Abstract
Epstein–Barr virus (EBV) transformed lymphoblastoid cell lines (LCLs) provide a conveniently accessible and renewable resource for functional genomic studies in humans. The ability to accumulate multidimensional data pertaining to the same individual cell lines, from complete genomic sequences to detailed gene regulatory profiles, further enhances the utility of LCLs as a model system. A lingering concern, however, is that the changes associated with EBV transformation of B cells reduce the usefulness of LCLs as a surrogate model for primary tissues. To evaluate the validity of this concern, we compared global gene expression and methylation profiles between CD20+ primary B cells sampled from six individuals and six independent replicates of transformed LCLs derived from each sample. These data allowed us to obtain a detailed catalog of the genes and pathways whose regulation is affected by EBV transformation. We found that the expression levels and promoter methylation profiles of more than half of the studied genes were affected by the EBV transformation, including enrichments of genes involved in transcription regulation, cell cycle and immune response. However, we show that most of the differences in gene expression levels between LCLs and B cells are of small magnitude, and that LCLs can often recapitulate the naturally occurring gene expression variation in primary B cells. Thus, our observations suggest that inference of the genetic architecture that underlies regulatory variation in LCLs can typically be generalized to primary B cells. In contrast, inference based on functional studies in LCLs may be more limited to the cell lines.
INTRODUCTION
Lymphoblastoid cell lines (LCLs), derived from B-lymphocytes, constitute an important surrogate model to study genotype–phenotype relationships in humans. Although LCLs were originally established as renewable sources of DNA (1), they are now being extensively used in studies of the genetic and epigenetic determinants of gene regulation (2–8), as well as for investigating host responses to different perturbations or treatments, such as gene knockdowns (9,10), radiation (11,12) and drugs (13–15).
In contrast to studies in model organisms, functional assays in LCLs are often the only feasible approach to conduct population-based studies in humans, because other samples are either impossible or extremely difficult to collect and maintain. As a result, many recent studies of regulatory variation in humans, including studies of human diseases, used LCLs as the model system. More generally, cell lines offer convenience and replicability, and the HapMap LCLs, in particular, represent the most complete catalog of human variation to date (16), with many of the genomes of these cell lines being fully sequenced as part of the 1000 Genomes project (17). Thus, LCLs are expected to continue to be an important model system for functional studies in humans, in particular for studies of regulatory variation.
However, cell lines often carry chromosomal abnormalities (18) may have pronounced batch effects related to preparation and/or growth rates (19), and the Epstein–Barr virus (EBV) transformation itself can alter the methylation status (20) and expression levels of a subset of genes (21). As a result, the notion that studies of the genetic and epigenetic basis of regulatory variation in LCLs can be used more generally to understand regulatory mechanisms in primary tissues is controversial. In particular, the extent to which gene regulation in LCLs recapitulates that of untransformed B cells is not well understood, because there are no genome-wide detailed catalogues of the regulatory effects associated with EBV transformation.
A number of recent studies focused on gene regulatory differences between LCLs and primary tissues, but to our knowledge, none of these studies used a design that allows one to directly estimate the independent effects of EBV transformation on gene expression levels and DNA methylation profiles. For example, Min et al. (22) investigated the effect of different sample processing approaches on gene expression measurements in LCLs and primary blood cells, including B cells. Their observations indicated that a large number of genes are differentially expressed between primary cells and cell lines. However, several confounders (including the use of multiple distinct RNA preparation protocols, which were their main focus) render this data set inappropriate for an investigation of the regulatory effects specifically due to EBV transformation. In turn, Sun et al. (23) investigated genome-wide differences in DNA methylation between LCLs and peripheral blood cells (PBCs). They identified 3723 autosomal DNA methylation sites that had significantly different methylation statuses across cell types. However, because B-lymphocytes comprise only ∼5% of the total PBCs, it is possible that many of the methylation differences observed by Sun et al. are bona fide differences between primary B cells and other PBCs, rather than shifts in methylation status due to the EBV transformation process itself.
To assess the effects of EBV transformation on gene regulation in B-lymphocytes, we collected gene expression and DNA methylation data from primary B cells and their corresponding LCLs. We used six independent biological replications of the EBV transformations in order to be able to explicitly take into account possible confounding effects. In addition to identifying regulatory differences between B cells and LCLs, we investigated whether the level of inter-individual variation in gene expression is maintained following EBV transformation, a property that is highly important in the contexts of expression quantitative trait loci (eQTL) studies in LCLs.
RESULTS
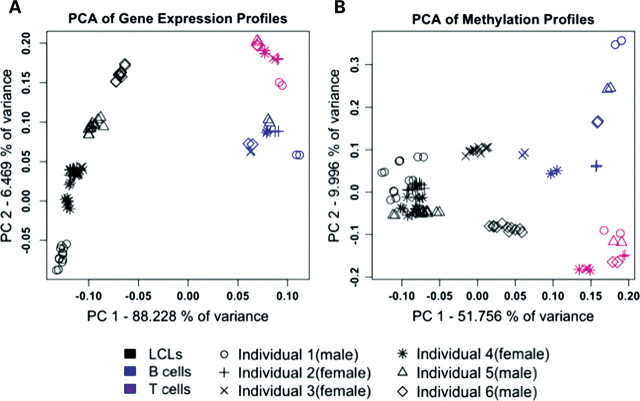
In order to assess the utility of LCLs as an in vitro model for studies of gene regulation, we compared methylation profiles and gene expression levels between primary and transformed B cells. Specifically, we used microarrays to estimate the expression levels of 25 160 genes, and characterize methylation status at 26 089 CpG sites (associated with 14 081 genes), in primary CD20+ B and CD3+ T cells from six individuals (three females, three males), as well as in newly transformed LCLs from these individuals, using six independent transformation replicates from each sample. The B cells, T cells and the newly derived LCLs were purified or transformed at the same time and from the same blood sample (see Materials and Methods for details on sample collection, transformations, sample processing and microarray data analysis; see Supplementary Material, Fig. S1 for an illustration of the study design and data sets S1 and S2 for the expression and methylation data for all genes). We performed multiple quality control analyses to confirm that the data quality is high (Supplementary Material, Figs S2–S11). As expected, gene expression estimates and methylation profiles in LCLs from independent biological transformation replicates from the same individuals (intra-individual variation) showed less variation than LCLs from different individuals (inter-individual variation; Supplementary Material, Figs S6 and S8). Interestingly, however, gene expression estimates and methylation profiles from all LCLs, regardless of the individual, were more highly correlated to each other than to data from either of the primary cell types (Supplementary Material, Figs S7 and S9). Indeed, a principal component analysis (PCA) indicates that cell type is a major source of variation in our data, and that the distinction between primary cells and the LCLs, based on either the gene expression or methylation data, is clear (Fig. 1A and B). These observations provided an initial indication of a biologically consistent and systematic effect of the transformation on global gene regulation.
