Abstract
Objective
Severe alcohol consumption can cause serious morbidity and death. Because the serotonin transporter (5-HTT) is an important regulator of neuronal 5-HT function, allelic differences at that gene may modulate the severity of alcohol consumption and predict therapeutic response to the 5-HT3 receptor antagonist, ondansetron.
Method
We randomized 283 alcoholics by genotype in the 5′-regulatory region of the 5-HTT gene (LL/LS/SS), with additional genotyping for another functional single nucleotide polymorphism (T/G), rs1042173, in the 3′-untranslated region, in a controlled double-blind trial. Subjects received ondansetron (4 μg/kg twice daily) or placebo for 11 weeks, plus standardized cognitive behavioral therapy.
Results
LL subjects who received ondansetron vs. placebo had fewer mean drinks per drinking day (DDD) and a higher percentage of days abstinent (PDA) (−1.62; p=0.007 and 11.27%; p=0.023). Within ondansetron recipients, DDD was lower and PDA higher in LL vs. LS/SS subjects (−1.53; p=0.005 and 9.73%; p=0.03). Ondansetron LL subjects also had fewer DDD and greater PDA than all other genotype and treatment groups combined (−1.45; p=0.002 and 9.65%; p=0.013). For both DDD and PDA, 5′-HTTLPR and rs1042173 variants interacted significantly (p=0.023 and 0.009). Ondansetron LL/TT had fewer DDD and a greater PDA than all other genotype and treatment groups combined (−2.63; p<0.0001 and 16.99%; p=0.002).
Conclusions
We propose a new pharmacogenetic approach using ondansetron to reduce the severity of alcohol consumption and improve abstinence in alcoholics.
INTRODUCTION
Severe alcohol consumption can cause serious health problems, morbidity, and death (1). Finding efficacious treatments that can decrease the severity of alcohol consumption among alcohol-dependent individuals is, therefore, an important scientific and health goal. Because the serotonin (5-HT) system is an important regulator of the severity of alcohol consumption (2), medications that affect the function of the 5-HT transporter (5-HTT), which plays an important role in the regulation of neuronal 5-HT function (3), appear to be a particularly promising treatment (4).
SLC6A4, which is located on chromosome 17q11.1–q12, is the only known gene encoding the 5-HTT in the human genome (5). Because the amino acid sequence and sensitivity of the human 5-HTT is common to all tissues, including blood cells and neurons (5, 6), it is reasonable to suspect that genetic variation that alters the function of the platelet 5-HTT might also be seen in neuronal cells.
The SLC6A4 promoter contains a functional polymorphic region (5′-regulatory region of the 5-HTT; 5′-HTTLPR) with a long form (L) that possesses an additional 44 base pairs that are absent in the short variant (S). The 5′-HTTLPR polymorphism has been investigated extensively for a possible association with a wide range of psychiatric disorders. For example, the S allele has been associated with anxiety-related personality traits (7, 8) and depression (9), whilst the L allele has been linked with obsessive-compulsive disorder (10) and improved antidepressant response (11). Variants of the 5-HTT gene also have been associated with substance abuse disorders including alcohol dependence (12). Thus, 5′-HTTLPR polymorphisms can influence various psychiatric conditions including alcohol dependence.
In healthy humans, the LL genotype, compared with the LS and SS genotypes, has been associated with higher transcription rates in lymphoblast cells, greater 5-HT uptake into platelets (13) and lymphoblasts (14), and increased binding of [123I]2 β-carboxymethoxy-3 β-(4-iodophenyl)tropane (β-CIT) in human raphe nuclei (15). In contrast to healthy humans, neuroimaging studies have shown that alcohol-dependent LL variants, compared with their healthy counterparts, have lower β-CIT neuronal binding to 5-HTTs in the raphe nuclei of the brain (15). Consistent with these findings in neuronal tissue, studies in platelets have shown that alcohol-dependent individuals with the LL genotype, compared with those with the SS genotype, have significantly less 5-HT uptake and reduced paroxetine binding capacity (3, 16). Also, among those with the L variant, greater severity of lifetime alcohol consumption has been associated with lower levels of 5-HT uptake and binding (3). These findings have suggested a gene-by-environment interaction whereby 5-HTT gene expression is suppressed by increased alcohol consumption via an unknown mechanism, which, in turn, perpetuates further drinking of alcohol (3). Thus, LL genotype in alcohol-dependent individuals has an important effect on the function of the 5-HTT and the severity of lifetime alcohol consumption.
We discovered that another functional allelic variant, the single nucleotide polymorphism (SNP) rs1042173 (T/G) in the 3′-untranslated region (3′-UTR) of the 5-HTT gene, can be associated with the severity of alcohol consumption. TT homozygotes, compared with G-carriers, had a significantly greater severity of alcohol consumption (17). Significantly, T-allele-transfected HeLa cells, compared with their G-allele counterparts, had lower expression levels for both 5-HTT mRNA and protein (17). Because the rs1042173 SNP is located at or near a potential binding site for several microRNAs (18), and variation at this location may alter expression levels by affecting the stability of mRNA (19, 20), it would be reasonable to propose that the TT genotype of the 3′-UTR SNP might increase the effect of the LL genotype of the 5′-HTTLPR to produce a more marked reduction in 5-HTT expression in alcohol-dependent individuals. Lowered 5-HTT expression and binding would, paradoxically, be associated with reduced rather than increased 5-HT intrasynaptic neurotransmission and with up-regulation of postsynaptic 5-HT receptors (4). This is because 5-HTTs in the raphe nuclei are somatodendritic, and, therefore, auto-regulatory mechanisms would have the effect of decreasing 5-HT firing rates (21) and up-regulating postsynaptic 5-HT receptors.
This relative hypo-serotonergic state with up-regulation of post-synaptic 5-HT receptors might explain why alcohol-dependent individuals with the LL genotype compared with the LS/SS genotype of the 5′-HTTLPR have an increased urge to use alcohol (22). This supports the prediction from our hypothesis that blockade of these up-regulated post-synaptic 5-HT receptors in alcohol-dependent individuals with the LL genotype of the 5′-HTTLPR by ondansetron would result in a marked reduction in the severity of alcohol consumption (4). Other plausible explanations for ondansetron’s anti-drinking effects may exist, including the blockade of ethanol-induced receptor sensitization in the 5-HT system (23), which we would expect to be greater in humans with the LL compared with the LS or SS genotype due to decreased expression of the 5-HTT gene (15).
Predicated on the hypothesis that possession of the TT genotype of rs1042173—like the LL genotype of the 5′-HTTLPR in alcohol-dependent individuals—suppresses 5-HTT expression and binding (albeit through different molecular mechanisms), it is reasonable to hypothesize further that ondansetron’s therapeutic effect would be greatest among alcohol-dependent individuals who possessed the combination of the LL and TT genotypes. We, therefore, tested two predictions of our hypotheses. First, ondansetron would have a differentially greater therapeutic effect to reduce the severity of alcohol consumption—i.e., drinks per drinking day, our primary efficacy variable—and to increase the percentage of days abstinent, our secondary outcome variable, among alcohol-dependent individuals with the LL genotype compared with S-carriers of the 5′-HTTLPR. Second, ondansetron’s therapeutic effect would be greatest among alcohol-dependent individuals who possessed both the LL genotype of the 5′-HTTLPR and the TT genotype of rs1042173 in the 3′-UTR.
