Abstract
Bardet–Biedl syndrome (BBS) is a syndromic form of retinal degeneration. Recently, homozygosity mapping with a consanguineous family with isolated retinitis pigmentosa identified a missense mutation in BBS3, a known BBS gene. The mutation in BBS3 encodes a single amino acid change at position 89 from alanine to valine. Since this amino acid is conserved in a wide range of vertebrates, we utilized the zebrafish model system to functionally characterize the BBS3 A89V mutation. Knockdown of bbs3 in zebrafish alters intracellular transport, a phenotype observed with knockdown of all BBS genes in the zebrafish, as well as visual impairment. Here, we find that BBS3 A89V is sufficient to rescue the transport delays induced by the loss of bbs3, indicating that this mutation does not affect the function of BBS3 as it relates to syndromic disease. BBS3L A89V, however, was unable to rescue vision impairment, highlighting a role for a specific amino acid within BBS3 that is necessary for visual function, but dispensable in other cell types. These data aid in our understanding of why patients with the BBS3 A89V missense mutation only present with isolated retinitis pigmentosa.
INTRODUCTION
Bardet–Biedl syndrome (BBS, OMIM 209900) is a genetically heterogeneous autosomal recessive disorder characterized by retinitis pigmentosa, obesity, polydactyly, renal abnormalities, hypogenitalism and cognitive impairment (1–4). Moreover, BBS is associated with an increased risk for hypertension, diabetes and heart defects (1,2,5). BBS patients present with early and progressive photoreceptor degeneration and are blind by the third decade of life (2,6–13). To date, 12 BBS (BBS1–12) genes are reported to individually cause BBS (14–27). Additionally, hypomorphic mutations in MKS1 and CEP290 have been associated with BBS, representing BBS13 and BBS14, respectively (28). The BBS genes belong to multiple protein families and function cannot be defined based on homology; however, recent advances in molecular pathophysiology and animal models have helped to elucidate why 14 different genes can lead to the same phenotype. Work in mouse, zebrafish, Caenorhabditis elegans and Chlamydomonas has provided multiple lines of evidence supporting a role for BBS proteins in cilia function and intraflagellar and/or intracellular transport (19,22,23,26,29–36). Although progress has been made in understanding the pathophysiology of BBS, there are major gaps in our understanding of the precise cellular function of the BBS proteins.
BBS3 (ARL6, ADP-ribosylation factor-like), a member of the Ras family of small GTP-binding proteins, was initially identified as a BBS gene through computational genomics and high-density single nucleotide polymorphism (SNP) genotyping (21,22). Several mutations (G2X, T31M, T31R, P108L, R122X, G169A and L170W) leading to BBS have been reported throughout BBS3 (21,22,37). Knockdown of bbs3 using an antisense oligonucleotide [Morpholino (MO)] results in two cardinal features of BBS in the zebrafish: reduced size of the ciliated Kupffer's Vesicle and delays in intracellular melanosome transport (35,38). These prototypical phenotypes are preset with knockdown of all BBS genes in the zebrafish (26,34,35,38). Recently, we identified a second longer eye-specific transcript of BBS3, BBS3L, which is required for retinal organization and function in both the mouse and zebrafish (38). Knockdown of either both bbs3 transcripts or bbs3L alone leads to vision impairment in zebrafish. To determine the functional requirement of each transcript, RNA encoding either human BBS3 or BBS3L was co-injected with the bbs3 aug MO, which targets both transcripts. We determined that human BBS3 RNA is sufficient to suppress the melanosome transport delays, but not the vision defect. In contrast, BBS3L RNA was sufficient to rescue the vision defect; however, it was unable to suppress the cardinal phenotypes of BBS seen in the zebrafish, supporting a retina specific role for BBS3L (38).
BBS is rare in the general population; however, the study of this disease can offer insight into normal retinal development as well as provide an understanding of the pathophysiology involved in non-syndromic forms of BBS. Homozygosity mapping of a consanguineous Saudi Arabian family has identified a missense mutation (A89V) in BBS3 that leads to non-syndromic retinitis pigmentosa (39,40). The identification of specific mutations in the same gene that results in either syndromic or non-syndromic retinitis pigmentosa will provide insight into tissue-specific functional regions of BBS3 in the retina. Moreover, understanding the functional domains of proteins involved in vision aids in our understanding of not only the disease state, but also normal vision development.
Here we report the functional characterization of the BBS3 missense mutation (A89V), which occurs in a highly conserved region of BBS3. The function of the BBS3 A89V mutation was evaluated by utilizing gene knockdown of bbs3 coupled with RNA rescue in the zebrafish. We examined the intracellular transport of melanosomes, a cardinal feature of BBS gene knockdown in the zebrafish, and visual function using a vision startle assay. The A89V mutation can suppress the melanosome transport defects, but not the vision impairment observed with the loss of bbs3. Thus, the missense mutation identified in patients with non-syndromic retinal degeneration has uncovered an amino acid in BBS3 that is necessary for vision. The A89V mutation is able to function in melanosome transport, demonstrating that the mutant form of the protein retains the ability to function in tissues typically affected by BBS.