Figure 1.
PCA of data from (A) the 48 803 gene expression probes, and (B) the 25 690 autosomal methylation probes, across all primary cells and LCLs.
The effect of cell transformation on gene expression levels
Of the 25 160 genes assayed, 14 373 (57%) were detected as expressed in at least one sample. We found that largely overlapping sets of genes were expressed in B and T cells (11 303 and 10 912 genes, respectively; with 83% overlap; Supplementary Material, Fig. S12). In turn, 2217 genes were expressed exclusively in LCLs (corresponding to 16.5% of genes expressed in LCLs; Supplementary Material, Fig. S12). Using GO functional annotation, we did not find any significant (after correction for multiple testing) enrichment among these 2217 genes. Nevertheless, among the top-ranked results, we found enrichment for genes involved in cytokine activity, signal transduction and receptor and immune response activity (Supplementary Material, Table S4).
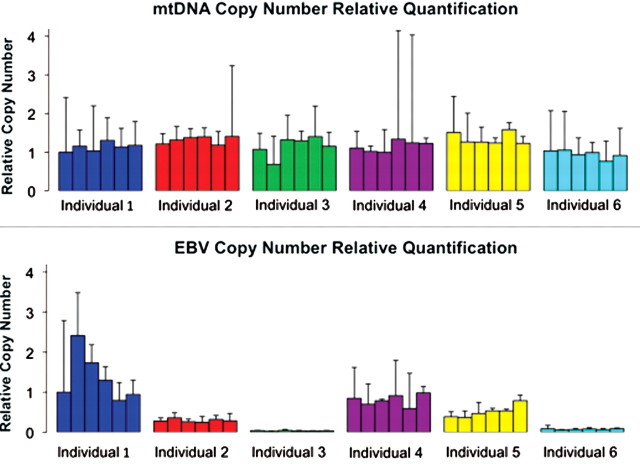
To better understand the effect of cell transformation on gene regulation, we first measured relative EBV and mtDNA copy numbers in each of the 36 LCL cultures obtained from the six individuals. Variation in mtDNA copy numbers in the LCLs was similarly low within and between individuals (Fig. 2A). On the other hand, inter-individual variation in EBV copy numbers was significantly greater than the intra-individual variation (Fig. 2B; P < 0.001), suggesting a possible genetic basis for susceptibility to EBV infection. A previous study suggested that differences in EBV copy numbers may significantly contribute to regulatory variation across cell lines (24). We revisited this question by focusing on gene expression data from LCLs. In our data, we identified only 160 genes (Supplementary Material, Table S1) whose expression levels were significantly associated with EBV copy numbers [false discovery rate (FDR) < 0.01; see Materials and Methods for details on modeling the gene expression data]. Consistent with the findings of Choy et al. (24), and with the known anti-apoptotic role of NF-κB signaling in LCLs (25,26), we inferred that 35 (21.9%) of these genes are direct or indirect regulatory targets of NF-κB signaling [using STRING protein–protein interaction database (27)].
Figure 2.
Relative EBV and mtDNA copy numbers in six unrelated individuals. Data from different individuals (for six LCLs from each individual) are plotted in different colors. The relative copy numbers were calculated using the ‘delta delta Ct’ method, using RNaseP as an endogenous reference gene. These bars are not statistical error per se; rather, they represent the range of possible copy number values defined by the standard error of the delta Ct's.
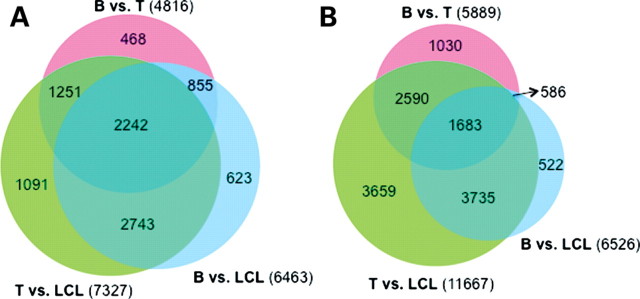
We next focused on differences in gene expression levels between cell types. To do so, we first regressed out the effect of EBV copy numbers from the estimates of gene expression levels in the LCLs. We then renormalized the residuals along with the gene expression estimates from the primary cells (see Materials and Methods for more details). We used likelihood ratio tests, within the framework of the linear models, to identify differentially expressed genes between cell types, considering data only from genes that were detected as expressed (see Materials and Methods). At an FDR < 0.01, we classified 4816 genes as differentially expressed between primary B and T cells (these correspond to 47.9% of the 10 059 genes detected as expressed in both cell types; Fig. 3A and Supplementary Material, Table S1). We similarly classified 7327 genes as differentially expressed between T cells and LCLs (corresponding to 70.4% of the 10 410 genes detected as expressed in both cell types), of which 3493 were also differentially expressed between B and T cells. These observations suggest that the majority (72.5%) of regulatory differences between B and T cells are maintained through the transformation (Fig. 3A and Supplementary Material, Table S1). Finally, we classified 6463 genes as differentially expressed between primary B cells and LCLs (60.7% of the 10 641 genes detected as expressed in both cell types; Fig. 3A and Supplementary Material, Table S1). While the numbers of genes detected as differentially expressed across cell types is large, most of the expression differences are of small magnitude (Supplementary Material, Fig. S13). For example, only 33 genes are differentially expressed between primary B cells and LCLs with a fold change of >1.5.