To test these hypotheses, we conducted a phase 2, 11-week randomized trial (after a 1-week single-blind placebo lead-in) in 283 alcohol-dependent men and women who all received weekly standardized cognitive behavioral therapy as their psychosocial treatment and either ondansetron (4 μg/kg twice daily) or placebo. Prospectively, subject randomization was stratified at enrollment by 5′-HTTLPR genotype, with additional genotyping for rs1042173 in the 3′-UTR and expression analysis for the 5-HTT gene after randomization.
METHOD
Participants
Briefly, our 283 enrollees: were mostly White males (73.1% male; 84.8% White, and the remainder, 15.2%, were Hispanic); had alcohol dependence but no axis I diagnosis other than nicotine dependence according to the Diagnostic and Statistical Manual of Mental Disorders, 4th edition (24); were aged 20 to 78 years (mean age, 44.7 years), and scored >8 on the Alcohol Use Disorders Identification Test (25), which assessed the severity of alcohol-related problems. Most (i.e., 211) of the participants were enrolled at the University of Texas Health Science Center at San Antonio and the remainder (i.e., 72 participants) at the University of Virginia. Enrollees were current drinkers of alcohol (i.e., not abstinent) and, therefore, not withdrawing from alcohol. Enrollees were required to present themselves for clinic attendance under the breath alcohol concentration limit of 0.02% to complete the rating scales. This breath alcohol concentration limit was achieved without significant withdrawal symptoms, as exemplified by the extremely low average values for both scores during the study that are provided in the Results section. Enrollees were physically healthy, not pregnant, and not using abused drugs at enrollment. All subjects provided informed consent to participate.
Enrollees were randomized using an urn (26) procedure after the screening visit to either treatment or placebo under each of 3 different genotypes (LL, LS, or SS genotypes of the 5′-HTTLPR). Genotyping and analysis of the cohort for rs1042173 (i.e., TT/TG/GG) were done post-randomization.
Study procedures
Testing took place at the University of Texas Health Science Center at San Antonio and the University of Virginia from 11/14/2000 to 12/17/2004 and from 5/13/2006 to 10/20/2008, respectively.
Subjects were enrolled in the study whilst they were still drinking alcohol (i.e., without a period of abstinence). At enrollment, we collected self-reported alcohol consumption based on the timeline follow-back method over the past 90 days (27), and did genotyping on all samples.
Other screening parameters, including those of physical health, breath alcohol concentration, and additional psychosocial measures associated with drinking behavior, were the same as—and were collected as—detailed in our previous trials (28, 29).
Eligible subjects were randomized at screen, entered a single-blind placebo period at week 1, and received their double-blind medication assignment (i.e., ondansetron 4 μg/kg twice daily or placebo) between weeks 2 and 12.
From weeks 1 to 12, subjects received weekly assessments of alcohol consumption using the timeline follow-back method, adverse events using the Systematic Assessment for Treatment Emergent Events (30), alcohol withdrawal using the revised Clinical Institute Withdrawal Assessment for Alcohol scale (31), urine drug testing, concomitant medication use, and pill count.
All subjects received weekly standardized, manual-driven (32, 33) cognitive behavioral therapy in groups of up to 8 subjects as their psychosocial treatment. Cognitive behavioral therapy is an integration of cognitive, behavioral, and social learning theory that enables alcoholic subjects to achieve and maintain abstinence by enhancing their ability to manage high-risk situations, which can trigger alcohol-seeking behavior (34, 35). Cognitive behavioral therapy was provided using a standardized manual and protocol, which was delivered by skilled Master’s- and doctoral-level psychologists. The same manual was used at both study sites. The cognitive behavioral therapy sessions were audiotaped with the subjects’ permission. A doctoral-level psychologist supervised the delivery of cognitive behavioral therapy by reviewing a random selection of about 10% of these tapes. Furthermore, the cognitive behavioral therapy supervisor met with the psychologists who delivered the treatment on a weekly basis. All those who delivered cognitive behavioral therapy at both sites were re-evaluated formally every 6 months. The overall supervisor for the delivery of cognitive behavioral therapy at both sites was the same individual throughout the study. A review of the supervisor’s notes on the delivery of cognitive behavioral therapy at both sites revealed no evidence of any potential differential “drift” at either location. These stringent procedures for the delivery of cognitive behavioral therapy ensured that it was a stable platform of psychosocial treatment against which to measure the added effect of the medication. At scheduled intervals, other measures, including those of physical health and other psychosocial measures, were collected as described previously (28, 29).
DNA extraction and genotyping
Genomic DNA was extracted from the blood of each subject at baseline with a Gentra Puregene® kit (QIAGEN Inc., Valencia, CA). The 5′-HTTLPR L/S alleles were determined as described earlier (3, 36) with custom-made primers. The alleles of rs1042173 were genotyped with pre-made TaqMan® genotyping assays (Applied Biosystems, Foster City, CA) as reported previously (17). Detailed information on primer/probe sequences and biological information on the two polymorphisms are summarized in Table 1. To test for potential population stratification between the treatment and placebo groups, we did genotyping for 24 ancestry-informative markers for all participant DNA samples to calculate percent ancestry. These markers have been used by many other research groups and demonstrated to have high-frequency differences for South American/European ancestry and European/West African ancestry (37, 38). Information on these markers is provided in Table S1 in the data supplement that accompanies the online edition of this article.
Table 1.
Biological Information on SLC6A4 Polymorphisms Examined in the Study
| Physical Position | Chromosome position (NCBI Genome build) | Alleles | MAF | p-Values for the deviation from HWEb | Primers and probe sequences / Context sequence ID of ABI primers and probes | Number of missing data points | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CEUa | Pooledb | Whiteb | Hispanicb | Pooled | White | Hispanic | |||||
| Promoter | 25,588,443: 25,588,485 (36.1) | L/S | 0.450 | 0.438 | 0.424 | 0.447 | 0.461 | 0.850 | 0.461 | Forward: TCCT CCGCTTTGGCG CCTCTTCC Reverse: TGGGGGTTGCAGGGGA GATCCTG |
0 |
| Exon 15 (3′-UTR) | 25,549,137 (36.3) | G/T | 0.433 | 0.430 | 0.440 | 0.424 | 0.457 | 0.544 | 0.827 | C_7473190_10 | 7 |
Abbreviations: L, long allele; S, short allele; MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium; ABI, Applied Biosystems (Foster City, CA); NCBI, National Center for Biotechnology Information; 3′-UTR, 3′-untranslated region.
European sample from HapMap project.
Data from this study.
The pooled sample consists of samples from subjects of Caucasian and Hispanic origin.
Assessment of Sample Admixture Between Treatment Groups
The program STRUCTURE (http://pritch.bsd.uchicago.edu/software/structure2_2.html) was used to assess population stratification as well as to estimate genetic ancestry proportions for each participant enrolled at the two centers. We then used the individual ancestry proportion estimates as covariates in all statistical models. Prior to obtaining the individual ancestry proportion estimates for our population, we assessed the number of parental populations (K) that captured most of the stratification in the population. For this, we analyzed the data set with K=2 through K=10, and the simulation parameters were set to 10,000 burn-ins and 10,000 Markov Chain Monte Carlo iterations. The K value with the highest probability to capture stratification was 3; hence, we obtained the ancestry proportion estimates by analyzing the data set assuming 3 parental populations (K=3) and the presence of population admixture. Self-reported ethnicity was not used as additional data in STRUCTURE analyses.
