accounting for effect direction and is robust to differences between phenotype measurement methods. Therefore, our findings were unlikely to be affected by differences in effect size due to the use of a different laboratory assay in MICROS compared with the two German studies (results not shown).
Investigation of the MTHFR-CLCN6-NPPA-NPPB locus
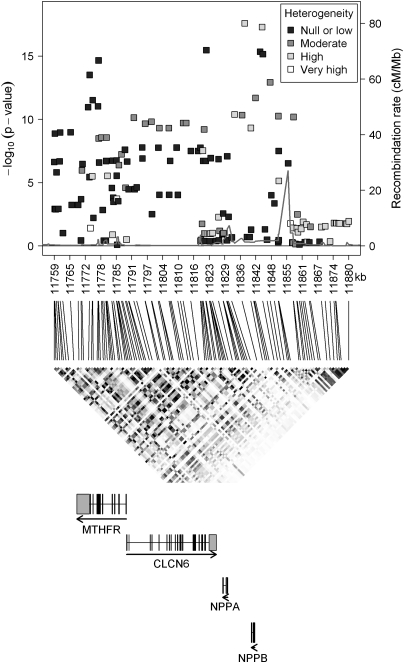
The four genes in the MTHFR-CLCN6-NPPA-NPPB locus are located within the 121 kb region on chromosome 1 (Fig. 2). To investigate whether different variants in this locus were independently associated with the NT-proBNP level, we selected all 128 SNPs available for a thorough analysis. Results of the combined analysis of the discovery and replication samples for these SNPs, together with information on LD and gene location, are given in Figure 2 (for additional details, see Supplementary Material, Table S1). The smallest P-values were obtained for two SNPs in the NPPB gene, rs6668352 (P-value = 1.7 × 10–18) and rs12406383 (P-value = 3.7 × 10–18), although both associations were characterized by strong heterogeneity between studies with I2 values of 53 and 56%, respectively (Fig. 2). Other SNPs located either in the promoter region of the NPPB gene (rs12406089, P-value = 3.5 × 10–16; rs6668659, P-value = 5.3 × 10–16), in the CLCN6 gene (rs1023252, P-value = 3.2 × 10–16) or in the MTHFR gene (rs12121543, P-value = 1.8 × 10–15; rs3818762, P-value = 3.1 × 10–14) showed strong associations, and the effects were homogeneous across studies.
Figure 2.
Results of the inverse-variance-weighted fixed-effect meta-analysis of the discovery and replication studies (combined analysis) on the MTHFR-CLCN6-NPPA-NPPB gene cluster on chromosome 1. Upper panel: Plot of –log10(P-values) against SNP physical position; the density of the colouring indicates the between-study heterogeneity of the effect size estimates, based on the I2 statistics. Heterogeneity is classified into four categories: null or low (I2≤ 25%), moderate (25% < I2≤ 50%), high (50% < I2≤ 75%) and very high (I2> 75%) (44). The recombination intensity was estimated from phased haplotypes in HapMap Release 22 (NCBI build 36) (45), and is reported as a light-grey line. Middle panel: LD (r2) in the HapMap-CEU population Release 22 (NCBI build 36). Colours vary between white (r2= 0) and black (r2= 1). Lower panel: Structure of the four genes in the locus according to the UCSC Table browser (46), which is based on NCBI build 36. This graph was in part created using utilities from the SNAP software (47).
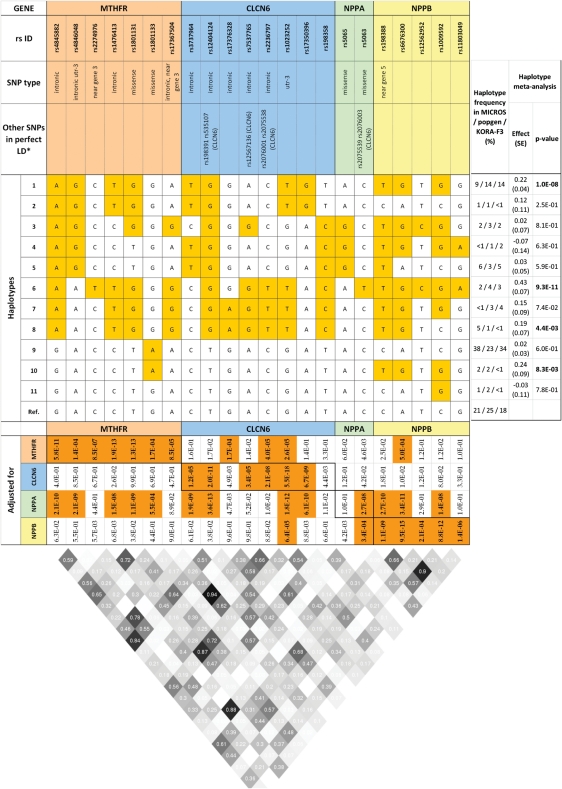
From the 128 SNPs available in the locus of interest, we selected 22 (highlighted in grey in Supplementary Material, Table S1) that were genotyped in at least one study and had high genotype imputation quality (Rsq > 0.6) in the others (see Materials and Methods and Supplementary Material, Table S2). To assess which of these SNPs were independently associated with NT-proBNP, we performed a conditional regression analysis of individual SNP, adjusting for the effect of SNPs in other genes by including one gene at a time into the model. Effects were then pooled over studies, using an inverse-variance-weighted fixed-effect meta-analysis, and results are presented in Figure 3. Only one SNP was found to be associated with NT-proBNP independently from the other genes’ variants, namely rs1023252 in the CLCN6 gene, which showed a strong trait association even after adjustment for SNPs in the NPPB gene.