Figure 3.
Venn diagrams of the numbers of (A) differentially expressed genes, and, (B) differentially methylated probes, between pairs of cell types. A similar plot, including only genes that are expressed in all three cell types, is available as Supplementary Material, Figure S14.
Among the subset of genes that are differentially expressed between the primary T and B cells, we found slight enrichments for genes associated with signal transduction and immune system (Supplementary Material, Table S5). As expected, among genes with the largest magnitude of expression difference between B and T cells were a number of immune system-related genes, including T-cell-specific (TRAT1, CD3G, CD28, CD3E, STAT4) and B cell-specific (VPREB3, FCRLA, CD19, BANK1, FCER2) genes (Supplementary Material, Table S1). When we considered genes that were classified as differentially expressed between either primary cell type and LCLs, the top-ranked results (while not significant after correction for multiple testing) implied an over-representation of genes involved in transcriptional regulation, signal transduction and catalytic activity (Supplementary Material, Tables S6 and S7).
The effect of transformation on variation in gene expression levels
The comparison of gene expression patterns within and between cell types also allowed us to examine whether inter-individual variation in gene expression levels is affected by EBV transformation. Characterization of the nature and extent of inter-individual variation in gene expression levels is an essential feature of eQTL mapping studies. Of the 10 641 genes expressed in both primary B cells and LCLs, we identified only 627 genes with a significantly different between-individual variation in gene expression levels across the two cell types (using an uncorrected P < 0.01, which is conservative with respect to our conclusions that between-individual variation in gene expression levels in the B cells was largely maintained in the LCLs). Among these 627 genes, the between-individual variation in gene expression levels was higher in B cells compared with LCLs in 550 (87.7%; Supplementary Material, Table S1). Thus, our analyses suggest that LCLs can often recapitulate the naturally occurring gene expression variation in primary B cells.
In the context of eQTL studies, genes with the highest variation across individuals are of particular interest because the power to map eQTLs for these genes is higher. We thus next focused on the 500 (roughly 5%) transcripts with the highest between-individual variation in gene expression levels in primary B cells. Of these, 140 (28%) had significantly different between-individual variation of gene expression levels across the two cell types (uncorrected P < 0.01), a much larger proportion compared with our genome-wide estimate of 5.9%. Nevertheless, these observations indicate that between-individual variation in gene expression levels, for the majority of genes, is generally maintained through the viral transformation.
The effect of transformation on promoter methylation profiles
To examine the effects of viral transformation on methylation profiles, we first assessed the effect of EBV copy number on DNA methylation status, using a similar approach to the one described above for the analysis of gene expression data (see Materials and Methods for details on modeling the methylation data). We identified 1823 CpG sites (associated with the promoters of 1681 genes) at which methylation status was significantly associated with EBV copy number (FDR < 0.01). Among these, there was an enrichment of genes associated with signal transduction and receptor activity, among others (P < 0.001; Supplementary Material, Table S8).
We next focused on differences in methylation patterns across cell types. To do so, we regressed out the effects of EBV copy numbers on methylation levels, and analyzed the methylation data using a probe-wise linear model with fixed effects for cell type and sex, as well as a fixed effect to account for correlation between individuals (see Materials and Methods). We identified 5889 probes (22.6%; associated with 4872 genes), which were differentially methylated between B and T cells (at an FDR < 0.01; Fig. 3B and Supplementary Material, Table S2). Similarly, 6526 probes (associated with 5011 genes) were differentially methylated between B cells and LCLs (FDR < 0.01; Fig. 3B). In contrast, a much larger number of probes, 11 667 (associated with 8443 genes), were differentially methylated between T cells and LCLs (FDR < 0.01; Fig. 3B), of which 4273 were also differentially methylated between B and T cells. These observations suggest that the majority (87.7%) of methylation differences between B and T cells are maintained through transformation.
Among genes whose promoters were differentially methylated between each of the primary cells and the LCLs, we found an over-representation of genes associated with protein binding, receptor activity and immune system (Supplementary Material, Tables S9 and S10). Genes whose promoters were differentially methylated between the primary B and T cells were enriched for pathways involved in protein binding and cytokine–chemokine activity (Supplementary Material, Table S11). The P-values associated with these enrichments (P < 0.05 in all cases) would not be considered significant after correction for multiple testing. Nevertheless, these functional enrichments are intuitive given the known immune-related functions of these cell types and the stimulation caused by the viral transformation. Finally, in accordance with the previous observations (23), we also found that among differentially methylated sites across cell types, the vast majority of the CpG sites were less methylated in LCLs compared with the primary cells (Supplementary Material, Figs S15 and S16).
Combined analysis of methylation and gene expression data
We performed a combined analysis of the methylation and gene expression data in order to examine whether changes in DNA methylation could account for differences in gene expression levels between B cells and LCLs. To do so, we focused on 7157 genes that were expressed in both B cells and LCLs, which were associated with at least one probed CpG site. We then regressed out methylation effects before analyzing the gene expression data from both cell types using a linear model (see Materials and Methods). We compared the evidence supporting a difference in the gene expression level between cell types with and without the correction for the methylation level.