Power and statistical analyses
Statistical Power
We did our power analysis as described in Section 8.3 of Cohen (39). For our primary outcome variable, drinks per drinking day, a total sample size of 280 (140 in each treatment arm) had a statistical power of 0.91 to detect an effect size of 0.25 in the interaction between treatment and genotype (LL vs. LS/SS), at a significance level of 0.05. Similarly, we had a statistical power of 0.91 to detect an effect size of 0.25 in the interaction between 5′-HTTLPR and rs1042173 genotypes. Statistical power was 0.91 to detect an effect size of 0.35 in the effect of combined genotypes and treatment. We, therefore, had adequate statistical power to test both predictions from our primary hypothesis.
Data Quality and Statistical Analysis
A database coordinator and statistician supervised data quality. Individual subject plots were checked for unusual values and for completeness. Because the primary efficacy variable, the weekly average of drinks per drinking day, is a direct quantification of the average amount of alcohol consumption done per drinking day within each week, it is a valid measure of the severity of alcohol consumption. Additionally, we have shown that the differential effects of the 5-HTT promoter and 3′-UTR regions are sensitive to alterations in drinks per drinking day as the measure of the severity of alcohol consumption (3, 17). The use of drinks per drinking day to measure the severity of alcohol consumption in this study has, therefore, both construct and experimental validity. We validated the calculation of drinks per drinking day as correct against the case records. Although not specific to our primary hypothesis, we chose a secondary outcome variable, percentage of days abstinent, to provide additional clinical support for our findings. Database checks for percentage of days abstinent were the same as those used for drinks per drinking day. Data were analyzed using SAS version 9.1 statistical software (40) according to the intent-to-treat principle; all subjects were included in the analysis.
Our analytic procedure was to use mixed-effects linear regression models, which can accommodate missing data at random, to study the effect of treatment and genotypes, as well as their interaction, on the efficacy measure of alcohol consumption. The models included random intercept and slope (for temporal trend) and were adjusted for covariates such as the participants’ average drinking levels prior to the study, age, gender, and ethnicity defined by ancestral markers (i.e., White and Hispanic). For ease of interpretation, we used two separate mixed-effects models to study the interaction between treatment and the two functional variants. In the first model, all 2-way interaction terms between genotypes (LL vs. LS/SS; TT vs. TG/GG) and treatment were included to evaluate the difference in mean values between the genotypes and treatment combinations of interest. In the second model, we compared the ondansetron LL/TT participants with those who had one of the three other genotypes (i.e., LL/G-carriers, S-carriers/TT, or S-carriers/G-carriers) and tested the hypothesis that ondansetron’s therapeutic effect could be greatest among those possessing the LL/TT genotype.
For our analyses, we made the assumption that data not present were missing at random for the outcomes of interest (drinks per drinking day and percentage of days abstinent). Under that assumption, mixed-effects models would yield consistent results, and are the preferred method to handle dropout compared with other approaches—e.g., last observation carried forward (41). Whilst missing at random is not testable directly (42), we investigated the relationship between dropout and longitudinal drinking outcomes by a joint random effects model (43) to simultaneously describe repeated measures of the weekly average of drinks per drinking day and percentage of days abstinent (by the mixed-effects model above) and time to dropout (by a Cox model). The random effects (random intercept and random slope) in the mixed-effects model were included in the Cox model to link the two models. The estimation was implemented in SAS Proc NLMIXED (44, 45). We found that after adjusting for risk factors, the time to dropout did not depend on the random effects from the longitudinal model of drinks per drinking day (p=0.89) and percentage of days abstinent (p=0.99), respectively, suggesting that dropout time is not correlated with heterogeneity in the weekly average of drinks per drinking day or percentage of days abstinent repeated measures; that is, dropout was not informative. The parameter estimates for the longitudinal outcomes from this joint model were similar to those in our present model that assumed that data were missing at random. This additional analysis provided evidence of the validity of the missing at random assumption.
In all statistical models tested here, we used a dominant genetic model based on the functionality of 5′-HTTLPR and rs1042173 alleles from previous studies as described in the Introduction. That is, we dichotomized the sample into LL vs. S-carriers and TT vs. G-carriers for the 5′-HTTLPR and the SNP rs1042173, respectively.
RESULTS
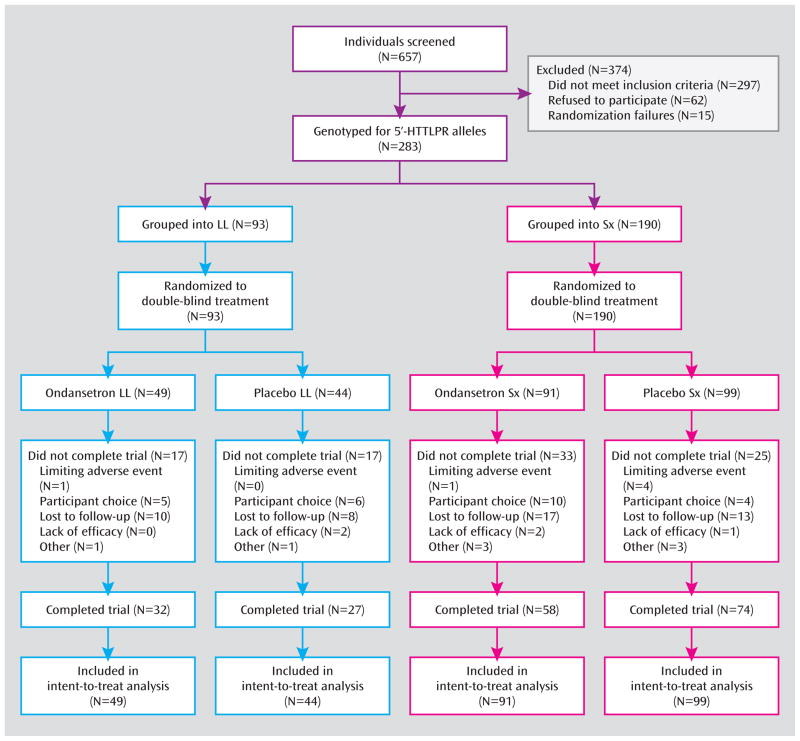
Subjects had an average age of 44.46 (SD 12.34) years, with 73.14% being male, 84.81% being White, and the remainder, 15.19%, being Hispanic. Their other demographic characteristics did not differ significantly by treatment and placebo for either of the 5′-HTTLPR genotype subgroups (Table 2). The study population satisfied Hardy-Weinberg requirements (see Table 1). Also, 66.4% of subjects completed all 12 weeks, with no significant group differences (Figure 1).
Table 2.