Figure 3.
Haplotype distribution of the 22 SNPs selected for haplotype analysis of the MTHFR-CLCN6-NPPA-NPPB gene cluster. Different colours were used to distinguish the four genes. In the ‘haplotype' panel, alleles that differ from the reference haplotype alleles are highlighted in yellow. In the ‘adjusted for' panel, the P-values of the conditional association analysis are reported. Significant associations (P-values ≤ 0.003, see Materials and Methods) are highlighted in bold on an orange background. In the diagonal cells, results of the unadjusted association between NT-proBNP and the SNP in the relative column are reported. Out of the diagonal, results from conditional analyses are reported, where association at each particular variant is adjusted for SNPs in the gene indicated on the left side of the figure. In the right panel, study-specific haplotype frequencies and results of the haplotype association analysis are reported, with significant P-values given in bold. In the bottom panel, the LD structure (r2) for the three studies is reported.
Another SNP, rs5063 in the NPPA gene, showed a trait association that was independent from that of SNPs in the NPPB gene, but not from that of SNPs in CLCN6 or MTHFR. Marker rs5063 is in high LD (r2> 0.98) with two SNPs in the CLCN6 gene, rs2075538 and rs2075539, which are adjacent to rs1023252 (see above). This implies that variants located in the CLCN6 and/or in the NPPA genes could be related to NT-proBNP independently of the NPPB gene. The association of SNPs in the MTHFR gene with NT-proBNP disappeared upon adjustment for SNPs in the NPPB or CLCN6 gene. Finally, adjustment for SNPs in the NPPA gene had no major influence on the association between NT-proBNP and SNPs in the other three genes.
Among the 22 SNPs more closely investigated in the four-gene cluster, rs1023252 in the CLCN6 was the best predictor of the NT-proBNP level, explaining 2.9% of the trait variance in MICROS, 2.2% in popgen, 0.5% in KORA-F3 and 2.1% in the overall sample. Among the SNPs in the NPPB domain, rs6676300 was the most predictive one, explaining 2.0, 2.1 and 0.3% of the trait variance, in MICROS, popgen and KORA-F3, respectively (1.7% overall). When including all SNPs from the same gene domain in the model, the largest proportion of variance was explained by SNPs in the CLCN6 domain (2.1% overall). SNPs in the NPPB domain explained 1.9%. When all SNPs in the whole locus in the same model were included, we were able to explain 2.4% of the total variance. Additional details are provided in Supplementary Material, Table S3. In general, we observed that the proportion of variance explained by the genetic variants of the gene cluster was smaller in the KORA-F3 study sample, compared with MICROS or popgen. We investigated whether the low variance explained in KORA-F3 might have been due to a problem of genotype measurement error due to the lower average imputation quality (median Rsq = 0.68 versus Rsq = 0.95 and 0.99 in MICROS and popgen, respectively). The data did not support this hypothesis since the variance explained by individual SNPs in KORA-F3 was very low also for SNPs which were either genotyped or imputed with high quality (see Supplementary Material, Tables S2 and S3). A more convincing explanation could be the older age of KORA-F3 participants, with age being a strong predictor of the NT-proBNP level. In fact, adding age and sex to the model with the 22 SNPs led to a variance explained in KORA-F3 similar to that of popgen (24.3 versus 24.9%).
Given that SNPs in the MTHFR, CLCN6 and NPPA genes provided additional information to predict the NT-proBNP level compared with studying variants in the NPPB gene alone, we further explored the associations between NT-proBNP and genetic variation in the MTHFR-CLCN6-NPPA-NPPB gene locus by performing a haplotype analysis of the 22 selected SNPs. Haplotype effects were again pooled across the three studies using an inverse-variance-weighted fixed-effect meta-analysis. Four haplotypes were consistently found to be associated with loge(NT-proBNP) in all studies, with pooled effect estimates ranging from 0.19 to 0.43 loge(pg/ml), and with P-values between 8.3 × 10−03 and 9.3 × 10–11 (Fig. 3). A fifth haplotype showed borderline association (effect = 0.15, P-value = 0.074) with loge(NT-proBNP). All five haplotypes were characterized by a G allele for SNP rs6676300, which is located near the promoter region of the NPPB gene. However, other haplotypes carrying this allele were not associated with the trait (Fig. 3), suggesting that more complex genotype combinations could affect the NT-proBNP level.
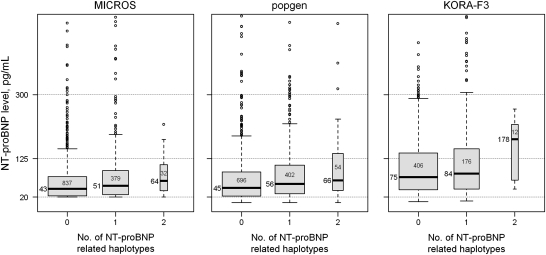
Figure 4 highlights the extent of the genetically derived differences in NT-proBNP concentrations showing the levels for subjects carrying one or two of the four significantly associated haplotypes. It can be seen that NT-proBNP levels increase with increasing number of haplotype copies. In MICROS, the median NT-proBNP level of individuals lacking two haplotypes was 43 pg/ml, compared with 64 pg/ml for individuals with two copies. The corresponding figures in the German samples were 45 versus 66 pg/ml (popgen) and 75 versus 178 pg/ml (KORA-F3), respectively. NT-proBNP variability also increased with an increasing number of haplotypes.
Figure 4.