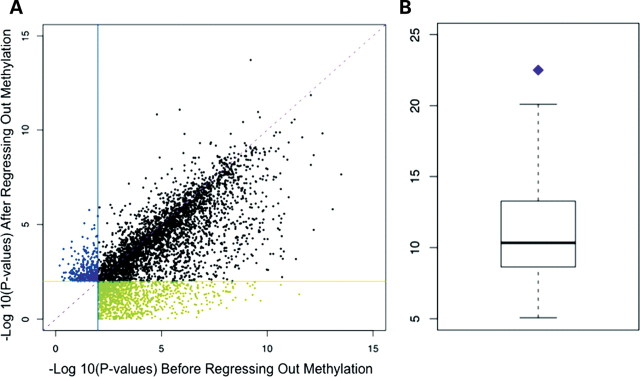
For the majority of genes found to be differentially expressed in either analysis (70.8%), the evidence for a difference in the expression level between cell types was roughly equally compelling (namely, with FDR < 0.01) regardless of whether methylation status was taken into account (Fig. 4A). For a small (6.7%), yet significant (by permutation; P = 0.037) subset of differentially expressed genes, we did not find evidence for a difference in the expression level between cell types when we considered the uncorrected expression level data, but after methylation levels were regressed out, we were able to reject the null hypothesis of no differences in the gene expression level between B cells and LCLs (at an FDR < 0.01). This result suggests that a subset of the observed alterations of methylation levels in LCLs is not random, but systematic and replicable.
Figure 4.
(A) Plot of the P-values obtained by testing the null hypothesis that there is no gene expression level difference between B cells and LCLs, using expression data before (x-axis) or after (y-axis) methylation levels were regressed. For 1058 genes (green), we found evidence for gene expression differences between cell types before (FDR < 0.01), but not after methylation levels were regressed out. For 314 genes (blue), we found evidence for gene expression differences between cell types after (FDR < 0.01), but not before methylation levels were regressed out. (B) A box plot of the number of genes for which evidence for gene expression differences between cell types before but not after methylation levels were regressed out is expected by chance alone (by permutation). The purple point indicates the observed proportion.
In turn, we found a higher proportion of differentially expressed genes (22.5%; permutation P = 0.022; Fig. 4B) for which the evidence for differences in the expression level between cell types was compelling (FDR < 0.01) before, but not after we regressed out the methylation levels. Assuming a causal relationship between promoter methylation and gene expression levels [a link supported by a large body of work (28–30)], our data suggest that changes in methylation profiles following EBV transformation can explain only a modest proportion of gene expression differences between the B cells and LCLs. Indeed, based on permutations (see Materials and Methods), we estimate that at most 12.2% of differences in gene expression levels between the two cell types may be explained, at least in part, by corresponding differences in promoter methylation status. That said, we cannot exclude the possibility that methylation at genomic regions outside those that were assayed by the array [for example, gene-body methylation (31)], may explain a higher proportion of the variation in gene expression levels between primary B cells and LCLs.
DISCUSSION
The goal of this work was to determine the suitability of LCLs as a model system for functional genomic studies. By carefully documenting the effects of EBV transformation on gene regulation, we aimed to contribute towards a resolution to the outstanding debate among human geneticists regarding the utility of LCLs as a surrogate model for primary tissues—B cells in particular. Our observations suggest that studies of the genetic architecture underlying variation in regulatory phenotypes in LCLs may be generalized, but that inference based on functional response phenotypes in LCLs should be interpreted with caution.
These conclusions, which we discuss in more details below, naturally depend on the exact context. We hope that our data will allow other investigators to decide, given their specific goals, whether the regulatory landscape in LCLs is sufficiently similar to that of B cells to warrant their use as a model system in each case. For that purpose, we provide all the data collected in this study, both as raw measurements (available at the GEO database) and as summaries of model estimates (Supplementary Material, Tables S1 and S2).
We note that the LCLs we generated for this study are newly derived cell lines. We therefore were unable to address at this time questions regarding the effect of age and passing generations on gene regulation in LCLs. To do so, in the next 5 years we will thaw these cells every 3–6 months, culture them, obtain updated genome-wide transcriptional profiles and freeze them again. We will use this scheme to study the effects that passing generations of cell line maintenance may have on gene regulation, thereby providing additional information on the utility of LCLs as a continuously renewable source for genomic studies.
The effects of EBV transformation
Our study design afforded us considerable power to detect regulatory differences between cell types. Thus, we were able to detect even small differences in gene expression and methylation levels. Using our approach, we classified the expression levels of 9111 genes as affected by the viral transformation, though most of these changes were of small magnitude. Overall, gene expression levels and methylation profiles in the LCLs were distinct from those of the primary B or T cells (Supplementary Material, Figs S7 and S9), reflecting the global cellular response to viral transformation. Nevertheless, most regulatory differences between the primary B and T cells were maintained in the LCLs, providing a strong indication that LCLs retain many of the specific characteristics of primary B cells.
A considerable subset (24.7%) of the genes whose regulation was affected by EBV transformation consists of genes whose expression levels were detected only in the LCLs. These are genes that were not expressed in the primary B cells, and whose expression was triggered by the viral transformation. This observation raises a possible caveat of our study regarding our choice to compare the transcriptional profiles of naive primary B cells to that of LCLs. It can be reasonably argued that LCLs, as a model system, reflect more faithfully the profile of activated B cells. However, regulatory differences due to the specific activating agent may be as abundant as the differences between resting and activated B cells (32), making it difficult to predict what primary cell stimulation might reflect most faithfully the EBV transformation. The correspondence of regulatory networks between activated B cells and LCLs will therefore have to be studied in each case separately.