Baseline Demographics and Psychopathological Characteristics
| Outcomes | Ondansetron (n=140) | Placebo (n=143) | ||||||
|---|---|---|---|---|---|---|---|---|
| LL (n=49) | LS/SS (n=91) | TT (n=42) | TG/GG (n=95) | LL (n=44) | LS/SS (n=99) | TT (n=48) | TG/GG (n=92) | |
| Age, y | 43.76 (13.00) | 44.92 (13.02) | 44.22 (13.92) | 44.57 (11.82) | 45.63 (12.55) | 44.67 (12.25) | 43.39 (11.33) | 46.11 (12.74) |
| Sex, no. (%) | ||||||||
| Male | 35 (71.43) | 67 (71.28) | 31 (73.81) | 67 (71.28) | 31 (70.45) | 74 (74.75) | 40 (83.33) | 63 (68.48) |
| Female | 14 (28.57) | 27 (28.72) | 11 (26.19) | 27 (28.72) | 13 (29.55) | 25 (25.52) | 8 (16.67) | 29 (31.52) |
| Race/ethnicity, no. (%) | ||||||||
| White | 45 (91.84) | 77 (81.91) | 37 (88.10) | 81 (86.17) | 37 (84.10) | 81 (82.82) | 37 (77.08) | 78 (84.78) |
| Hispanic | 4 (8.16) | 14 (14.89) | 5 (11.90) | 13 (13.83) | 7 (15.9) | 18 (18.18) | 11 (22.92) | 14 (15.22) |
| Self-reported drinks/drinking day† | 9.64 (4.04) | 9.48 (4.81) | 10.70 (4.08) | 9.01 (4.65) | 8.86 (5.11) | 9.81 (4.53) | 10.22 (5.96) | 9.15 (3.84) |
| Self-reported percentage of days abstinent† | 46.27 (3.58) | 36.55 (3.17) | 42.36 (3.84) | 40.46 (2.91) | 35.00 (3.76) | 38.31 (2.89) | 41.56 (3.57) | 31.75 (3.10) |
| Breath alcohol concentration, % | 0.002 (0.005) | 0.002 (0.004) | 0.003 (0.006) | 0.001 (0.001) | 0.002 (0.004) | 0.002 (0.004) | 0.001 (0.003) | 0.002 (0.005) |
| CIWA-Ar score | 1.38 (1.72) | 1.69 (2.16) | 1.46 (1.96) | 1.85 (2.23) | 1.71 (2.03) | 1.78 (1.98) | 1.39 (1.67) | 2.47 (2.38) |
| Age of alcoholism onset, y | 30.78 (11.97) | 30.81 (13.89) | 38.79 (12.66) | 31.64 (13.64) | 32.0 (12.1) | 30.7 (12.7) | 31.23 (12.67) | 31.34 (12.56) |
| AUDIT score | 25.0 (5.65) | 23.58 (5.73) | 23.18 (5.54) | 25.74 (5.57) | 23.66 (6.19) | 23.18 (5.83) | 23.29 (6.02) | 23.21 (5.93) |
| Social class, no. (%)‡ | ||||||||
| 1–3 | 21 (48.84) | 33 (38.82) | 21 (56.76) | 32 (36.78) | 19 (47.50) | 43 (49.43) | 41 (50.62) | 41 (51.00) |
| 4–6 | 21 (48.84) | 45 (52.94) | 15 (40.54) | 48 (55.17) | 18 (45.00) | 39 (44.83) | 36 (44.44) | 36 (44.00) |
| 7–9 | 1 (2.33) | 7 (8.24) | 1 (2.70) | 7 (8.05) | 3 (7.50) | 5 (5.75) | 4 (4.94) | 4 (5.00) |
| Weight, kg | 83.4 (19.00) | 79.5 (17.00) | 81.81 (17.56) | 80.46 (18.32) | 82.5 (18.20) | 83.8 (16.40) | 81.14 (16.39) | 84.94 (16.90) |
Abbreviations: CIWA-Ar, revised Clinical Institute Withdrawal Assessment for Alcohol scale; AUDIT, Alcohol Use Disorders Identification Test.
Values are expressed as mean (SD) unless otherwise indicated. All values were collected at the screening visit.
Reflects mean values during the 90-day period preceding the screening visit.
Defined by Hollingshead and Redlich (55).
Figure 1.
Flow diagram of alcohol-dependent participants in a randomized controlled trial of ondansetron.
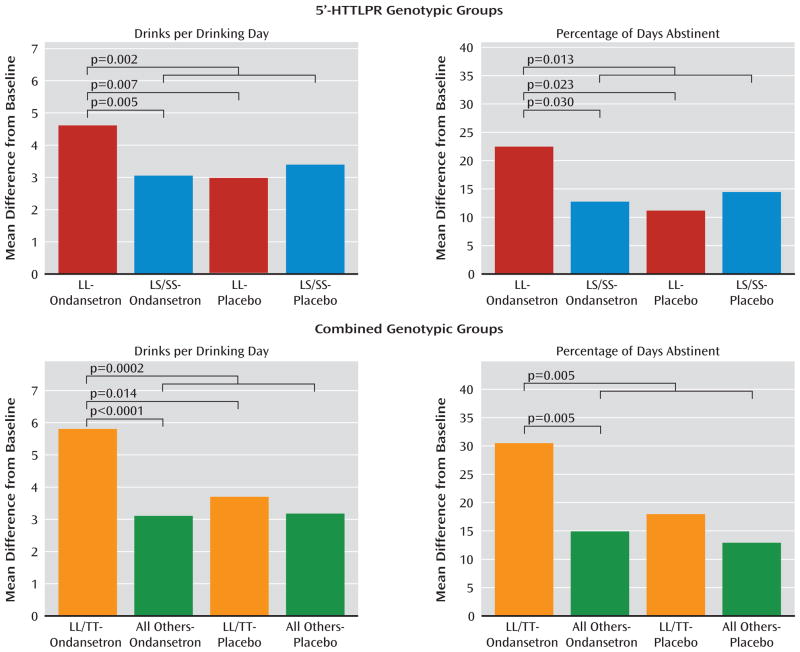
In our first model, for drinks per drinking day and percentage of days abstinent—two drinking measures with a statistically significant (p<0.05) correlation coefficient of −0.35—there was a significant interaction between 5′-HTTLPR genotype and treatment (F=6.68; p=0.010 and F=4.33; p=0.038). Ondansetron reduced drinks per drinking day and increased percentage of days abstinent in LL compared with LS/SS subjects (mean difference, −1.53; 95% CI −2.59 to −0.47; p=0.005; effect size=0.47, and 9.73; 95% CI 0.95 to 18.50; p=0.03; effect size=0.29; see Figure 2). The reduction in drinks per drinking day and increase in percentage of days abstinent by ondansetron compared with placebo was significant in subjects with the LL genotype (mean difference, −1.62; 95% CI −2.79 to −0.46; p=0.007; effect size=0.56, and 11.27; 95% CI 1.55 to 21.00; p=0.023; effect size=0.41; see Figure 2). Also, the ondansetron LL group had fewer drinks per drinking day and more percentage of days abstinent than all the other genotype and treatment groups combined (mean difference, −1.45; 95% CI −2.37 to −0.54; p=0.002; effect size=0.45, and 9.65; 95% CI 2.05 to 17.25; p=0.013; effect size=0.32; see Figure 2). For both drinks per drinking day and percentage of days abstinent, significant outcomes emerged in the third week of the study.
Figure 2.
Severity of alcohol drinking and abstinence rates among genotypic variants in the serotonin transporter gene before and during treatment with ondansetron or placebo.
Numbers of participants in the genotypic groups are listed in Table 2. 5′-HTTLPR = 5′-regulatory region of the serotonin transporter gene.