Study-specific distribution of (untransformed) NT-proBNP levels by number of significantly associated haplotypes, carried by the subject. 0: no copies; 1, 2: one or two copies of any significant haplotype. The number of individuals per category is indicated within the boxes; the median NT-proBNP level (pg/ml) is reported close to the black horizontal line in the middle of the box.
DISCUSSION
Our GWA analysis of serum NT-proBNP levels identified an association with a cluster of four genes, MTHFR, CLCN6, NPPA and NPPB, and this result was replicated in a meta-analysis of two independent German samples. Conditional analyses highlighted a signal in CLCN6 independent from the NPPB signal. Four haplotypes spanning these genes were found to be significantly associated with NT-proBNP. These haplotypes showed a dosage effect in that the NT-proBNP level tended to increase with an increasing number of copies carried by an individual.
In an earlier candidate gene study of the BNP pathway, polymorphisms in the NPPB gene were found to be associated with a considerable variation in the BNP level, explaining up to 6% of the trait variability in patients undergoing elective cardiac catheterization (19). Two polymorphisms of the NPPB gene, rs198389 and rs198388, have been reported as being associated with NT-proBNP levels in 164 normo-albuminuric type 1 diabetic (13). In another study, a 4-polymorphism haplotype in the NPPA gene turned out to be associated with the NT-proBNP level of healthy individuals, although this association was not seen in patients with either heart failure or acute coronary syndrome (14). In contrast, Vassalle et al. (15) found an association between an ScaI polymorphism in the NPPA gene (corresponding to rs5065) and ANP, BNP and NT-proBNP levels in their sample of severe heart failure patients. In our study, we could not replicate the trait association of rs5065 but found an association with a nearby SNP rs5063 that was partially independent from the NPPB variants. Associations between ANP and BNP level, respectively, and polymorphisms in the NPPA and NPPB genes were also reported by Newton-Cheh et al. (16), in a study including the Framingham Heart Study, the Finrisk1997 Study and the Malmö Diet and Cancer Study (MDCS). We confirmed the association between rs632793 in the NPPA domain and NT-proBNP observed in the MDCS study, with effect size of very similar magnitude (0.13 log pg/ml, P-value = 1 × 10−09 in MDCS versus 0.16, P-value = 3.6 × 10−08 in our study). The same SNP was also strongly associated with the BNP in all other studies. We could not replicate the association with rs5068 found in MDCS. The CLCN6 SNP rs1023252 was not associated with the BNP in the Framingham Heart Study, but the association with NT-proBNP was not evaluated.
Evidence from all these earlier studies supports, in a very consistent way, an association between polymorphisms in the NPPB and NPPA genes and NT-proBNP levels. However, the four genes in the MTHFR-CLCN6-NPPA-NPPB locus are in LD with each other, and so, investigation of any of these genes should control for the effects of the other genes. For example, a study on Korean individuals reported a strong association between the MTHFR C688T variant and plasma BNP levels (20), but there was no control for NPPA or NPPB gene variants. Thus, the suggested prognostic value of the MTHFR C677T variant for BNP concentration could reflect, indirectly, variants located in one of the other genes in the locus.
A similar situation has been observed when investigating blood pressure. Newton-Cheh et al. (16) showed that polymorphisms in the NPPB and NPPA genes associated with BNP levels were also associated with blood pressure and hypertension. In a meta-analysis of GWA studies on blood pressure, the same authors observed that the SNP most strongly associated with systolic blood pressure, among all available variants in the four-gene locus, was located in the MTHFR gene but discussed the difficulty to dissect which of the genes in the locus could be responsible for blood pressure variations (21).
Conditional analyses, where the association between a genetic variant in one gene is adjusted for variants located in the other genes, can help understand the complex picture by disentangling the effects of correlated gene variants. Haplotype analysis can, on the other hand, provide a comprehensive view of the complexity of the gene cluster. The results of our conditional analyses suggest that the association between NT-proBNP and variants in the MTHFR gene observed in our GWA analysis might reflect the trait association of variants in the CLCN6 and NPPB genes. On the contrary, the association of the SNP rs1023252 in the CLCN6 gene appeared to be mostly independent from variants in all nearby genes, including NPPB. Three other variants in complete LD with each other (rs5063 in NPPA and rs2075538 and rs2075539 in CLCN6) were associated with NT-proBNP levels independently from SNPs in NPPB.
The independency of the signals in the CLCN6 gene shown by our conditional analyses suggests a role of this gene in the regulation of NT-proBNP, which has not been hypothesized previously. CLCN6 gene function has been poorly understood so far. What is known is that the gene encodes one of several voltage-gated chloride channels (CLC-6), which is predominantly located in intracellular membranes and is widely expressed in various tissues (22). Gene targeting and transgenic animal models have been used to delineate the functional role of cardiac chloride channels in the context of health and disease. In general, it has been shown that chloride channels may contribute to cardiac arrhythmogenesis, myocardial hypertrophy, heart failure and cardioprotection against ischaemia/reperfusion (23). Although this evidence points towards association with cardiovascular phenotypes, no direct link between BNP or NT-proBNP and voltage-gated chloride channels, including CLC-6, has been shown so far. The findings from our study suggest the hypothesis of a role of the CLCN6 gene in the regulation of NT-proBNP either directly or indirectly by modifying NPPB gene expression. However, we cannot exclude the possibility that the CLCN6 variants identified in our study, in particular rs1023252, may be simply markers in LD with an unobserved causal variant located inside the NPPB gene domain, or indeed may be more directly involved in long-range regulation of NPPB expression.