Regardless of this caveat, the effect of the viral transformation is clearly systematic. Indeed, this conclusion is robust, regardless of the choice to consider all 9111 differentially expressed genes between LCLs and primary B cells, or to focus only on the subset of 6463 differentially regulated genes that were detected as expressed in both cell types. Although most of the observed differences in expression levels across cell types are small, it is reasonable to assume that the state of most regulatory networks in LCLs is considerably different than that of primary B cells. This raises the possibility that studies of gene regulatory networks in LCLs may not always be informative with respect to the corresponding response of primary B cells, let alone that of unrelated primary tissues. The discrepancy between LCLs and B cells may be particularly pronounced if studies focus perturbations that affect pathways and biological processes that are enriched with regulatory differences between the cell types, such as immune and stress response, cell cycle and receptor activity. More generally, we suggest that specific pilot studies should be performed, for each perturbation of interest, comparing the regulatory response of LCLs to that of primary tissues before larger efforts are embarked upon.
LCLs as a model system for studies of epigenetic regulation
We found a relatively small number of genes (160) whose expression levels were correlated with EBV copy numbers in the LCLs. In contrast, the methylation profiles at the putative promoters of 1681 genes were significantly correlated with EBV copy numbers. The reported numbers partly reflect our power to detect these correlations, which depend on the sample size and the error properties of each measurement (gene expression or methylation levels). A more robust observation, which takes into account differences in measurement properties, is that we find a correlation of EBV copy numbers with either expression or methylation levels for 2 and 26% of genes whose expression or promoter methylation levels, respectively, were affected by the viral transformation. In addition to the striking EBV copy number effect, we found that methylation levels in LCLs are systematically lower than in B cells [consistent with previous reports (23)].
These observations suggest that LCLs may not be a useful model system for studies of epigenetic regulation (at least not by promoter DNA methylation). However, we also found that overall methylation patterns in LCLs retain many of the specific characteristics of primary B cells. In addition, the proportion of gene expression differences between B cells and LCLs that can potentially be explained by differences in promoter methylation is, at most, modest (12.2%, when we ignore the direction of the correlation; namely, assume that even positive correlations between methylation and gene expression levels imply causality). Thus, our interpretation of these observations is that when EBV copy numbers are taken into account, LCLs can be used to study regulation by DNA methylation, with the caveat that the subset of regulatory interaction that can be studied will be smaller than in primary tissues. Providing some measure of support for this notion, a recent study of methylation QTLs in LCLs found that inter-individual variation in gene expression levels can often be explained, at least in part, by differences in promoter methylation (33).
LCLs may be a faithful model for eQTL mapping studies
Though the regulation of thousands of genes was affected by the viral transformation, the between-individual variation in gene expression levels seen in the primary B cells was generally maintained in the LCLs in the majority of the cases. These results suggest that for most genes, eQTLs found in LCLs may faithfully reflect the genetic architecture underlying regulatory variation in primary B cells. Indeed, several studies empirically tested this notion and provided evidence that the majority of eQTLs seen in LCLs can be recovered in primary tissues, including in non-blood-related tissue types (34–37). The most recent study (37) estimated that roughly 70% of cis eQTLs identified in LCLs could be replicated in primary skin tissue. Thus, though gene regulation is often cell-type specific (and varies temporally as well), an accumulating body of work suggests that certain components of variation in gene expression phenotypes are often shared across tissues, and are also maintained in LCLs. Based on our observations [and those of others (35–37)], we suggest that LCLs are indeed a useful model of the genetic architecture underlying regulatory variation in primary tissues, especially in primary B cells.
MATERIALS AND METHODS
Subjects and cell preparations
A unit of blood was obtained from six unrelated healthy individuals of Caucasian ethnicity (age range: 20–45). All samples were collected from October 2009 to January 2010 by Research Blood Components LLC (Brighton, MA, USA), under the company's IRB. Subjects were not on any medications and were not fasting at the time of the blood draw. Unpurified buffy coats were prepared from whole blood by centrifugation at 200g for 10 min at room temperature and then shipped (at room temperature) to the University of Chicago.
At the University of Chicago, we purified peripheral blood mononuclear cells (PBMCs) by further processing the buffy coats with density gradient centrifugation using ficoll-hypaque. PBMCs were counted on a hemocytometer. For each individual, we derived eight independent cultures of LCLs by EBV-mediated transformation, using the protocol provided by Coriell Cell repositories. We cultured the cell lines for 4–6 weeks until the cultures contained roughly 20 million cells. In addition, we isolated CD20+ B cells and CD3+ T cells from PBMCs by positive selection, using magnetic anti-CD20 and anti-CD3 mAb-coated microbeads (MACs, Miltenyi Biotec, Auburn, CA, USA). The newly derived LCLs are available by request.
RNA and DNA extractions
For each sample (primary cells and LCL cultures), we extracted total RNA and DNA using the QIAGEN RNeasy Plus Mini Kit or QIAamp DNA Blood Mini Kit, respectively. For RNA samples, the concentration was determined on a Nanodrop ND-100 Spectrophotometer (NanoDrop Technologies, Rockland, DE, USA) and the quality was measured using an Agilent 2100 Bioanalyzer lab chip (Agilent Technologies, Santa Clara, CA, USA). For each individual, we selected six (out of the eight) highest quality LCL samples based on RNA integrity number scores (Supplementary Material, Fig. S2 and Table S3) for gene expression profiling. For DNA samples, the concentration was determined both by Nanodrop and PicoGreen assays (Molecular Probes, Eugene, OR, USA).