RESULTS
BBS3 conservation and BBS3L mutant expression
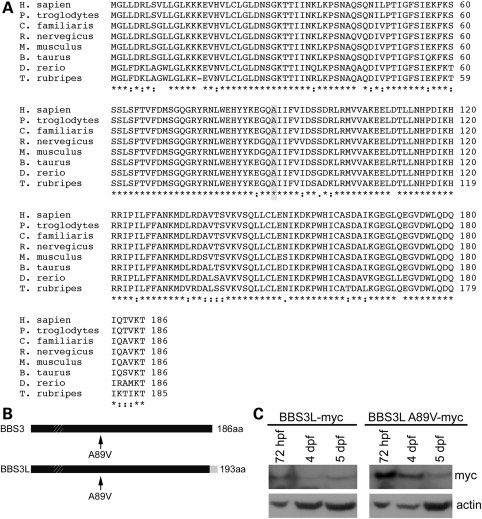
Previous homozygosity mapping with a consanguineous family from Saudi Arabia identified a novel missense mutation (A89V) in BBS3 that results in non-syndromic retinitis pigmentosa (39,40). Comparison across vertebrate species with available full-length BBS3 sequences demonstrates that the A89V mutation occurs in an evolutionarily conserved region of the protein, suggesting that the alanine at position 89 may have an essential function within the protein (Fig. 1A). The location of the mutation upstream of the C-terminus would impact both BBS3 and BBS3L, as this region is identical between the two isoforms (Fig. 1B). Additionally, the missense mutation is located downstream of the phosphate binding loop (P-loop), which is important for binding triphosphates of the GTP nucleotide (41). To test whether BBS3L A89V is stably expressed, we performed western blot analysis on embryos injected with either human myc-tagged BBS3L or BBS3L A89V RNA. We found that similar to BBS3L, BBS3L A89V was present through 5 days post-fertilization (dpf) (Fig. 1C). Thus, the BBS3 A89V missense mutation is in an evolutionally conserved region of the protein and would be found in both BBS3 and BBS3L. Moreover, the mutation does not impact BBSL expression, indicating that the mutation does not inhibit protein expression.
Figure 1.
BBS3 conservation and protein expression. (A) Multi-species alignment of BBS3 demonstrating the conservation among vertebrates. Shaded box highlights the location of the A89V mutation. Asterisks (*) indicate identical amino acids, while colons (:) and periods (.) represent conserved amino acids. (B) Schematic depicting the location of the A89V mutation in human BBS3 and BBS3L isoforms. Hatched box depicts the location of the P-loop motif and the grey box on the C-terminus of BBS3L denotes the region of difference between the two isoforms. (C) Western blot analysis of staged zebrafish embryos injected with either human BBS3L or BBS3L A89V myc-tagged RNA. Both proteins are present through 5 dpf. Actin served as a control.
BBS3 A89V functions in melanosome transport
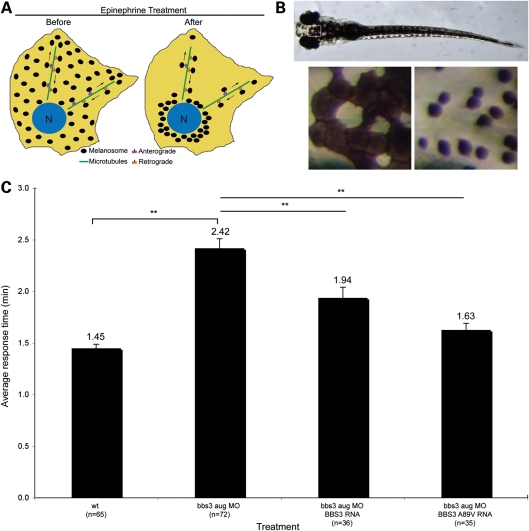
Knockdown of bbs3 using a MO that targets both transcripts (bbs3 aug MO) results in intracellular melanosome transport delay, a phenotype related to syndromic disease and shared among all BBS genes (26,34,35,38). In response to light or hormonal stimuli, zebrafish alter their skin pigmentation through intracellular melanosome transport within the melanophores (42–45). To test the rate of cellular trafficking, 6-day-old zebrafish embryos were dark adapted and treated with epinephrine to induce retrograde melanosome transport (Fig. 2A). Retrograde transport results in the movement of the dispersed melanosomes to a perinuclear location (Fig. 2B). Rescue experiments were performed by co-injecting RNA encoding human BBS3 or BBS3 A89V with the bbs3 aug MO. Wild-type embryos demonstrate rapid melanosome aggregation, averaging 1.45 min, whereas knockdown of bbs3 leads to a statistically significant delay in transport [analysis of variance (ANOVA) with Tukey, P < 0.01] (Fig. 2C and Table 1). We previously demonstrated that human BBS3, but not human BBS3L, can suppress the melanosome transport delays, thus we focused on BBS3 to investigate the role of the A89V mutation in syndromic disease (38). Similar to BBS3 RNA, co-injection of BBS3 A89V RNA with the bbs3 aug MO can restore transport times to wild-type levels (Fig. 2C and Table 1). These results demonstrate that the BBS3 A89V missense mutation can function to suppress the cardinal BBS phenotype of intracellular melanosome transport. This is consistent with the observation that the human patients harboring the BBS3 A89V mutation do not present with BBS-related phenotypes, such as obesity, polydactyly, renal anomalies or cognitive impairment.