There was a significant interaction between the rs1042173 and 5′-HTTLPR genotypes for both drinks per drinking day and percentage of days abstinent (F=5.21; p=0.023 and F=6.85; p=0.009). Using the likelihood ratio test, the p-value for adding three additional terms associated with rs1042173 genotype in the model was 0.025 for drinks per drinking day and 0.019 for percentage of days abstinent. Hence, the rs1042173 genotype added to the effect of 5′-HTTLPR genotype on drinks per drinking day and percentage of days abstinent. Also, we found that there was a significant main effect of the combined genotypes and treatment for both drinks per drinking day and percentage of days abstinent (F=4.14; p=0.001 and F=2.61; p=0.023). The largest drinks per drinking day reduction and percentage of days abstinent increase was seen when the ondansetron recipients with the LL/TT genotype were compared with all the other participants who received placebo (mean difference, −2.67; 95% CI −4.01 to −1.33; p<0.0001; effect size=0.86, and 17.98; 95% CI 6.83 to 29.14; p=0.002; effect size=0.65). Notably, however, there was a significant reduction in drinks per drinking day and increase in percentage of days abstinent for the ondansetron LL/TT group compared with all others who received ondansetron (mean difference, −2.57; 95% CI −3.93 to −1.21; p=0.0002; effect size=0.92, and 15.50; 95% CI 4.14 to 26.86; p=0.008; effect size=0.62). The difference in drinks per drinking day between LL/TT ondansetron and LL/TT placebo recipients was also significant (mean difference, −2.06; 95% CI −3.72 to −0.39; p=0.015; effect size=0.72). Perhaps because of the relatively small sample size for a dichotomous variable, the difference in percentage of days abstinent between LL/TT ondansetron and LL/TT placebo recipients was marginal but not significant (mean difference, 12.52; 95% CI −1.43 to 26.47; p=0.079; effect size=0.51).
Notably, the effects of ondansetron to reduce drinks per drinking day and increase percentage of days abstinent were also greater in those with the LL/TT genotype compared with LL/G-carriers (mean difference, −2.34; 95% CI −3.98 to −0.71; p=0.005; effect size=0.82, and 15.25; 95% CI 1.51 to 28.98; p=0.03; effect size=0.62), the concomitant possession of the TT genotype appears to increase treatment response among those with the LL genotype.
The therapeutic effect sizes for LL and the LL/TT combination for ondansetron, compared with all the other genotype and treatment combinations, were 0.45 and 0.89, respectively, for drinks per drinking day, and were 0.32 and 0.63, respectively, for percentage of days abstinent. Effect sizes of 0.2, 0.5, and 0.8 are small, medium, and large, respectively (39).
There was no significant difference in the overall drug use rate between any of the treatment groups (p=0.28), and the overall rate was 58.0%. The three most commonly used drugs were nicotine, cannabis, and cocaine (53.0%, 17.7%, and 4.9%, respectively).
For all subjects, we observed a mean: pill-taking rate of 62.7%; breath alcohol concentration of less than the lowest detectable value of 0.01%, and alcohol withdrawal score on the revised Clinical Institute Withdrawal Assessment for Alcohol scale (31) of 0.86. There were no significant group differences on these parameters (data not shown).
No adverse event occurred with significantly greater frequency for ondansetron recipients than for placebo recipients, or vice versa (all p values > 0.05) except fatigue (p=0.019). The percentages of individuals in the ondansetron vs. placebo groups with at least one occurrence of the five most commonly reported adverse events were as follows: insomnia (20.5% vs. 22.3%), headache (20.9% vs. 19.4%), appetite disturbance (18.0% vs. 20.1%), fatigue (18.0% vs. 11.7%), and diarrhea (13.1% vs. 15.2%). No untoward medical consequence occurred that resulted in death, was life-threatening, required inpatient hospitalization, or resulted in persistent or significant disability or incapacity.
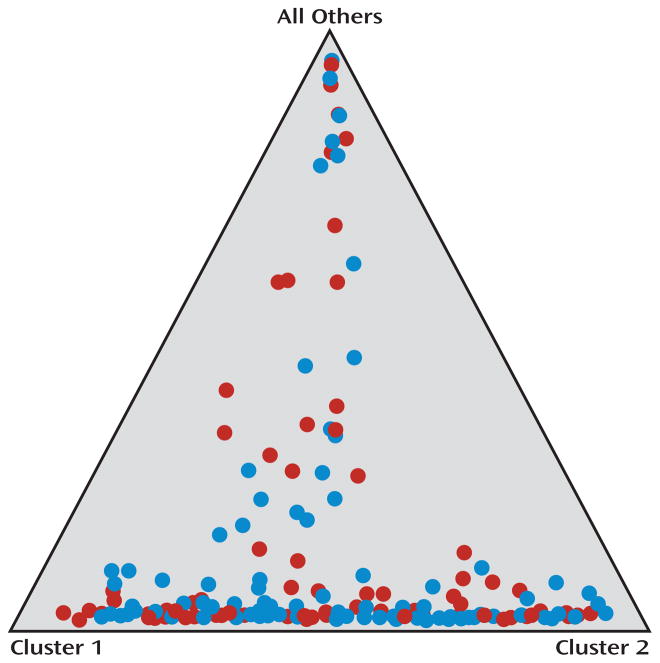
Population stratification for all subjects included in the intent-to-treat sample, using the 24 ancestry-informative SNP markers, showed that there was no significant difference in population structure between the ondansetron and placebo groups. The range of average proportions for the 2 clusters (i.e., genetic ancestry) was 0.15 to 0.43 in the ondansetron group and 0.18 to 0.40 in the placebo group, and the mean value of alpha was 0.091 (see Figure 3).
Figure 3.
Summary of clustering results for the studied population
Each individual is represented by a colored circle. Red circles represent those who received placebo, and blue circles represent those who received ondansetron. Data were plotted assuming three parental populations, and each circle shows the mean estimated ancestry for an individual in the sample. The differences between the placebo and ondansetron groups in mean proportions of ancestry for each of the three parental populations were not significant.
DISCUSSION
Our findings showed that ondansetron is a promising therapeutic agent for the treatment of severe alcohol consumption among alcohol-dependent individuals with the LL genotype of the 5′-HTTLPR. These findings are supported by a pilot human laboratory study where ondansetron suppressed alcohol self-administration in the LL compared with the LS/SS genotype significantly among non-treatment-seeking alcoholics (46).
From a qualitative clinical perspective, the relationship between drinking level and health consequences is not linear. For men, who constituted 73% of our subject population, high-risk drinking of alcohol, which occurs at or above 5 drinks/drinking day, is associated with severe health consequences, including accidental injuries (47), deaths from external sources (48), being a target of aggression or committing an aggressive act (49), and numerous medical, legal, and occupational problems (50). Therefore, using that clinical criterion, subjects with the LL or LL/TT genotype treated with placebo remained in the high-risk drinking category. In contrast, subjects with the LL or LL/TT genotype treated with ondansetron were, on average, no longer in the high-risk drinking category. Thus, subjects with the LL or LT/TT genotype who received ondansetron, compared with their counterparts who got placebo, had a qualitatively greater clinical improvement. Furthermore, the clinical importance of our findings was underscored by demonstrating that both LL and the combination of LL/TT genotypes also were significant predictors to increasing the percentage of days abstinent for those who received ondansetron compared with placebo.