To capture the effect of the combination of variants in the MTHFR-CLCN6-NPPA-NPPB gene cluster, we estimated haplotypes using the 22 best SNPs available in the locus (i.e. those that were genotyped in at least one study and had high imputation quality in the others), and we were able to estimate reliable haplotypes. Four different haplotypes were associated with higher NT-proBNP levels, consistently across the three studies. Although these haplotypes were not solely tagged by SNPs in or around NPPB, the G allele of rs6676300, located near the promoter of NPPB, was present in all four haplotypes. This could support rs6676300 as the best proxy of a functional SNP for NPPB expression. Haplotypes carrying the rs6676300 G allele not associated with NT-proBNP were also observed, although it is possible that this lack of association may be due to the low power of these low-frequency haplotypes. On the other hand, rs6676300 showed only modest association with BNP in the Framingham Heart Study (16), which was not robust to adjustment for nearby SNPs.
The genetically derived differences in NT-proBNP levels were substantial in all three of our studies: individuals carrying two copies of the associated haplotypes showed an increase in the median NT-proBNP level between 20 and 100 pg/ml in different populations, compared with the reference group carrying no copies of these four haplotypes. This variation represents an important finding, particularly if we consider that the proportion of individuals carrying two copies of the four haplotypes was substantial in our populations: 2.0, 2.6 and 4.7% in KORA-F3, MICROS and popgen, respectively. These findings call for further research to evaluate a possible predictive role of these haplotypes in the identification of patients at risk of heart failure.
The availability of SNPs imputed on the same HapMap–CEU data release was at the same time a strength and a limitation for our fine-mapping analysis. The strength was that the imputation based on the same map enabled a direct comparison of results across studies. The limitation was that some of the imputed SNPs had suboptimal imputation quality, which translates into measurement error affecting both SNP-based and haplotype-based analyses. This non-differential measurement error is known to bias the results towards the null (24) and might have had some impact on our findings from both the conditional and the haplotype analysis.
Although not surviving Bonferroni correction for claiming replication, two SNPs in the MEIS1 and NR2F1 (also known as COUP-TF1) genes showed promising findings, with replication P-values reaching nominal significance and P-values of the combined analysis being smaller than those of the discovery analysis (Table 2). The biological plausibility of these findings (25–27) makes them worth further investigation.
From a methodological point of view, our study highlighted an interesting problem in the analysis of quantitative traits, namely laboratory assay limit of detection (LOD). Although censored data due to the presence of an LOD are not uncommon in GWA analyses of quantitative traits, an adequate modelling of such data has rarely been performed. In our MICROS GWA study, nearly one-fifth of individuals had NT-proBNP values below the LOD, which was addressed by modelling the logarithm of NT-proBNP with a Tobit model (28), assuming a Weibull distribution of the trait. Compared with the naïve approach of excluding samples below the LOD, or of imputing missing values by a fixed NT-proBNP level (LOD or LOD/2), the Tobit method can protect not only against bias (29) but also against a considerable loss of power. In fact, the results from our MICROS study were consistent with those from the replication studies, where data censoring was not present due to the use of a different laboratory method.
In conclusion, our study identifies variants within the MTHFR-CLCN6-NPPA-NPPB gene cluster that associate with regulation of NT-proBNP levels. The association of SNPs in the CLCN6 gene surviving the adjustment for NPPB and NPPA variants suggests a potentially independent role of CLCN6 in the regulation of NT-proBNP levels. The identification of haplotypes in this gene cluster with substantial effects on NT-proBNP levels calls for future research to further clarify the role of this locus in the regulation of NT-proBNP.
MATERIALS AND METHODS
Written informed consent was obtained from all participants in the studies involved in this work. All study protocols were approved by the institutional ethical review committees of the participating centres.
Individual study description
The MICROS study (discovery)
The MICROS study is a cross-sectional genetic study performed in South Tyrol (Italy) in 2001–03 (17). Participants were recruited from three villages (Stelvio, Vallelunga and Martello) located in the Italian Alps, in a German-speaking region bordering with Austria and Switzerland that experienced a prolonged period of isolation from surrounding populations. Information on the participants’ health status was collected through a standardized questionnaire. Laboratory data were obtained through standard blood analyses. All DNA samples were genotyped on Illumina Infinium HumanHap300 v2 SNP bead microarrays according to the manufacturer's instructions. Genotypes were obtained using the proprietary Bead Studio software. A total of 1334 individuals were suitable for analysis after data cleaning: call rate <98%, minor allele frequency (MAF) <1% and Hardy–Weinberg equilibrium (HWE) test P-value <10−06. NT-proBNP levels were measured in 1325 subjects using chemiluminescence immunoassay at the Institute for Clinical Chemistry and Laboratory Medicine, University of Regensburg, Germany.
The popgen biobank (replication)
Data on German healthy individuals were obtained from the ‘control' group of the popgen biobank (30). Genotyping was carried out as part of the GWAS initiative of the German National Genome Research Network (NGFN) and was performed by an Affymetrix service facility (South San Francisco, CA, USA), using the Affymetrix Genome-Wide Human SNP Array 6.0 (1000K) (Santa Clara, CA, USA). Genotype calling was carried out using Affymetrix's Birdseed v2 algorithm with the default quality thresholds. Samples with >5% missing genotypes, showing excess genetic dissimilarity to the remaining subjects, or with evidence of a cryptic relatedness to other study participants, were removed. These quality control measures left 1213 samples for inclusion in the replication cohort. All sex assignments could be verified by reference to the proportion of heterozygous SNPs on the X chromosome. NT-proBNP was assessed in 1152 serum samples with the elecsys NT-proBNP assay (Roche Diagnostics) at the Institute for Clinical Chemistry in Kiel, Germany.