Figure 2.
BBS3 A89V functions in melanosome transport. (A) Schematic illustrating the retrograde movement of melanosomes within the melanocyte from the periphery before epinephrine treatment to the perinuclear region after epinephrine treatment. (B) Dorsal view of a dark-adapted wild-type 6-day-old zebrafish embryo. Boxed head region is magnified below the full embryo. The left-hand image shows melanocytes prior to epinephrine treatment, while the right-hand panel depicts the same melanocytes at the endpoint of retrograde transport. (C) The graph summarizes the average epinephrine-induced response times in minutes for each experimental group. Both human BBS3 and BBS3 A89V RNAs are able to suppress the transport time seen with knockdown of bbs3. Sample size (n) is denoted on the X-axis. **P < 0.01, ANOVA with Tukey. Data presented as the mean ± SEM.
Table 1.
Melanosome transport (MT) times and vision assay responses
| Treatment | MT (min) | n | Vision (# responses) | n |
|---|---|---|---|---|
| wt | 1.45a | 65 | 3.77 | 64 |
| bbs3 aug MO | 2.42b | 72 | 1.91b | 75 |
| bbs3 aug MO +hBBS3 RNA | 1.94a,b | 36 | 1.00b | 15 |
| bbs3 aug MO +hBBS3 A89V RNA | 1.63a | 35 | 1.05b | 42 |
| bbs3 aug MO +hBBS3L RNA | NA | NA | 3.46 | 26 |
| bbs3 aug MO +hBBS3L A89V RNA | NA | NA | 1.60b | 25 |
aANOVA and Tukey test, P < 0.01 when compared with bbs3 aug MO.
bANOVA and Tukey test, P < 0.01 when compared with wt.
BBS3L A89V does not function in vision
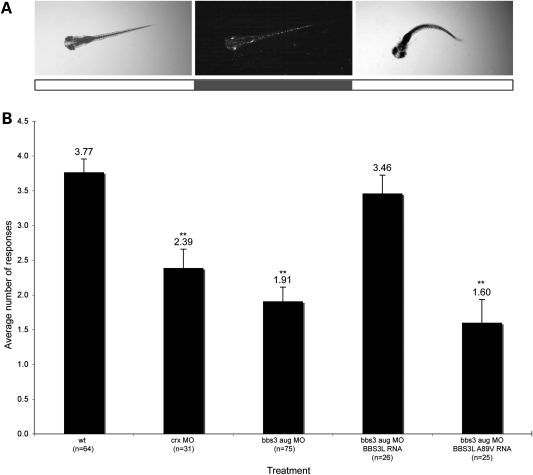
Both the mouse and zebrafish model systems have demonstrated that BBS3L is necessary for proper retinal function. Additionally, rescue experiments in the zebrafish have shown that human BBS3L is sufficient to suppress the bbs3 aug MO-induced vision defect (38). Since patients with the BBS3 A89V mutation present with only retinitis pigmentosa, we sought to functionally test the role of this mutation in vision by co-injecting BBS3L and BBS3L A89V RNA in bbs3 knockdown embryos. Visual function was evaluated by using a natural escape response that is elicited when zebrafish embryos are exposed to rapid changes in light intensity (38,46). In the vision startle assay, an embryos response to five short blocks in bright light are monitored and recorded. Visually responsive embryos change swimming directions in response to short blocks of bright light (Fig. 3A). Wild-type embryos respond on average 3.77 times (Fig. 3B). Cone-rod homeobox (crx) gene knockdown was used as a control for vision impairment as crx is necessary for photoreceptor formation in the zebrafish (47,48). crx knockdown embryos respond an average of 2.39 times, indicative of vision impairment (Fig. 3B) (ANOVA with Tukey, P < 0.01). bbs3 aug MO-injected embryos show a statistically significant reduction in the number of responses when compared with controls (Fig. 3B) (ANOVA with Tukey, P < 0.01) (38). Co-injection of BBS3L RNA with the bbs3 aug MO restored visual responsiveness back to wild-type levels, indicating that BBS3L RNA was sufficient to rescue vision (Fig. 3B). Conversely, BBS3L A89V RNA was not able to restore visual function back to wild-type levels as embryos responded on average 1.60 times (Fig. 3B). Western blot analysis confirms expression of BBS3L A89V through 5 dpf, when the vision assay is performed (Fig. 1C). The inability of BBS3L A89V RNA to restore vision provides strong functional support that this missense mutation leads to non-syndromic retinitis pigmentosa.