Relative EBV and mtDNA copy number
We determined the relative EBV and mitochondrial DNA copy numbers of the LCL DNA samples using TaqMan quantitative PCR (qPCR) assays. The mtDNA copy number analysis assayed a 151 bp fragment of the MT-CYB gene, and the EBV copy number assay interrogated a 72 bp fragment of the IR1 gene of the virus. As an internal reference, we multiplexed an RNaseP TaqMan assay with EBV or mtDNA assays. We observed approximately equal amplification efficiencies (38) for each assay in the multiplex reaction (Supplementary Material, Fig. S17). The relative EBV and mtDNA copy numbers were calculated using the ‘delta delta Ct method’ (38). Additionally, we compared the amplification curves using DNA from the LCLs to that of a DNA template from the Namalwa cell line (ATCC CRL-1432), which was determined by fluorescence in-situ hybridization to have two integrated EBV copies per diploid genome (39). Using the qPCR results from the Namalwa cell line as standard, and by applying a conversion factor of 6.6 pg DNA per diploid genome, we confirmed that all our newly derived LCLs carry EBV DNA. We estimated that the newly derived LCL with the lowest EBV copy number (Fig. 2) carries 38 copies of EBV per diploid genome.
Gene expression profiling
The gene expression study design is illustrated in Supplementary Material, Figure S1. The study included 96 RNA samples: 8 different cell types (B cell + T cell + 6 independent LCLs per individual) × 6 individuals × 2 technical replicates (each RNA sample hybridized in duplicate). All samples were sent at the same time to the Southern California Genotyping Consortium for hybridization on HumanHT-12 v3 Expression BeadChip arrays (Illumina Inc.), which contain 48 803 probes targeting 25 160 unique RefSeq genes. The sample order was randomized across chips so that differences between individuals or cell types would not be confounded with differences in chip. Following cDNA synthesis, hybridization, scanning and image processing, probe intensity measurements were sent back to the University of Chicago. The intensity estimates were then log-transformed and quantile normalized using the ‘lumi’ package in R (all analyses were conducted in R v2.10.1). To remove probes for targets that were likely not expressed, we filtered out all probes that did not have a detection P-value <0.01 in at least one sample. Finally, gene-level estimates of expression were obtained by taking the median value for all probes for that gene.
Assessing the effect of EBV copy number on gene expression levels
In order to identify genes whose expression levels were associated with the EBV genome copy number, we focused on the expression estimates of the 13 440 genes detected as expressed in LCLs. We used the following gene-specific linear mixed-effects model to assess the EBV copy number effect on gene expression levels. For each gene, if Yil represents the normalized gene expression value for cell line l of individual i, we assume that:
where:
Here, μ is an overall mean expression value for a given gene. δsex(i) and θl are fixed effects for the sex of individual i and the EBV copy number of cell line l, respectively. γi is a random effect accounting for correlation between cell lines from the same individual, and is assumed to be N(0, σ2) distributed. In order to test the effect of EBV copy number on gene expression levels, we compared the fit of different parameterizations of this model [a null model where EBV copy number has no effect (i.e. θl = 0) and an alternative where EBV copy number does have an effect (i.e. θl ≠ 0)] by calculating a likelihood ratio test statistic and determining the P-values based on a χ2 distribution with one degree of freedom. We calculated maximum likelihoods for the different parameterizations using ‘lme’ in the nlme library and corrected P-values for multiple testing using the FDR approach of Benjamini and Hochberg (40).
Identifying gene expression differences between cell types
For each comparison, we focused on the genes detected as expressed in both of the compared cell types, renormalizing the gene expression values based on the overlapping set of detected genes for the respective comparison. When comparing gene expression levels between primary cells and LCLs, we first regressed out the effect of EBV copy number from the estimates of gene expression levels in the LCLs using a linear model as described above, except that we did not include a sex term. The EBV-corrected expression values were determined by adding the gene-specific residuals of this model back to the overall mean of each gene. We then normalized the primary cell data along with the EBV-corrected expression values for the LCLs, and averaged the expression values of the technical replicates. We identified the differentially expressed genes between cell types using likelihood ratio tests in the context of the following gene-specific fixed-effects linear model. If we now let Yic represent the normalized gene expression value for cell type c (B, T or LCL) of individual i, we assume that:
where:
Here, μ represents the mean expression level for gene g across all samples in the comparison, and θc, δsex(i) and αi are fixed effects for cell type, sex and individual, respectively. Again we assess how well different parameterizations of the model fit the data. Here, the null (H0) is that θc = 0, while the alternative (Ha) is that θc ≠ 0. We calculated the likelihood ratio statistics using ‘lm’ and obtained corrected P-values as described above.
Analysis of the effect of viral transformation on variation in gene expression levels
In order to assess the effect of viral transformation on variation in gene expression levels, we focused on genes expressed in both primary B cells and in the LCLs. Following background correction, log-transformation and quantile normalization, we regressed out the effect of EBV copy numbers as previously described. We used an F-test to compare the variances of six B cell populations and six LCL populations (one randomly chosen from each individual—we repeated the process multiple times and the results were consistent in all cases).
DNA methylation profiling
The methylation study design is identical to the expression study design described above, and also includes two technical replicates. We randomized the order of the 96 DNA samples across the 12-sample format chips and had methylation profiling done at the Southern California Genotyping Consortium using Illumina Infinium HumanMethylation27 BeadChip arrays, which have 27 578 probes targeting CpGs in the proximal promoter of 14 475 CCDS regions, 110 miRNAs and ∼200 genes chosen for their association with cancer or imprinting. Probes on this array were previously re-mapped (Bell, J.T. et al., unpublished data) to the human genome (hg18) using MAQ (41) and BLAT (42). We therefore focused only on the 26 690 probes that were mapped to a unique location in the genome. We obtained successful methylation profiling for 95 of the 96 samples and excluded the failed sample from the subsequent analysis. We also excluded data from 601 methylation probes with missing information in one or more individuals. To analyze methylation profiles across individuals and between cell types, we used the same framework of linear modeling described for the gene expression analysis, using Illumina β values as measures of methylation levels.