The KORA-F3 survey (replication)
The KORA-F3 surveys for genetic research, described in detail elsewhere (31,32), were initiated as part of the MONICA (Monitoring of Trends of Cardiovascular Diseases) multi-centre study. The third KORA survey (KORA-S3) is a population-based sample from the general population of the south German city of Augsburg and surrounding counties recruited in 1994–95. A subsample consisting of 1644 individuals from this survey with 10-year follow-up information available (KORA-F3) was successfully genotyped with the 500K Affymetrix Chip (per-sample call rate >93%). All participants were German citizens of European origin. NT-proBNP was assessed in deep frozen (−80°C) serum samples with the elecsys NT-proBNP assay (Roche Diagnostics) in 594 samples. The analytical range is 5–35 000 pg/ml and the coefficient of variation is <10%.
Genotype imputation
Using the Phase II CEU HapMap individuals as a reference panel (release 22, build 36), genotypes were imputed for each study (MICROS, popgen and KORA-F3) for the entire set of polymorphic HapMap SNPs (>2.5 million SNPs), using a hidden Markov model as implemented in the MACH software version 1.0.15/16 (33) (http://www.sph.umich.edu/csg/abecasis/MACH/). At each marker, imputation results were summarized as an ‘allele dosage', defined as the expected number of copies of the minor allele, which can vary between 0 and 2. The Rsq statistic was estimated at each SNP as the ratio of the observed variance of the allele dosage (imputed value) to the expected variance under HWE (34). Consequently, Rsq reflects possible deviations from HWE and imputation quality. SNPs with Rsq ≤ 0.3, indicating low-quality imputation, were discarded. Information on genotyping platforms and imputation performance is summarized in Supplementary Material, Table S4.
Statistical analysis
In the discovery study, the diagnostic assay for assessing the NT-proBNP level had an LOD equal to 20 pg/ml. Consequently, the NT-proBNP level had to be considered a left-censored phenotype. Given that 18% of observations were below the LOD, the use of conventional statistical methods could have led to biased estimates in a regression analysis (35). We therefore fitted a Tobit model to the loge(NT-proBNP) values, making the assumption that loge(NT-proBNP) had a Weibull density function (28). The LOD was loge-transformed accordingly. The model was adjusted for age, sex and study village. Briefly, under the assumption of the data distribution, the Tobit procedure computes the probability that a value is censored and uses this probability to guide the estimation of the regression coefficients via maximum likelihood. Estimates obtained with the likelihood-based method are less biased than those obtained with other methods (29), thereby ensuring normality of the regression residuals. To account for any inter-individual relatedness of MICROS participants, a polygenic linear model was fitted to estimate the inverse of the variance/covariance matrix, based on the genomic kinship matrix (36) and using the GenABEL package (37). GWA between residuals of the Tobit model and SNP dose levels was assessed by means of an approximate score test statistic (38), using probABEL software (http://mga.bionet.nsc.ru/~yurii/ABEL/) and assuming an additive model of inheritance. To account for the number of independent tests that could be performed on a (potentially) infinitely dense genetic map, the threshold for claiming genome-wide significance was set to 7.2 × 10−08 (18). To assess the possible impact of pharmacological treatment on our GWA results, three sensitivity analyses were performed, each time excluding a given group of individuals by considering the possible effect of such drugs on NT-proBNP levels, according to Troughton et al. (39): (i) reduction (n = 214, 16% of individuals); (ii) increase (n = 72, 5%); (iii) alteration, that is, either reduction or increase (n = 286, 22%). The excluded drugs with a potential ‘stimulating' effect on NT-proBNP levels were: antihypertensives, antithrombotic agents, platelet aggregation inhibitors (aspirin), selective beta-2-adrenoreceptor agonists (Salbutard), digoxin and beta-blocking agents. Drugs that could reduce the NT-proBNP level included agents acting on the renin–angiotensin system (65% of ACE-inhibitors), anti-gout treatment (Allopulinor), lipid-modifying agents, vasodilator-nitrates, sex hormones, anti-arrhythmics, diuretics, peripherical vasodilators and alpha- and beta-blocking agents (Carvedilol).
Replication analysis
Replication of our most promising findings was attempted in two independent samples from the general population of two German regions, which share much of their ethnic background with the South Tyrol population in Italy. From the discovery GWA analysis, SNPs with P-values <10−05 were selected, provided that at least one more SNP with P-value <10−04 was located within 100 kb. From these SNPs, independent loci were identified based on LD, with independence defined by pairwise r2 <0.2. One SNP per locus was chosen and, after accounting for multiple testing, SNPs with power to replicate <80% were excluded. Power estimates were obtained with QUANTO (40) (http://hydra.usc.edu/GxE/), assuming a replication sample size of 1746 individuals (sum of popgen and KORA-F3), an additive genetic model and HWE.
In popgen and KORA-F3, SNP dose levels were tested for an association with loge(NT-proBNP), using an ordinary linear regression model, adjusted for age and sex, using ProbABEL for popgen and MACH2QTL (http://www.sph.umich.edu/csg/abecasis/MACH/) for KORA-F3. Regression coefficients and SEs from individual replication studies were pooled using an inverse-variance-weighted fixed-effect meta-analysis, using the metafor R package (http://www.wvbauer.com/downloads.html). Associations were considered replicated if the direction in the replication analysis was the same as in the discovery analysis and the one-sided P-value was <0.002, corresponding to 0.05 divided by the number of SNPs tested, n = 25 (Bonferroni correction).