Figure 3.
The BBS3L A89V mutation is not functional in vision. (A) Still images of a 5-day-old zebrafish embryo illustrating the characteristic escape response elicited by a sudden change in light intensity. The white boxes below the stills indicate lights on, while the grey box indicates lights off. For visualization of the embryo in the dark, image contrast manipulation was performed in Adobe Photoshop. (B) Graphical representation of the vision startle response data. crx gene knockdown was used as a control for visual impairment. Human BBS3L can rescue the vision defect, whereas human BBS3L A89V is not able to rescue the vision defect in bbs3 knockdown embryos. The sample size (n) is noted on the X-axis. **P < 0.01, ANOVA with Tukey. Data presented as the mean ± SEM.
DISCUSSION
The present study utilizes the zebrafish to examine the function of BBS3 A89V in intracellular transport, a phenotype associated with the knockdown of BBS genes, and vision. This mutation was identified in a consanguineous Saudi Arabian family that presented with only retinitis pigmentosa (39,40). The polymorphism phenotyping program, PolyPhen, predicts that the BBS3 A89V missense mutation is a benign non-synonomous SNP; however, the alanine at position 89 is highly conserved in vertebrates, suggesting that this amino acid has an essential function. Using RNA rescue experiments, we demonstrate that unlike BBS3L RNA, the BBS3L A89V RNA does not rescue the vision defect observed with the loss of bbs3. Although the A89V mutation is not functional in vision, BBS3 A89V RNA is able to suppress the cardinal zebrafish BBS phenotype of melanosome transport. Taken together, these data demonstrate that the BBS3 A89V mutation identified in patients with non-syndromic retinal degeneration is critical and specific for the vision defect.
This study highlights the importance of functionally evaluating mutations that fall within different splice variants of a single gene for disease or tissue-specific relevance. It is estimated that alternative splicing affects 94% of multi-exon human genes (49,50). Multiple splice isoforms with potentially diverse functions can be generated from a single gene, thus contributing to phenotypic complexity in disease. Recently, a retina specific splice variant was identified for another BBS gene, BBS8 (51). A splice-site mutation was identified in BBS8 that resulted in the skipping of a retina specific exon (51). This mutation results in affected individuals presenting with non-syndromic retinitis pigmentosa, similar to what was seen with patients harboring the BBS3 A89V missense mutation (51). While the BBS3 A89V mutation does not fall within in a splice site, it is located such that it impacts both the canonical BBS3 isoform as well as the eye-specific BBS3L isoform. Thus, mutations targeting a gene with multiple isoforms can have different affects on disease presentation.
Our finding that the BBS3 A89V missense mutation is specific for vision demonstrates that this region is important for proper function of the protein in the eye. While no known domains are predicated to map to this region of BBS3, it is possible that the A89V mutation abrogates the interaction of BBS3 with a retina specific modifier and/or retina specific binding partner. Mutations in another BBS gene, CEP290, also result in phenotypic variation. This gene has been implicated in numerous diseases, in particular BBS and isolated blindness (52). While this gene has been implicated in several syndromic diseases, a mouse model for Cep290, the rd16 mouse in which a few exons of the gene are missing, only manifests early onset retinal degeneration (53). Moreover, additional splice isoforms are predicted for Cep290 and western blot analysis supports the notion that there are retina specific isoforms (53). These observations for BBS8, Cep290 and now BBS3 indicate that mutations need to be evaluated in the context of both isoform and tissue specificity. Moreover, this study highlights the importance of functionally testing human disease mutations to further elucidate their underlying pathophysiology in disease.
MATERIALS AND METHODS
Ethics statement
The University Animal Care and Use Committee at the University of Iowa approved all animal work in this study.
Animal care
Adult zebrafish were maintained under standard conditions and embryos collected from natural spawnings (54). Embryos were staged using previously described criteria (55).
Conservation
Using the Ensemble genome (release 59), orthologs to BBS3 were identified by performing BLAST algorithms with the human BBS3 sequence (ENST00000335979). Sequences were aligned using ClustalW (56).
Morpholino injections
Antisense MOs were air pressure injected into one- to four-cell-staged embryos at a concentration of 12 ng for bbs3 and 1 ng for crx.
MO sequences:
bbs3_aug (35,38): 5′-AGCTTGTCAAAAAGCCCCATTTGCT-3′;
crx (48): 5′-ATGTAGGACATCATTCTTGGGACGG-3′.