Combined analysis of gene expression and methylation data
In order to examine whether DNA methylation changes between cell types could explain gene expression level changes between B cells and LCLs, we focused on 7157 genes that were expressed in both B cells and LCLs and were represented by probes on the methylation array. We regressed out methylation levels from gene expression using an equivalent procedure to the one described for regressing EBV copy number effects from LCL expression measures. In other words, we fit a linear model to the data that includes a methylation term and then used the overall mean gene expression level plus the residuals of the model fit as the gene expression estimates.
Subsequently, we identified genes differentially expressed between B cells and LCLs using the gene expression data either before or after correcting for methylation levels. Here, as above, we used likelihood ratio tests to determine differentially expressed genes. This analysis allowed us to assess the fraction of differentially expressed genes potentially explained by differences in DNA methylation by determining the fraction of genes differentially expressed in either analysis for which the evidence for differences in expression levels between cell types was compelling (FDR < 0.01) before, but not after we regressed out the methylation levels (FDR > 0.01). For the purposes of the gene-wise joint analysis, we assumed that each CpG represented on the methylation array could act independently on the gene to which it is assigned. Therefore, if a gene had multiple probes and only a subset of those probes had the effect of creating or eliminating compelling evidence for differential expression, we considered this to be the ‘dominant effect' on the gene. Consequently, the effect of methylation on each gene was only counted once, according to this hierarchy. Similarly, the plot in Figure 4A displays the effect of a single probe chosen randomly from the subset producing the ‘dominant effect' on gene expression. We used permutations to assess the statistical significance of our results. To do this, we permuted the assignment of methylation arrays and repeated the analysis. We performed 1000 permutations and the P-value was calculated as the fraction of permutations with at least as large a fraction of genes fitting the patterns of interest (e.g. significant before but not after regressing out methylation).
Analysis of enrichment in functional categories
We used a web-based Gene Ontology application, DAVID (43), to examine enrichment of biological functional annotations among different classes of genes (e.g. genes whose expression or methylation levels are correlated with EBV copy number). Complete results from these analyses are available in Supplementary Material, Tables S4–S12.
Electronic database information
The gene expression and methylation data are available at the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under series accession number GSE26212.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by NIH grants HL092206 to Y.G. and C.O., and HL085197 to C.O.
ACKNOWLEDGEMENTS
We thank Z. Gauhar, L. Barreiro, A. Pai, A. Di Rienzo, J. Maranville, and all members of the Gilad lab for discussions and/or for comments on the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Carl B., Kroll H., Bux J., Bein G., Santoso S. B-lymphoblastoid cell lines as a source of reference DNA for human platelet and neutrophil antigen genotyping. Transfusion. 2000;40:62–68. doi: 10.1046/j.1537-2995.2000.40010062.x. doi:10.1046/j.1537-2995.2000.40010062.x. [DOI] [PubMed] [Google Scholar]
- 2.Cheung V.G., Spielman R.S., Ewens K.G., Weber T.M., Morley M., Burdick J.T. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. doi:10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. doi:10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veyrieras J.B., Kudaravalli S., Kim S.Y., Dermitzakis E.T., Gilad Y., Stephens M., Pritchard J.K. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. doi:10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. doi:10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D., et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. doi:10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. doi:10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen A., Rauch T.A., Pfeifer G.P., Hu V.W. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB J. 2010;24:3036–3051. doi: 10.1096/fj.10-154484. doi:10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mei Y.P., Zhu X.F., Zhou J.M., Huang H., Deng R., Zeng Y.X. siRNA targeting LMP1-induced apoptosis in EBV-positive lymphoma cells is associated with inhibition of telomerase activity and expression. Cancer Lett. 2006;232:189–198. doi: 10.1016/j.canlet.2005.02.010. doi:10.1016/j.canlet.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Badhai J., Frojmark A.S., Razzaghian H.R., Davey E., Schuster J., Dahl N. Posttranscriptional down-regulation of small ribosomal subunit proteins correlates with reduction of 18S rRNA in RPS19 deficiency. FEBS Lett. 2009;583:2049–2053. doi: 10.1016/j.febslet.2009.05.023. doi:10.1016/j.febslet.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu N., Qin Y., Fridley B.L., Hou J., Kalari K.R., Zhu M., Wu T.Y., Jenkins G.D., Batzler A., Wang L. Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res. 2010;20:1482–1492. doi: 10.1101/gr.107672.110. doi:10.1101/gr.107672.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Correa C.R., Cheung V.G. Genetic variation in radiation-induced expression phenotypes. Am. J. Hum. Genet. 2004;75:885–890. doi: 10.1086/425221. doi:10.1086/425221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watters J.W., Kraja A., Meucci M.A., Province M.A., McLeod H.L. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc. Natl Acad. Sci. USA. 2004;101:11809–11814. doi: 10.1073/pnas.0404580101. doi:10.1073/pnas.0404580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan S., Bleibel W.K., Huang R.S., Shukla S.J., Wu X., Badner J.A., Dolan M.E. Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer Res. 2007;67:5425–5433. doi: 10.1158/0008-5472.CAN-06-4431. doi:10.1158/0008-5472.CAN-06-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang R.S., Duan S., Shukla S.J., Kistner E.O., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Dolan M.E. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am. J. Hum. Genet. 2007;81:427–437. doi: 10.1086/519850. doi:10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montpetit A., Chagnon F. The haplotype map of the human genome: a revolution in the genetics of complex diseases. Med. Sci. (Paris) 2006;22:1061–1067. doi: 10.1051/medsci/200622121061. [DOI] [PubMed] [Google Scholar]