To improve the precision of effect estimates, the effect estimates of the SNPs submitted to replication were pooled across the three studies (MICROS, popgen and KORA-F3) by means of an inverse-variance-weighted fixed-effect meta-analysis. To account for potential differences in the effect sizes due to the use of different laboratory assays in MICROS compared with the German studies, a sensitivity analysis was performed by meta-analysing P-values, using a sample-size-weighted Z-score method, which accounts for effect direction, implemented in METAL (41).
Fine mapping of the MTHFR-CLCN6-NPPA-NPPB gene cluster association
To dissect the effects of single variants in the trait-associated locus on chromosome 1 (map positions 11 759 037 to 11 851 482), the original list of 128 SNPs in the region was reduced to 29 high-quality SNPs that satisfied all of the following criteria: (i) present in all the three studies as either genotyped or imputed; (ii) genotyped in at least one of the three studies; (iii) if not genotyped, a high imputation quality (Rsq > 0.6); (iv) P-value of association ≤0.05 in at least one of the three studies (non-null effect size). The list was further reduced after removing seven SNPs that were in perfect LD with other SNPs from the list (r2> 0.98). The final selection included 22 SNPs (see Supplementary Material, Table S2, for the complete list of SNPs along with their imputation quality). The number of SNPs genotyped versus imputed was 15 versus 7, 12 versus 10 and 7 versus 15, in MICROS, popgen, and KORA-F3, respectively. Of these 22 SNPs, 7 were located in the MTHFR gene, 8 in the CLCN6 gene, 2 in the NPPA gene and 5 in the NPPB gene. To assess which SNPs were independently associated with NT-proBNP, we fitted the following four linear regression models to the loge(NT-proBNP) value for each of the 22 SNPs, using the allele dosage of the SNP as an influential variable:
adjusting for age and sex alone;
adjusting for age, sex, plus all SNPs belonging to the second gene in the cluster;
adjusting for age, sex, plus all SNPs belonging to the third gene in the cluster;
adjusting for age, sex, plus all SNPs belonging to the fourth gene in the cluster.
To account for inter-individual relatedness in the MICROS study, linear mixed models were fitted to the residuals obtained from the regression of NT-proBNP on age and sex. Results from all studies were pooled using an inverse-variance-weighted fixed-effect meta-analysis. Given that the 22 SNP considered were in LD, adjustment for multiple comparisons was performed according to Nyholt (42) based on the observed LD pattern within each population. The resulting threshold for statistical significance was 0.003.
The same 22 SNPs were included in study-specific haplotype analyses, using the haplo.stats R package (43) (http://mayoresearch.mayo.edu/mayo/research/schaid_lab/software.cfm), assuming an additive model of inheritance. Results were pooled using an inverse-variance-weighted fixed-effects meta-analysis.
If not specified otherwise, data management, data analysis, programming and the creation of graphs were all performed using R 2.9.2 (R Development Core Team 2009) (http://www.R-project.org).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
MICROS: In South Tyrol, the study was funded by the Ministero della Salute (RF-PAB-2006-342336), the Ministry of Health and Department of Educational Assistance, University and Research of the Autonomous Province of Bozen/Bolzano, the South Tyrolean Sparkasse Foundation and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). Popgen: The popgen study was supported by the German Ministry of Education and Research (BMBF) through the National Genome Research Network (NGFN). It is currently funded by the Ministry of Science, Commerce and Transportation of the State of Schleswig-Holstein. KORA-F3: The work of KORA is supported by the German Federal Ministry of Education and Research (BMBF) in the context of the German National Genome Research Network (NGFN-2 and NGFN-plus) and within the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ. The genetic epidemiological work was funded by an NIH subcontract from the Children's Hospital, Boston, USA (H.E.W., I.M.H., prime grant 1 R01 DK075787-01A1), the German National Genome Research Net NGFN2 and NGFNplus (H.E.W. 01GS0823) and the Munich Center of Health Sciences (H.E.W.). NT-proBNP measurements were funded by the Kompetenznetz Herzinsuffizienz (German Heart Failure Network), the Federal Ministry of Education and Research (BMBF), and NT-proBNP kits were provided by Roche Diagnostics (Mannheim, Germany). The KORA research platform and the MONICA Augsburg studies were initiated and financed by the Helmholtz Research Center München for Environmental Health and the German Federal Ministry of Education and Research and the State of Bavaria. Funding to pay the Open Access publication charges for this article was provided by European Academy Bozen/Bolzano (EURAC), Bolzano, Italy.