Interestingly, the concomitant possession of the TT genotype of rs1042173 among those with the LL genotype of the 5′-HTTLPR enhanced the treatment response to ondansetron. Obviously, the magnitude of this increased effect cannot be determined directly within the same individual because the LL and TT genotypes are located within the same 5-HTT gene. It can, however, be inferred indirectly from the finding that the treatment effect of ondansetron among those with the combined LL and TT genotypes is significantly greater than that of an undifferentiated group of those with the LL genotype who would have different allelic frequencies of the TT, TG, or GG genotype, and the combined LL/TT group that received ondansetron had the best outcome. Nevertheless, despite our finding of a statistical interaction between the LL and TT genotypes, the exact molecular mechanism for this added therapeutic effect remains to be determined.
The operative issue in the clinical practice of pharmacogenetics is to identify and treat those who will respond the best to a particular medication. Hence, our findings promulgate the clinical approach of identifying alcohol-dependent individuals who are likely to respond to ondansetron based on their genotype analysis at the 5-HTT gene (i.e., LLs with or without the TT genotype) and then providing them with the medication.
Importantly, our primary outcome variable tested a specific scientific hypothesis related to ondansetron’s effects on the severity of alcohol consumption in alcohol-dependent individuals who varied by 5-HTT genotype. This specific pharmacogenetic hypothesis was proposed prior to the conduct of the present study (4), and developed through systematic experimental studies (3, 17). Testing a specific hypothesis is a conservative scientific approach because it avoids the presumption that a particular biological or genetic effect can be extrapolated to explain different patterns of drinking behavior. Consistent with this approach, we restricted the test of the effect of ondansetron response to just one other secondary variable—percentage of days abstinent—to provide additional information to clinicians. Our findings underscore the clinical importance of our results because they demonstrate that both LL and the combination of LL/TT genotypes predicted therapeutic response to ondansetron by increasing the percentage of days abstinent relative to placebo.
Notably, even though we enrolled into this clinical trial currently drinking alcohol-dependent individuals who had to have a breath alcohol concentration of no greater than 0.02% for clinical attendance, the breath alcohol concentration level reported in the trial was extremely low, and no significant withdrawal symptoms were reported. This phenomenon shows that alcohol-dependent individuals are able to moderate or taper their drinking level, perhaps as a result of the behavioral contingency of monitoring their drinking level (51). Also, this finding demonstrates the feasibility of providing treatment to alcohol-dependent individuals at the typical point of maximum crisis—when they are actively drinking alcohol and asking for help—and suggests that prior detoxification is not always a necessary prerequisite for the treatment of alcohol-dependent individuals who can be managed within an outpatient setting.
Our study was limited by five factors. First, we did not have equal numbers of individuals with different ethnicity. Hence, we could not test formally for a specific effect of ethnicity on genetic profile; however, the general trend and magnitude of the differences reported were similar for Whites and Hispanics (data not shown). Additionally, we performed an analysis for genetic structure with 24 unlinked ancestry-informative markers, which has been reported to be as effective in estimating continental population ancestry as are analyses with more than 90 ancestry-informative markers (38), and these ancestry proportion estimates were used as covariates in all our statistical models in place of self-defined ethnicity. The ancestry proportions, estimated by the program STRUCTURE in each of the three parental population groups, were symmetric between samples in the ondansetron and placebo groups (data not shown). Furthermore, we did not find in our statistical analyses that ancestry proportions were a significant covariate. Second, not all alcohol-dependent individuals were treated successfully with ondansetron—just those with a specific allelic constitution of the 5-HTT gene. Hence, more research is needed to find alcoholics with other genetic polymorphisms who will respond significantly to alternative medications. Third, there might be other genetic variants within the 5-HTT gene that could increase further the specificity of ondansetron’s treatment response; such relationships can only be elucidated through larger-scale pharmacogenomic studies. Fourth, the molecular mechanisms associated with the differential effect in the severity of alcohol consumption on L and S variants of the 5′-HTTLPR and T and G alleles of the 3′-UTR are currently not well understood; thus, additional in vitro and in vivo studies are needed. Fifth, whilst we considered the possibility that comorbid depression or anxiety-related disorders that have shown an association with serotonergic genotypes could confound our findings, we think this unlikely since this cohort excluded such subjects.
We minimized Type I error by performing inferential analysis only to test pre-specified pharmacogenetic hypotheses on a single primary endpoint, and by doing only pairwise comparisons within different genotype groups when there was a significant interaction effect between treatment and genotype.
Predicated on our earlier report that early-onset alcoholics were more likely than late-onset alcoholics to respond therapeutically to ondansetron treatment (28), we considered the possibility that age of onset could be a proxy for serotonin transporter genotype, or vice versa. Early-onset alcoholics differ from late-onset alcoholics by having an earlier age of alcohol-related problems, typically before the age of 25 years (although this can vary by definition), and increased rates of impulse-dyscontrol-related disorders (52). Notably, in the present cohort, there was no significant association between serotonin transporter genotype and age of onset (data not shown), and those with the LL or LL/TT genotype responded to ondansetron irrespective of age of onset. Despite the lack of a significant association between age of onset and genotype, and contrary to expectation (53), the slightly stronger trend was for those who had the LL genotype and were late-onset alcoholics to respond better to ondansetron than the LL early-onset alcoholics (data not shown). This also was unexpected because the preponderance of the literature suggests that it is the S allele that is more likely to be associated with an early onset of alcoholism (54). Importantly, as exemplified by the contradictory reports in the literature, age of onset is difficult to standardize, especially across different populations and ethnicities, and there have been over 150 years of debate as to the constellation and stability of alcohol-related problems that should be used to make this characterization (52). In contrast, the measurement of genetic variation is extremely accurate, and can be used reliably over time and across populations. A pharmacogenetic approach might, therefore, be a practical and useful method in general practice to identify those who should or should not receive ondansetron.
In sum, the results of our study have enabled us to integrate a unique scientific hypothesis, and a systematic series of basic research and human laboratory studies, with new clinical knowledge to demonstrate the promise of a novel pharmacogenetic approach to reduce the severity of alcohol consumption and increase abstinence among alcoholics with specific polymorphisms of the 5-HTT gene.
Supplementary Material
Acknowledgments
We are grateful to the National Institute on Alcohol Abuse and Alcoholism for its generous support through grants 7 U10 AA011776-10, 7 R01 AA010522-12, 2 R01 AA010522-13, 5 R01 AA013964-05, 5 R01 AA014628-04, and 5 R01 AA012964-06 awarded to B. A. Johnson, grant 5 K23 AA000329-06 awarded to N. Ait-Daoud, and grant RC1 AA019274 awarded to L. Liu, and to the National Institute on Drug Abuse for its support of M. D. Li through grants DA012844 and DA013783. We also thank Robert H. Cormier, Jr., BA, and Ann Richards, BA, for their assistance with manuscript preparation.
Footnotes
FINANCIAL DISCLOSURES
B. A. Johnson has served as a consultant to Johnson & Johnson (Ortho-McNeil Janssen Scientific Affairs, LLC), Transcept Pharmaceuticals, Inc., D&A Pharma, Organon, ADial Corporation, Psychological Education Publishing Company (PEPCo LLC), and Eli Lilly and Company. J. D. Roache has a contract with Eli Lilly and Company. M. D. Li serves as a scientific advisor to ADial Corporation. N. Ait-Daoud, C. Seneviratne, M. A. Javors, X.-Q. Wang, L. Liu, J. K. Penberthy, and C. C. DiClemente report no competing interests. The University of Virginia has applied for patents regarding the use of medications to treat alcoholism.