- 17.Hayden E.C. International genome project launched. Nature. 2008;451:378–379. doi: 10.1038/451378b. doi:10.1038/451378b. [DOI] [PubMed] [Google Scholar]
- 18.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. doi:10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akey J.M., Biswas S., Leek J.T., Storey J.D. On the design and analysis of gene expression studies in human populations. Nat. Genet. 2007;39:807–808. doi: 10.1038/ng0707-807. author reply 808-9) doi:10.1038/ng0707-807. [DOI] [PubMed] [Google Scholar]
- 20.Hannula K., Lipsanen-Nyman M., Scherer S.W., Holmberg C., Hoglund P., Kere J. Maternal and paternal chromosomes 7 show differential methylation of many genes in lymphoblast DNA. Genomics. 2001;73:1–9. doi: 10.1006/geno.2001.6502. doi:10.1006/geno.2001.6502. [DOI] [PubMed] [Google Scholar]
- 21.Carter K.L., Cahir-McFarland E., Kieff E. Epstein-Barr virus-induced changes in B-lymphocyte gene expression. J. Virol. 2002;76:10427–10436. doi: 10.1128/JVI.76.20.10427-10436.2002. doi:10.1128/JVI.76.20.10427-10436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Min J.L., Barrett A., Watts T., Pettersson F.H., Lockstone H.E., Lindgren C.M., Taylor J.M., Allen M., Zondervan K.T., McCarthy M.I. Variability of gene expression profiles in human blood and lymphoblastoid cell lines. BMC Genomics. 2010;11:96. doi: 10.1186/1471-2164-11-96. doi:10.1186/1471-2164-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y.V., Turner S.T., Smith J.A., Hammond P.I., Lazarus A., Van De Rostyne J.L., Cunningham J.M., Kardia S.L. Comparison of the DNA methylation profiles of human peripheral blood cells and transformed B-lymphocytes. Hum. Genet. 2010;127:651–658. doi: 10.1007/s00439-010-0810-y. doi:10.1007/s00439-010-0810-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choy E., Yelensky R., Bonakdar S., Plenge R.M., Saxena R., De Jager P.L., Shaw S.Y., Wolfish C.S., Slavik J.M., Cotsapas C., et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. doi:10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cahir McFarland E.D., Izumi K.M., Mosialos G. Epstein-Barr virus transformation: involvement of latent membrane protein 1-mediated activation of NF-kappaB. Oncogene. 1999;18:6959–6964. doi: 10.1038/sj.onc.1203217. doi:10.1038/sj.onc.1203217. [DOI] [PubMed] [Google Scholar]
- 26.Sylla B.S., Hung S.C., Davidson D.M., Hatzivassiliou E., Malinin N.L., Wallach D., Gilmore T.D., Kieff E., Mosialos G. Epstein-Barr virus-transforming protein latent infection membrane protein 1 activates transcription factor NF-kappaB through a pathway that includes the NF-kappaB-inducing kinase and the IkappaB kinases IKKalpha and IKKbeta. Proc. Natl Acad. Sci. USA. 1998;95:10106–10111. doi: 10.1073/pnas.95.17.10106. doi:10.1073/pnas.95.17.10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen L.J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. doi:10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kass S.U., Goddard J.P., Adams R.L. Inactive chromatin spreads from a focus of methylation. Mol. Cell Biol. 1993;13:7372–7379. doi: 10.1128/mcb.13.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kass S.U., Landsberger N., Wolffe A.P. DNA methylation directs a time-dependent repression of transcription initiation. Curr. Biol. 1997;7:157–165. doi: 10.1016/s0960-9822(97)70086-1. doi:10.1016/S0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 30.Keshet I., Yisraeli J., Cedar H. Effect of regional DNA methylation on gene expression. Proc. Natl Acad. Sci. USA. 1985;82:2560–2564. doi: 10.1073/pnas.82.9.2560. doi:10.1073/pnas.82.9.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maunakea A.K., Nagarajan R.P., Bilenky M., Ballinger T.J., D'Souza C., Fouse S.D., Johnson B.E., Hong C., Nielsen C., Zhao Y., et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. doi:10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirakawa A.K., Liao F., Zhang H.H., Hedrick M.N., Singh S.P., Wu D., Farber J.M. Pathway-selective suppression of chemokine receptor signaling in B cells by LPS through downregulation of PLC-beta2. Cell Mol. Immunol. 2010;7:428–439. doi: 10.1038/cmi.2010.46. doi:10.1038/cmi.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibbs J.R., van der Brug M.P., Hernandez D.G., Traynor B.J., Nalls M.A., Lai S.L., Arepalli S., Dillman A., Rafferty I.P., Troncoso J., et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. doi:10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bullaughey K., Chavarria C.I., Coop G., Gilad Y. Expression quantitative trait loci detected in cell lines are often present in primary tissues. Hum. Mol. Genet. 2009;18:4296–4303. doi: 10.1093/hmg/ddp382. doi:10.1093/hmg/ddp382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M., et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. doi:10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. doi:10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding J., Gudjonsson J.E., Liang L., Stuart P.E., Li Y., Chen W., Weichenthal M., Ellinghaus E., Franke A., Cookson W., et al. Gene expression in skin and lymphoblastoid cells: refined statistical method reveals extensive overlap in cis-eQTL signals. Am. J. Hum. Genet. 2010;87:779–789. doi: 10.1016/j.ajhg.2010.10.024. doi:10.1016/j.ajhg.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. doi:10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Lawrence J.B., Villnave C.A., Singer R.H. Sensitive, high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell. 1988;52:51–61. doi: 10.1016/0092-8674(88)90530-2. doi:10.1016/0092-8674(88)90530-2. [DOI] [PubMed] [Google Scholar]
- 40.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 41.Li H., Ruan J., Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. doi:10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. doi:10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]