ACKNOWLEDGEMENTS
We owe a debt of gratitude to all participants of the three studies. For the MICROS study, we thank primary care practitioners Raffaela Stocker, Stefan Waldner, Toni Pizzecco, Josef Plangger and Ugo Marcadent and the personnel of the Hospital of Silandro (Department of Laboratory Medicine) for their collaboration in the project. We thank Dr Maurizio F. Facheris for his assistance with the classification of medical treatments as reported by study participants, and Clemens Egger and Yuri D'Elia for their invaluable help with the data management. For the KORA-F3 study, we thank the Genome Analysis Center (GAC) of the Helmholtz Zentrum München chaired by J. Adamski, where the genotyping was performed. For the popgen biobank, we acknowledge the support received through the DFG excellence cluster ‘Inflammation at Interfaces’.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Nakagawa O., Ogawa Y., Itoh H., Suga S., Komatsu Y., Kishimoto I., Nishino K., Yoshimasa T., Nakao K. Rapid transcriptional activation and early mRNA turnover of brain natriuretic peptide in cardiocyte hypertrophy. Evidence for brain natriuretic peptide as an emergency cardiac hormone against ventricular overload. J. Clin. Invest. 1995;96:1280–1287. doi: 10.1172/JCI118162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Bold A.J., Ma K.K., Zhang Y., de Bold M.L., Bensimon M., Khoshbaten A. The physiological and pathophysiological modulation of the endocrine function of the heart. Can. J. Physiol. Pharmacol. 2001;79:705–714. [PubMed] [Google Scholar]
- 3.Clerico A., Emdin M. Diagnostic accuracy and prognostic relevance of the measurement of the cardiac natriuretic peptides: a review. Clin. Chem. 2004;50:33–50. doi: 10.1373/clinchem.2003.024760. [DOI] [PubMed] [Google Scholar]
- 4.Jortani S.A., Prabhu S.D., Valdes R., Jr. Strategies for developing biomarkers of heart failure. Clin. Chem. 2004;50:265–278. doi: 10.1373/clinchem.2003.027557. [DOI] [PubMed] [Google Scholar]
- 5.Mueller T., Gegenhuber A., Poelz W., Haltmayer M. Head-to-head comparison of the diagnostic utility of BNP and NT-proBNP in symptomatic and asymptomatic structural heart disease. Clin. Chim. Acta. 2004;341:41–48. doi: 10.1016/j.cccn.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 6.Mueller T., Gegenhuber A., Poelz W., Haltmayer M. Biochemical diagnosis of impaired left ventricular ejection fraction—comparison of the diagnostic accuracy of brain natriuretic peptide (BNP) and amino terminal proBNP (NT-proBNP) Clin. Chem. Lab. Med. 2004;42:159–163. doi: 10.1515/CCLM.2004.029. [DOI] [PubMed] [Google Scholar]
- 7.Costello-Boerrigter L.C., Boerrigter G., Redfield M.M., Rodeheffer R.J., Urban L.H., Mahoney D.W., Jacobsen S.J., Heublein D.M., Burnett J.C. Amino-terminal pro-B-type natriuretic peptide and B-type natriuretic peptide in the general community: determinants and detection of left ventricular dysfunction. J. Am. Coll. Cardiol. 2006;47:345–353. doi: 10.1016/j.jacc.2005.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emdin M., Passino C., Prontera C., Fontana M., Poletti R., Gabutti A., Mammini C., Giannoni A., Zyw L., Zucchelli G., et al. Comparison of brain natriuretic peptide (BNP) and amino-terminal ProBNP for early diagnosis of heart failure. Clin. Chem. 2007;53:1289–1297. doi: 10.1373/clinchem.2006.080234. [DOI] [PubMed] [Google Scholar]
- 9.Clerico A., Fontana M., Zyw L., Passino C., Emdin M. Comparison of the diagnostic accuracy of brain natriuretic peptide (BNP) and the N-terminal part of the propeptide of BNP immunoassays in chronic and acute heart failure: a systematic review. Clin. Chem. 2007;53:813–822. doi: 10.1373/clinchem.2006.075713. [DOI] [PubMed] [Google Scholar]
- 10.Nowatzke W.L., Cole T.G. Stability of N-terminal pro-brain natriuretic peptide after storage frozen for one year and after multiple freeze-thaw cycles. Clin. Chem. 2003;49:1560–1562. doi: 10.1373/49.9.1560. [DOI] [PubMed] [Google Scholar]
- 11.Wang T.J., Larson M.G., Levy D., Benjamin E.J., Corey D., Leip E.P., Vasan R.S. Heritability and genetic linkage of plasma natriuretic peptide levels. Circulation. 2003;108:13–16. doi: 10.1161/01.CIR.0000081657.83724.A7. [DOI] [PubMed] [Google Scholar]
- 12.Benjamin E.J., Dupuis J., Larson M.G., Lunetta K.L., Booth S.L., Govindaraju D.R., Kathiresan S., Keaney J.F., Jr., Keyes M.J., Lin J.P., et al. Genome-wide association with select biomarker traits in the Framingham Heart Study. BMC Med. Genet. 2007;9(Suppl. 1):11. doi: 10.1186/1471-2350-8-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lajer M., Tarnow L., Jorsal A., Parving H.H. Polymorphisms in the B-type natriuretic peptide (BNP) gene are associated with NT-proBNP levels but not with diabetic nephropathy or mortality in type 1 diabetic patients. Nephrol. Dial. Transplant. 2007;22:3235–3239. doi: 10.1093/ndt/gfm360. [DOI] [PubMed] [Google Scholar]
- 14.Weber M., Burian M., Dragutinovic I., Moellmann H., Nef H., Elsaesser A., Mitrovic V., Hamm C., Geisslinger G. Genetic polymorphism of the type A human natriuretic peptide receptor (NPR-A) gene contributes to the interindividual variability in the BNP system. Eur. J. Heart Fail. 2008;10:482–489. doi: 10.1016/j.ejheart.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Vassalle C., Andreassi M.G., Prontera C., Fontana M., Zyw L., Passino C., Emdin M. Influence of ScaI and natriuretic peptide (NP) clearance receptor polymorphisms of the NP system on NP concentration in chronic heart failure. Clin. Chem. 2007;53:1886–1890. doi: 10.1373/clinchem.2007.088302. [DOI] [PubMed] [Google Scholar]