DNA constructs and RNA synthesis
The A89V mutation was generated by introducing the appropriate nucleotide change into C-terminally myc-tagged human BBS3 or BBS3L constructs using the Quick Change II site-directed mutagenesis kit (Stratagene).
Primers:
BBS3 A89V-F: 5′-GGAACACTATTATAAAGAAGGCCAAG-3′;
BBS3 A89V-R: 5′-CACTACTATCAATGACAAAAATAATAA-3′.
RNA was synthesized using the mMessage mMachine transcription kit (Ambion) and injected into 1–2 cell embryos at a concentration of 8 pg.
Melanosome transport assay
The melanosome transport assay was performed as previously described (26,34,35). Briefly, epinephrine (500 μl/ml, Sigma E437) treatment was applied to dark-adapted 6-day-old zebrafish embryos. The movement of the melanosomes from the periphery to the perinuclear region was timed and recorded. A stereoscope equipped with a Zeiss Axiocam camera was used to image live embryos.
Vision startle response
Zebrafish embryos change swimming directions in response to rapid changes in light intensity. Prior to performing the vision startle assay, 5 dpf embryos are light adapted for 1 h. Embryos were exposed to five trials of rapid changes in light intensity spaced at 30 s intervals and the response, a distinct C-bend, scored (38,57). crx gene knockdown was used as a control for vision impairment. To ensure motility, embryos were probed with a blunt needle on their flank, invoking the same response observed in the vision assay.
Statistical analysis
Statistics for both the melanosome transport and vision startle assay were calculated using the one-way ANOVA paired with the Tukey honestly significant difference test.
Western blot
Embryos were injected with C-terminal myc-tagged human BBS3L (8pg) and BBS3L A89V (8pg) RNA at the 1–2 cell stage. Embryos (n = 15) were collected at the following time points: 72 hpf, 4 dpf and 5 dpf. Collected embryos were homogenized in lysis buffer [20 mm Tris; 100 mm NaCl; 1 mm EDTA (ethylenediaminetetraacetic acid); 0.5% Triton X-100; 0.5% SDS (sodium dodecyl sulfate)] with protease inhibitor [0.1 mm PMSF (phenylmethanesulfonyl fluoride, Roche); 10 μg/ml Leupeptin (Roche)] and the supernatant collected. Whole cell lysates were run and transferred using the X Cell SureLock Mini-Cell System under reduced conditions (Invitrogen). Samples were mixed with NuPAGE LDS Sample Buffer and NuPAGE Reducing Agent (Invitrogen), heated at 70°C for 10 min and run on a pre-cast 4–12% NuPAGE Novex Bis-Tris gel (Invitrogen). Protein was transferred to a PVDF membrane (polyvinylidene fluoride, Amersham). The membrane was blocked in 5% milk (Carnation) for 1 h and probed with either mouse monoclonal anti-Myc (1:2000, 9B11, Cell Signaling) or rabbit polyclonal anti-actin (1:2000, Sigma) antibody overnight at 4°. Horseradish peroxidase-conjugated species-specific secondary antibodies (1:10,000, Jackson ImmunoResearch) were used for detection of primary antibodies.
FUNDING
This work is supported by the Foundation fighting blindness (V.C.S.), National Institutes of Health Grants (R01CA112369 to D.C.S., REY110298 to V.C.S. and REY017168 to V.C.S.). Funding to pay the Open Access publication charges for this article was provided by Howard Hughes Medical Institution.
ACKNOWLEDGEMENTS
The authors thank L. Baye, J. Beck, S. Swaminathan and T. Westfall for technical assistance and D. Aguiar Crouch for administrative assistance. V.C.S. is an Investigator of the Howard Hughes Medical Institute.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Harnett J.D., Green J.S., Cramer B.C., Johnson G., Chafe L., McManamon P., Farid N.R., Pryse-Phillips W., Parfrey P.S. The spectrum of renal disease in Laurence-Moon-Biedl syndrome. N. Engl. J. Med. 1988;319:615–618. doi: 10.1056/NEJM198809083191005. [DOI] [PubMed] [Google Scholar]
- 2.Green J.S., Parfrey P.S., Harnett J.D., Farid N.R., Cramer B.C., Johnson G., Heath O., McManamon P.J., O'Leary E., Pryse-Phillips W. The cardinal manifestations of Bardet-Biedl syndrome, a form of Laurence-Moon-Biedl syndrome. N. Engl. J. Med. 1989;321:1002–1009. doi: 10.1056/NEJM198910123211503. [DOI] [PubMed] [Google Scholar]