Trial Registration: clinicaltrials.gov Identifier: NCT00382642
References
- 1.Cargiulo T. Understanding the health impact of alcohol dependence. Am J Health Syst Pharm. 2007;64(5 Suppl 3):S5–S11. doi: 10.2146/ajhp060647. [DOI] [PubMed] [Google Scholar]
- 2.Szumlinski KK, Diab ME, Friedman R, Henze LM, Lominac KD, Bowers MS. Accumbens neurochemical adaptations produced by binge-like alcohol consumption. Psychopharmacology. 2007;190:415–431. doi: 10.1007/s00213-006-0641-7. [DOI] [PubMed] [Google Scholar]
- 3.Johnson BA, Javors MA, Roache JD, Seneviratne C, Bergeson SE, Ait-Daoud N, Dawes MA, Ma JZ. Can serotonin transporter genotype predict serotonergic function, chronicity, and severity of drinking? Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:209–216. doi: 10.1016/j.pnpbp.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson BA. Serotonergic agents and alcoholism treatment: rebirth of the subtype concept—an hypothesis. Alcohol Clin Exp Res. 2000;24:1597–1601. [PubMed] [Google Scholar]
- 5.Ramamoorthy S, Bauman AL, Moore KR, Han H, Yang-Feng T, Chang AS, Ganapathy V, Blakely RD. Antidepressant- and cocaine-sensitive human serotonin transporter: molecular cloning, expression, and chromosomal localization. Proc Natl Acad Sci U S A. 1993;90:2542–2546. doi: 10.1073/pnas.90.6.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esterling LE, Yoshikawa T, Turner G, Badner JA, Bengel D, Gershon ES, Berrettini WH, Detera-Wadleigh SD. Serotonin transporter (5-HTT) gene and bipolar affective disorder. Am J Med Genet. 1998;81:37–40. [PubMed] [Google Scholar]
- 7.D'Souza UM, Craig IW. Functional polymorphisms in dopamine and serotonin pathway genes. Hum Mutat. 2006;27:1–13. doi: 10.1002/humu.20278. [DOI] [PubMed] [Google Scholar]
- 8.Williams LM, Gatt JM, Schofield PR, Olivieri G, Peduto A, Gordon E. 'Negativity bias' in risk for depression and anxiety: brain-body fear circuitry correlates, 5-HTT-LPR and early life stress. Neuroimage. 2009;47:804–814. doi: 10.1016/j.neuroimage.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 10.Bloch MH, Landeros-Weisenberger A, Sen S, Dombrowski P, Kelmendi B, Coric V, Pittenger C, Leckman JF. Association of the serotonin transporter polymorphism and obsessive-compulsive disorder: systematic review. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:850–858. doi: 10.1002/ajmg.b.30699. [DOI] [PubMed] [Google Scholar]
- 11.Huezo-Diaz P, Uher R, Smith R, Rietschel M, Henigsberg N, Marusic A, Mors O, Maier W, Hauser J, Souery D, Placentino A, Zobel A, Larsen ER, Czerski PM, Gupta B, Hoda F, Perroud N, Farmer A, Craig I, Aitchison KJ, McGuffin P. Moderation of antidepressant response by the serotonin transporter gene. Br J Psychiatry. 2009;195:30–38. doi: 10.1192/bjp.bp.108.062521. [DOI] [PubMed] [Google Scholar]
- 12.Li MD, Burmeister M. New insights into the genetics of addiction. Nat Rev Genet. 2009;10:225–231. doi: 10.1038/nrg2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenberg BD, Tolliver TJ, Huang SJ, Li Q, Bengel D, Murphy DL. Genetic variation in the serotonin transporter promoter region affects serotonin uptake in human blood platelets. Am J Med Genet. 1999;88:83–87. [PubMed] [Google Scholar]
- 14.Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Muller CR, Hamer DH, Murphy DL. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 15.Heinz A, Jones DW, Mazzanti C, Goldman D, Ragan P, Hommer D, Linnoila M, Weinberger DR. A relationship between serotonin transporter genotype and in vivo protein expression and alcohol neurotoxicity. Biol Psychiatry. 2000;47:643–649. doi: 10.1016/s0006-3223(99)00171-7. [DOI] [PubMed] [Google Scholar]
- 16.Javors MA, Seneviratne C, Roache JD, Ait-Daoud N, Bergeson SE, Walss-Bass MC, Akhtar FZ, Johnson BA. Platelet serotonin uptake and paroxetine binding among allelic genotypes of the serotonin transporter in alcoholics. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29:7–13. doi: 10.1016/j.pnpbp.2004.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seneviratne C, Huang W, Ait-Daoud N, Li MD, Johnson BA. Characterization of a functional polymorphism in the 3' UTR of SLC6A4 and its association with drinking intensity. Alcohol Clin Exp Res. 2009;33:332–339. doi: 10.1111/j.1530-0277.2008.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 19.Battersby S, Ogilvie AD, Blackwood DH, Shen S, Muqit MM, Muir WJ, Teague P, Goodwin GM, Harmar AJ. Presence of multiple functional polyadenylation signals and a single nucleotide polymorphism in the 3' untranslated region of the human serotonin transporter gene. J Neurochem. 1999;72:1384–1388. doi: 10.1046/j.1471-4159.1999.721384.x. [DOI] [PubMed] [Google Scholar]
- 20.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Little KY, McLaughlin DP, Zhang L, Livermore CS, Dalack GW, McFinton PR, DelProposto ZS, Hill E, Cassin BJ, Watson SJ, Cook EH. Cocaine, ethanol, and genotype effects on human midbrain serotonin transporter binding sites and mRNA levels. Am J Psychiatry. 1998;155:207–213. doi: 10.1176/ajp.155.2.207. [DOI] [PubMed] [Google Scholar]
- 22.Ait-Daoud N, Roache JD, Dawes MA, Liu L, Wang X-Q, Javors MA, Seneviratne C, Johnson BA. Can serotonin transporter genotype predict craving in alcoholism? Alcohol Clin Exp Res. 2009;33:1329–1335. doi: 10.1111/j.1530-0277.2009.00962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umathe SN, Bhutada PS, Raut VS, Jain NS, Mundhada YR. The 5-HT3 receptor antagonist, ondansetron, blocks the development and expression of ethanol-induced locomotor sensitization in mice. Behav Pharmacol. 2009;20:78–83. doi: 10.1097/FBP.0b013e3283242ff4. [DOI] [PubMed] [Google Scholar]
- 24.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4. Washington, D.C: American Psychiatric Association; 1994. [Google Scholar]
- 25.Bohn MJ, Babor TF, Kranzler HR. The Alcohol Use Disorders Identification Test (AUDIT): validation of a screening instrument for use in medical settings. J Stud Alcohol. 1995;56:423–432. doi: 10.15288/jsa.1995.56.423. [DOI] [PubMed] [Google Scholar]
- 26.Stout RL, Wirtz PW, Carbonari JP, Del Boca FK. Ensuring balanced distribution of prognostic factors in treatment outcome research. J Stud Alcohol Suppl. 1994;12:70–75. doi: 10.15288/jsas.1994.s12.70. [DOI] [PubMed] [Google Scholar]