- 16.Newton-Cheh C., Larson M.G., Vasan R.S., Levy D., Bloch K.D., Surti A., Guiducci C., Kathiresan S., Benjamin E.J., Struck J., et al. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat. Genet. 2009;41:348–353. doi: 10.1038/ng.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pattaro C., Marroni F., Riegler A., Mascalzoni D., Pichler I., Volpato C.B., Dal Cero U., De Grandi A., Egger C., Eisendle A., et al. The genetic study of three population microisolates in South Tyrol (MICROS): study design and epidemiological perspectives. BMC Med. Genet. 2007;5:8–29. doi: 10.1186/1471-2350-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudbridge F., Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 2008;32:227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanfear D.E., Stolker J.M., Marsh S., Rich M.W., McLeod H.L. Genetic variation in the B-type natiuretic peptide pathway affects BNP levels. Cardiovasc. Drugs Ther. 2007;21:55–62. doi: 10.1007/s10557-007-6007-5. [DOI] [PubMed] [Google Scholar]
- 20.Cho S.E., Hong K.S., Shin G.J., Chung W.S. The methylenetetrahydrofolate reductase C677T gene mutation is associated with hyperhomocysteinemia, cardiovascular disease and plasma B-type natriuretic peptide levels in Korea. Clin. Chem. Lab. Med. 2006;44:1070–1075. doi: 10.1515/CCLM.2006.194. [DOI] [PubMed] [Google Scholar]
- 21.Newton-Cheh C., Johnson T., Gateva V., Tobin M.D., Bochud M., Coin L., Najjar S.S., Zhao J.H., Heath S.C., Eyheramendy S., et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki T., Rai T., Hayama A., Sohara E., Suda S., Itoh T., Sasaki S., Uchida S. Intracellular localization of ClC chloride channels and their ability to form hetero-oligomers. J. Cell. Physiol. 2006;206:792–798. doi: 10.1002/jcp.20516. [DOI] [PubMed] [Google Scholar]
- 23.Duan D. Phenomics of cardiac chloride channels: the systematic study of chloride channel function in the heart. J. Physiol. 2009;587:2163–2177. doi: 10.1113/jphysiol.2008.165860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gustafson P. Measurement Error and Misclassification in Statistics and Epidemiology: Impact and Bayesian Adjustments. Chapman and Hall/CRC Press; 2003. Boca Raton, FL, USA. [Google Scholar]
- 25.Stankunas K., Shang C., Twu K.Y., Kao S.C., Jenkins N.A., Copeland N.G., Sanyal M., Selleri L., Cleary M.L., Chang C.P. Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ. Res. 2008;103:702–709. doi: 10.1161/CIRCRESAHA.108.175489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo L., Lynch J., Nakamura K., Fliegel L., Kasahara H., Izumo S., Komuro I., Agellon L.B., Michalak M. COUP-TF1 antagonizes Nkx2.5-mediated activation of the calreticulin gene during cardiac development. J. Biol. Chem. 2001;276:2797–2801. doi: 10.1074/jbc.C000822200. [DOI] [PubMed] [Google Scholar]
- 27.Mesaeli N., Nakamura K., Zvaritch E., Dickie P., Dziak E., Krause K.H., Opas M., MacLennan D.H., Michalak M. Calreticulin is essential for cardiac development. J. Cell. Biol. 1999;144:857–868. doi: 10.1083/jcb.144.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tobin J. Estimation of relationships for limited dependent variables. Econometrica. 1958;26:24–36. [Google Scholar]
- 29.Wang H., Fygenson M. Inference for censored quantile regression models in longitudinal studies. Ann. Stat. 2009;37:756–781. [Google Scholar]
- 30.Krawczak M., Nikolaus S., von Eberstein H., Croucher P.J., El Mokhtari N.E., Schreiber S. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype–phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 31.Wichmann H.E., Gieger C., Illig T. MONICA/KORA Study Group. KORA-gen—resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl. 1):26–30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 32.Heid I.M., Boes E., Müller M., Kollerits B., Lamina C., Coassin S., Gieger C., Döring A., Klopp N., Frikke-Schmidt R., et al. Genome-wide association analysis of high-density lipoprotein cholesterol in the population-based KORA study sheds new light in intergenic regions. Circ. Cardiovasc. Genet. 2008;1:10–20. doi: 10.1161/CIRCGENETICS.108.776708. [DOI] [PubMed] [Google Scholar]
- 33.Li Y., Willer C.J., Sanna S., Abecasis G.R. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang L., Li Y., Singleton A.B., Hardy J.A., Abecasis G., Rosenberg N.A., Scheet P. Genotype-imputation accuracy across worldwide human populations. Am. J. Hum. Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lubin J.H., Colt J.S., Camann D., Davis S., Cerhan J.R., Severson R.K., Bernstein L., Hartge P. Epidemiologic evaluation of measurement data in the presence of detection limits. Environ. Health. Perspect. 2004;112:1691–1696. doi: 10.1289/ehp.7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amin N., van Duijn C.M., Aulchenko Y.S. A genomic background based method for association analysis in related individuals. PLoS One. 2007;2:1274. doi: 10.1371/journal.pone.0001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aulchenko Y.S., Ripke S., Isaacs A., van Duijn C.M. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 38.Chen W.M., Abecasis G.R. Family-based association tests for genome wide association scans. Am. J. Hum. Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Troughton R.W., Richards A.M., Yandle T.G., Frampton C.M., Nicholls M.G. The effects of medications on circulating levels of cardiac natriuretic peptides. Ann. Med. 2007;39:242–260. doi: 10.1080/07853890701232057. [DOI] [PubMed] [Google Scholar]
- 40.Gaudermann W.J. Sample size requirements for association studies of gene–gene interaction. Am. J. Epidemiol. 2002;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 41.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nyholt D.R. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lake S., Lyon H., Silverman E., Weiss S., Laird N., Laird N.M., Schaid D.J. Estimation and tests of haplotype–environment interaction when linkage phase is ambiguous. Hum. Hered. 2002;55:56–65. doi: 10.1159/000071811. [DOI] [PubMed] [Google Scholar]
- 44.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O'Donnell C.J., de Bakker P. I. W. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.