- 3.Bardet G. On congenital obesity syndrome with polydactyly and retinitis pigmentosa (a contribution to the study of clinical forms of hypophyseal obesity). 1920. Obes. Res. 1995;3:387–399. doi: 10.1002/j.1550-8528.1995.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 4.Biedl A. A pair of siblings with adiposo-genital dystrophy. 1922. Obes. Res. 1995;3:404. doi: 10.1002/j.1550-8528.1995.tb00167.x. [DOI] [PubMed] [Google Scholar]
- 5.Elbedour K., Zucker N., Zalzstein E., Barki Y., Carmi R. Cardiac abnormalities in the Bardet-Biedl syndrome: echocardiographic studies of 22 patients. Am. J. Med. Genet. 1994;52:164–169. doi: 10.1002/ajmg.1320520208. [DOI] [PubMed] [Google Scholar]
- 6.Riise R. Visual function in Laurence-Moon-Bardet-Biedl syndrome. A survey of 26 cases. Acta Ophthalmol. Suppl. 1987;182:128–131. doi: 10.1111/j.1755-3768.1987.tb02610.x. [DOI] [PubMed] [Google Scholar]
- 7.Leys M.J., Schreiner L.A., Hansen R.M., Mayer D.L., Fulton A.B. Visual acuities and dark-adapted thresholds of children with Bardet-Biedl syndrome. Am. J. Ophthalmol. 1988;106:561–569. doi: 10.1016/0002-9394(88)90586-7. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson S.G., Borruat F.X., Apathy P.P. Patterns of rod and cone dysfunction in Bardet-Biedl syndrome. Am. J. Ophthalmol. 1990;109:676–688. doi: 10.1016/s0002-9394(14)72436-5. [DOI] [PubMed] [Google Scholar]
- 9.Fulton A.B., Hansen R.M., Glynn R.J. Natural course of visual functions in the Bardet-Biedl syndrome. Arch. Ophthalmol. 1993;111:1500–1506. doi: 10.1001/archopht.1993.01090110066026. [DOI] [PubMed] [Google Scholar]
- 10.Carmi R., Elbedour K., Stone E.M., Sheffield V.C. Phenotypic differences among patients with Bardet-Biedl syndrome linked to three different chromosome loci. Am. J. Med. Genet. 1995;59:199–203. doi: 10.1002/ajmg.1320590216. [DOI] [PubMed] [Google Scholar]
- 11.Beales P.L., Warner A.M., Hitman G.A., Thakker R., Flinter F.A. Bardet-Biedl syndrome: a molecular and phenotypic study of 18 families. J. Med. Genet. 1997;34:92–98. doi: 10.1136/jmg.34.2.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riise R., Andreasson S., Borgastrom M.K., Wright A.F., Tommerup N., Rosenberg T., Tornqvist K. Intrafamilial variation of the phenotype in Bardet-Biedl syndrome. Br. J. Ophthalmol. 1997;81:378–385. doi: 10.1136/bjo.81.5.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heon E., Westall C., Carmi R., Elbedour K., Panton C., Mackeen L., Stone E.M., Sheffield V.C. Ocular phenotypes of three genetic variants of Bardet-Biedl syndrome. Am. J. Med. Genet. A. 2005;132:283–287. doi: 10.1002/ajmg.a.30466. [DOI] [PubMed] [Google Scholar]
- 14.Katsanis N., Beales P.L., Woods M.O., Lewis R.A., Green J.S., Parfrey P.S., Ansley S.J., Davidson W.S., Lupski J.R. Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat. Genet. 2000;26:67–70. doi: 10.1038/79201. [DOI] [PubMed] [Google Scholar]
- 15.Slavotinek A.M., Stone E.M., Mykytyn K., Heckenlively J.R., Green J.S., Heon E., Musarella M.A., Parfrey P.S., Sheffield V.C., Biesecker L.G. Mutations in MKKS cause Bardet-Biedl syndrome. Nat. Genet. 2000;26:15–16. doi: 10.1038/79116. [DOI] [PubMed] [Google Scholar]
- 16.Nishimura D.Y., Searby C.C., Carmi R., Elbedour K., Van Maldergem L., Fulton A.B., Lam B.L., Powell B.R., Swiderski R.E., Bugge K.E., et al. Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2) Hum. Mol. Genet. 2001;10:865–874. doi: 10.1093/hmg/10.8.865. [DOI] [PubMed] [Google Scholar]
- 17.Mykytyn K., Braun T., Carmi R., Haider N.B., Searby C.C., Shastri M., Beck G., Wright A.F., Iannaccone A., Elbedour K., et al. Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat. Genet. 2001;28:188–191. doi: 10.1038/88925. [DOI] [PubMed] [Google Scholar]
- 18.Mykytyn K., Nishimura D.Y., Searby C.C., Shastri M., Yen H.J., Beck J.S., Braun T., Streb L.M., Cornier A.S., Cox G.F., et al. Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat. Genet. 2002;31:435–438. doi: 10.1038/ng935. [DOI] [PubMed] [Google Scholar]
- 19.Ansley S.J., Badano J.L., Blacque O.E., Hill J., Hoskins B.E., Leitch C.C., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 20.Badano J.L., Ansley S.J., Leitch C.C., Lewis R.A., Lupski J.R., Katsanis N. Identification of a novel Bardet-Biedl syndrome protein, BBS7, that shares structural features with BBS1 and BBS2. Am. J. Hum. Genet. 2003;72:650–658. doi: 10.1086/368204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang A.P., Nishimura D., Searby C., Elbedour K., Carmi R., Ferguson A.L., Secrist J., Braun T., Casavant T., Stone E.M., et al. Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3) Am. J. Hum. Genet. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan Y., Esmail M.A., Ansley S.J., Blacque O.E., Boroevich K., Ross A.J., Moore S.J., Badano J.L., May-Simera H.,, Compton D.S., et al. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat. Genet. 2004;36:989–993. doi: 10.1038/ng1414. [DOI] [PubMed] [Google Scholar]