- 27.Sobell LC, Sobell MB. Timeline follow-back: a technique for assessing self-reported alcohol consumption. In: Litten RZ, Allen JP, editors. Measuring Alcohol Consumption: Psychosocial and Biochemical Methods. Totowa, NJ: Humana Press Inc; 1992. pp. 41–72. [Google Scholar]
- 28.Johnson BA, Roache JD, Javors MA, DiClemente CC, Cloninger CR, Prihoda TJ, Bordnick PS, Ait-Daoud N, Hensler J. Ondansetron for reduction of drinking among biologically predisposed alcoholic patients: a randomized controlled trial. JAMA. 2000;284:963–971. doi: 10.1001/jama.284.8.963. [DOI] [PubMed] [Google Scholar]
- 29.Johnson BA, Rosenthal N, Capece JA, Wiegand F, Mao L, Beyers K, McKay A, Ait-Daoud N, Addolorato G, Anton RF, Ciraulo DA, Kranzler HR, Mann K, O'Malley SS, Swift RM Topiramate for Alcoholism Advisory Board, Topiramate for Alcoholism Study Group. Improvement of physical health and quality of life of alcohol-dependent individuals with topiramate treatment. Arch Intern Med. 2008;168:1188–1199. doi: 10.1001/archinte.168.11.1188. [DOI] [PubMed] [Google Scholar]
- 30.Levine J, Schooler NR. SAFTEE: a technique for the systematic assessment of side effects in clinical trials. Psychopharmacol Bull. 1986;22:343–381. [PubMed] [Google Scholar]
- 31.Sullivan JT, Sykora K, Schneiderman J, Naranjo CA, Sellers EM. Assessment of alcohol withdrawal: the revised clinical institute withdrawal assessment for alcohol scale (CIWA-Ar) Br J Addict. 1989;84:1353–1357. doi: 10.1111/j.1360-0443.1989.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 32.U.S. Department of Health and Human Services. NIH Pub. No. 92–1895. Washington, D.C: USDHHS; 1992. Cognitive-Behavioral Coping Skills Therapy Manual. [Google Scholar]
- 33.Monti PM, Abrams DB, Kadden R, Cooney N. Treating Alcohol Dependence: A Coping Skills Training Guide. New York: Guilford Press; 1989. [Google Scholar]
- 34.Marlatt GA, Gordon JR. Relapse Prevention: Maintenance Strategies in the Treatment of Addictive Behaviors. New York: Guilford Press; 1985. [Google Scholar]
- 35.Bandura A. Self-efficacy: toward a unifying theory of behavioral change. Psychol Rev. 1977;84:191–215. doi: 10.1037//0033-295x.84.2.191. [DOI] [PubMed] [Google Scholar]
- 36.Wendland JR, Martin BJ, Kruse MR, Lesch KP, Murphy DL. Simultaneous genotyping of four functional loci of human SLC6A4, with a reappraisal of 5-HTTLPR and rs25531. Mol Psychiatry. 2006;11:224–226. doi: 10.1038/sj.mp.4001789. [DOI] [PubMed] [Google Scholar]
- 37.Mao X, Bigham AW, Mei R, Gutierrez G, Weiss KM, Brutsaert TD, Leon-Velarde F, Moore LG, Vargas E, McKeigue PM, Shriver MD, Parra EJ. A genomewide admixture mapping panel for Hispanic/Latino populations. Am J Hum Genet. 2007;80:1171–1178. doi: 10.1086/518564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, Kittles R, Alarcon-Riguelme ME, Gregersen PK, Belmont JW, De La Vega FM, Seldin MF. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2. Hillsdale, NJ: L. Erlbaum Associates; 1988. [Google Scholar]
- 40.SAS Institute Inc. SAS version 9.1. Cary, NC: SAS Institute Inc; 2003. [Google Scholar]
- 41.Hamer RM, Simpson PM. Last observation carried forward versus mixed models in the analysis of psychiatric clinical trials. Am J Psychiatry. 2009;166:639–641. doi: 10.1176/appi.ajp.2009.09040458. [DOI] [PubMed] [Google Scholar]
- 42.Little R, Rubin DB. Statistical Analysis with Missing Data. 2. New York: Wiley; 2002. [Google Scholar]
- 43.Wulfsohn MS, Tsiatis AA. A joint model for survival and longitudinal data measured with error. Biometrics. 1997;53:330–339. [PubMed] [Google Scholar]
- 44.Vonesh EF, Greene T, Schluchter MD. Shared parameter models for the joint analysis of longitudinal data and event times. Stat Med. 2006;25:143–163. doi: 10.1002/sim.2249. [DOI] [PubMed] [Google Scholar]
- 45.Liu L. Joint modeling longitudinal semi-continuous data and survival, with application to longitudinal medical cost data. Stat Med. 2009;28:972–986. doi: 10.1002/sim.3497. [DOI] [PubMed] [Google Scholar]
- 46.Kenna GA, Zywiak WH, McGeary JE, Leggio L, McGeary C, Wang S, Grenga A, Swift RM. A within-group design of nontreatment seeking 5-HTTLPR genotyped alcohol-dependent subjects receiving ondansetron and sertraline. Alcohol Clin Exp Res. 2009;33:315–323. doi: 10.1111/j.1530-0277.2008.00835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cherpitel CJ, Bond J, Ye Y. Alcohol and injury: a risk function analysis from the Emergency Room Collaborative Alcohol Analysis Project (ERCAAP) Eur Addict Res. 2006;12:42–52. doi: 10.1159/000088582. [DOI] [PubMed] [Google Scholar]
- 48.Dawson DA. Alcohol and mortality from external causes. J Stud Alcohol. 2001;62:790–797. doi: 10.15288/jsa.2001.62.790. [DOI] [PubMed] [Google Scholar]
- 49.Dawson DA. Alcohol, drugs, fighting and suicide attempt/ideation. Addict Res. 1997;5:451–472. [Google Scholar]
- 50.Wechsler H, Nelson TF. Relationship between level of consumption and harms in assessing drink cut-points for alcohol research: commentary on “Many college freshmen drink at levels far beyond the binge threshold” by White et al. Alcohol Clin Exp Res. 2006;30:922–927. doi: 10.1111/j.1530-0277.2006.00124.x. [DOI] [PubMed] [Google Scholar]
- 51.Penberthy JK, Ait-Daoud N, Breton M, Kovatchev B, DiClemente CC, Johnson BA. Evaluating readiness and treatment seeking effects in a pharmacotherapy trial for alcohol dependence. Alcohol Clin Exp Res. 2007;31:1538–1544. doi: 10.1111/j.1530-0277.2007.00448.x. [DOI] [PubMed] [Google Scholar]
- 52.Johnson BA, Cloninger CR, Roache JD, Bordnick PS, Ruiz P. Age of onset as a discriminator between alcoholic subtypes in a treatment-seeking outpatient population. Am J Addict. 2000;9:17–27. doi: 10.1080/10550490050172191. [DOI] [PubMed] [Google Scholar]
- 53.Kenna GA. Pharmacotherapy of alcohol dependence: targeting a complex disorder. Drug Discov Today Ther Strateg. 2005;2:71–78. [Google Scholar]
- 54.Hallikainen T, Saito T, Lachman HM, Volavka J, Pohjalainen T, Ryynänen O-P, Kauhanen J, Syvälahti E, Hietala J, Tiihonen J. Association between low activity serotonin transporter promoter genotype and early onset alcoholism with habitual impulsive violent behavior. Mol Psychiatry. 1999;4:385–388. doi: 10.1038/sj.mp.4000526. [DOI] [PubMed] [Google Scholar]
- 55.Hollingshead AB, Redlich FC. Social Class and Mental Illness: A Community Study. New York: Wiley; 1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.