- 23.Li J.B., Gerdes J.M., Haycraft C.J., Fan Y., Teslovich T.M., May-Simera H., Li H., Blacque O.E., Li L., Leitch C.C., et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 24.Nishimura D.Y., Swiderski R.E., Searby C.C., Berg E.M., Ferguson A.L., Hennekam R., Merin S., Weleber R.G., Biesecker L.G., Stone E.M., et al. Comparative genomics and gene expression analysis identifies BBS9, a new Bardet-Biedl syndrome gene. Am. J. Hum. Genet. 2005;77:1021–1033. doi: 10.1086/498323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoetzel C., Laurier V., Davis E.E., Muller J., Rix S., Badano J.L., Leitch C.C., Salem N., Chouery E., Corbani S., et al. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat. Genet. 2006;38:521–524. doi: 10.1038/ng1771. [DOI] [PubMed] [Google Scholar]
- 26.Chiang A.P., Beck J.S., Yen H.J., Tayeh M.K., Scheetz T.E., Swiderski R.E., Nishimura D.Y., Braun T.A., Kim K.Y., Huang J., et al. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11) Proc. Natl Acad. Sci. USA. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoetzel C., Muller J., Laurier V., Davis E.E., Zaghloul N.A., Vicaire S., Jacquelin C., Plewniak F., Leitch C.C., Sarda P., et al. Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am. J. Hum. Genet. 2007;80:1–11. doi: 10.1086/510256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leitch C.C., Zaghloul N.A., Davis E.E., Stoetzel C., Diaz-Font A., Rix S., Alfadhel M., Lewis R.A., Eyaid W., Banin E., et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat. Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 29.Kulaga H.M., Leitch C.C., Eichers E.R., Badano J.L., Lesemann A., Hoskins B.E., Lupski J.R., Beales P.L., Reed R.R., Katsanis N. Loss of BBS proteins causes anosmia in humans and defects in olfactory cilia structure and function in the mouse. Nat. Genet. 2004;36:994–998. doi: 10.1038/ng1418. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura D.Y., Fath M., Mullins R.F., Searby C., Andrews M., Davis R., Andorf J.L., Mykytyn K., Swiderski R.E., Yang B., et al. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc. Natl Acad. Sci. USA. 2004;101:16588–16593. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mykytyn K., Mullins R.F., Andrews M., Chiang A.P., Swiderski R.E., Yang B., Braun T., Casavant T., Stone E.M., Sheffield V.C. Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc. Natl Acad. Sci. USA. 2004;101:8664–8669. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fath M.A., Mullins R.F., Searby C., Nishimura D.Y., Wei J., Rahmouni K., Davis R.E., Tayeh M.K., Andrews M., Yang B., et al. Mkks-null mice have a phenotype resembling Bardet-Biedl syndrome. Hum. Mol. Genet. 2005;14:1109–1118. doi: 10.1093/hmg/ddi123. [DOI] [PubMed] [Google Scholar]
- 33.Davis R.E., Swiderski R.E., Rahmouni K., Nishimura D.Y., Mullins R.F., Agassandian K., Philp A.R., Searby C.C., Andrews M.P., Thompson S., et al. A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc. Natl Acad. Sci. USA. 2007;104:19422–19427. doi: 10.1073/pnas.0708571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yen H.J., Tayeh M.K., Mullins R.F., Stone E.M., Sheffield V.C., Slusarski D.C. Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer's vesicle cilia function. Hum. Mol. Genet. 2006;15:667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- 35.Tayeh M.K., Yen H.J., Beck J.S., Searby C.C., Westfall T.A., Griesbach H., Sheffield V.C., Slusarski D.C. Genetic interaction between Bardet-Biedl syndrome genes and implications for limb patterning. Hum. Mol. Genet. 2008;17:1956–1967. doi: 10.1093/hmg/ddn093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blacque O.E., Reardon M.J., Li C., McCarthy J., Mahjoub M.R., Ansley S.J., Badano J.L., Mah A.K., Beales P.L., Davidson W.S., et al. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereiro I., Valverde D., Pineiro-Gallego T., Baiget M., Borrego S., Ayuso C., Searby C., Nishimura D. New mutations in BBS genes in small consanguineous families with Bardet-Biedl syndrome: detection of candidate regions by homozygosity mapping. Mol. Vis. 2010;16:137–143. [PMC free article] [PubMed] [Google Scholar]
- 38.Pretorius P.R., Baye L.M., Nishimura D.Y., Searby C.C., Bugge K., Yang B., Mullins R.F., Stone E.M., Sheffield V.C., Slusarski D.C. Identification and functional analysis of the vision-specific BBS3 (ARL6) long isoform. PLoS Genet. 2010;6:e1000884. doi: 10.1371/journal.pgen.1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abu Safieh L., Aldahmesh M., Shamseldin H., Hashem M., Shaheen R., Alkuraya H., Hazzaa S., Al-Rajhi A., Alkuraya F. Clinical and molecular characterization of Bardet-Biedl syndrome in consanguineous populations: the power of homozygosity mapping. J. Med. Genet. 2010;47:236–241. doi: 10.1136/jmg.2009.070755. [DOI] [PubMed] [Google Scholar]
- 40.Aldahmesh M.A., Safieh L.A., Alkuraya H., Al-Rajhi A., Shamseldin H., Hashem M., Alzahrani F., Khan A.O., Alqahtani F., Rahbeeni Z., et al. Molecular characterization of retinitis pigmentosa in Saudi Arabia. Mol. Vis. 2009;15:2464–2469. [PMC free article] [PubMed] [Google Scholar]
- 41.Wiens C.J., Tong Y., Esmail M.A., Oh E., Gerdes J.M., Wang J., Tempel W., Rattner J.B., Katsanis N., Park H.W., et al. The Bardet-Biedl syndrome-associated small GTPase ARL6 (BBS3) functions at or near the ciliary gate and modulates Wnt signalling. J. Biol. Chem. 2010;285:16218–16230. doi: 10.1074/jbc.M109.070953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skold H.N., Aspengren S., Wallin M. The cytoskeleton in fish melanophore melanosome positioning. Microsc. Res. Tech. 2002;58:464–469. doi: 10.1002/jemt.10164. [DOI] [PubMed] [Google Scholar]
- 43.Barral D.C., Seabra M.C. The melanosome as a model to study organelle motility in mammals. Pigment. Cell Res. 2004;17:111–118. doi: 10.1111/j.1600-0749.2004.00138.x. [DOI] [PubMed] [Google Scholar]
- 44.Marks M.S., Seabra M.C. The melanosome: membrane dynamics in black and white. Nat. Rev. Mol. Cell Biol. 2001;2:738–748. doi: 10.1038/35096009. [DOI] [PubMed] [Google Scholar]
- 45.Blott E.J., Griffiths G.M. Secretory lysosomes. Nat. Rev. Mol. Cell Biol. 2002;3:122–131. doi: 10.1038/nrm732. [DOI] [PubMed] [Google Scholar]
- 46.Easter S.S., Jr., Nicola G.N. The development of vision in the zebrafish (Danio rerio) Dev. Biol. 1996;180:646–663. doi: 10.1006/dbio.1996.0335. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y., Shen Y., Rest J.S., Raymond P.A., Zack D.J. Isolation and characterization of a zebrafish homologue of the cone rod homeobox gene. Invest. Ophthalmol. Vis. Sci. 2001;42:481–487. [PubMed] [Google Scholar]
- 48.Shen Y.C., Raymond P.A. Zebrafish cone-rod (crx) homeobox gene promotes retinogenesis. Dev. Biol. 2004;269:237–251. doi: 10.1016/j.ydbio.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 49.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 50.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riazuddin S.A., Iqbal M., Wang Y., Masuda T., Chen Y., Bowne S., Sullivan L.S., Waseem N.H., Bhattacharya S., Daiger S.P., et al. A splice-site mutation in a retina-specific exon of BBS8 causes nonsyndromic retinitis pigmentosa. Am. J. Hum. Genet. 2010;86:805–812. doi: 10.1016/j.ajhg.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coppieters F., Lefever S., Leroy B.P., De Baere E. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum. Mutat. 2010;31:1097–1108. doi: 10.1002/humu.21337. [DOI] [PubMed] [Google Scholar]
- 53.Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S.K., Cheng H., Scott A., Hurd R.E., et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio) Eugene, OR: University of Oregon Press; 1993. [Google Scholar]
- 55.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 56.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 57.Nishimura D.Y., Baye L.M., Perveen R., Searby C.C., Avila-Fernandez A., Pereiro I., Ayuso C., Valverde D., Bishop P.N., Manson F.D., et al. Discovery and functional analysis of a retinitis pigmentosa gene, C2ORF71. Am. J. Hum. Genet. 2010;86:686–695. doi: 10.1016/j.ajhg.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]