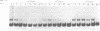Abstract
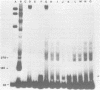
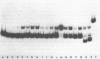
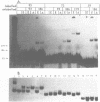
We have observed that ethanol precipitation and subsequent drying of small (less than 400 bp) radiolabelled DNA fragments is able to induce a transition to a form that migrates aberrantly on acrylamide gels. This unusual form has increased sensitivity to S1 nuclease, decreased sensitivity to restriction enzymes, and a concentration dependence for the reversion to the duplex form. Apparently, DNA denatures upon dehydration so that redissolving at low dilution will allow the collapse of DNA fragments into single-stranded hairpin structures. These structures are stable enough at low dilution to prevent complete reannealing of single stranded species. These single stranded species show strong binding to unidentified proteins present in nuclear extracts. This may give rise to misleading interpretations of mobility shift assays, especially if the single-stranded conformers have a similar mobility to the duplex fragment, which can occur in fragments that are 50-100 bp long. Evidence is presented that DNA, in general, denatures upon dehydration, but that hindrances to rotation in the solid state may prevent long fragments from dissociating.
Full text
PDF


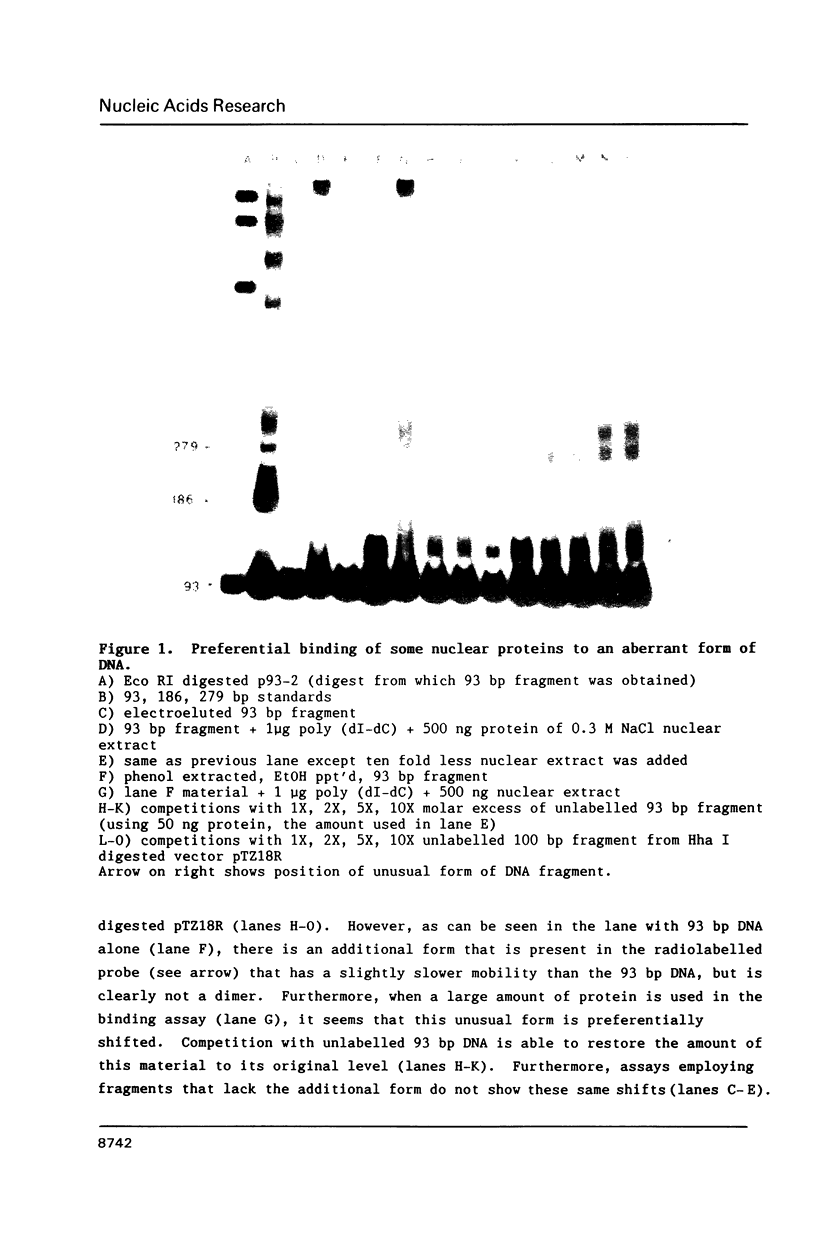





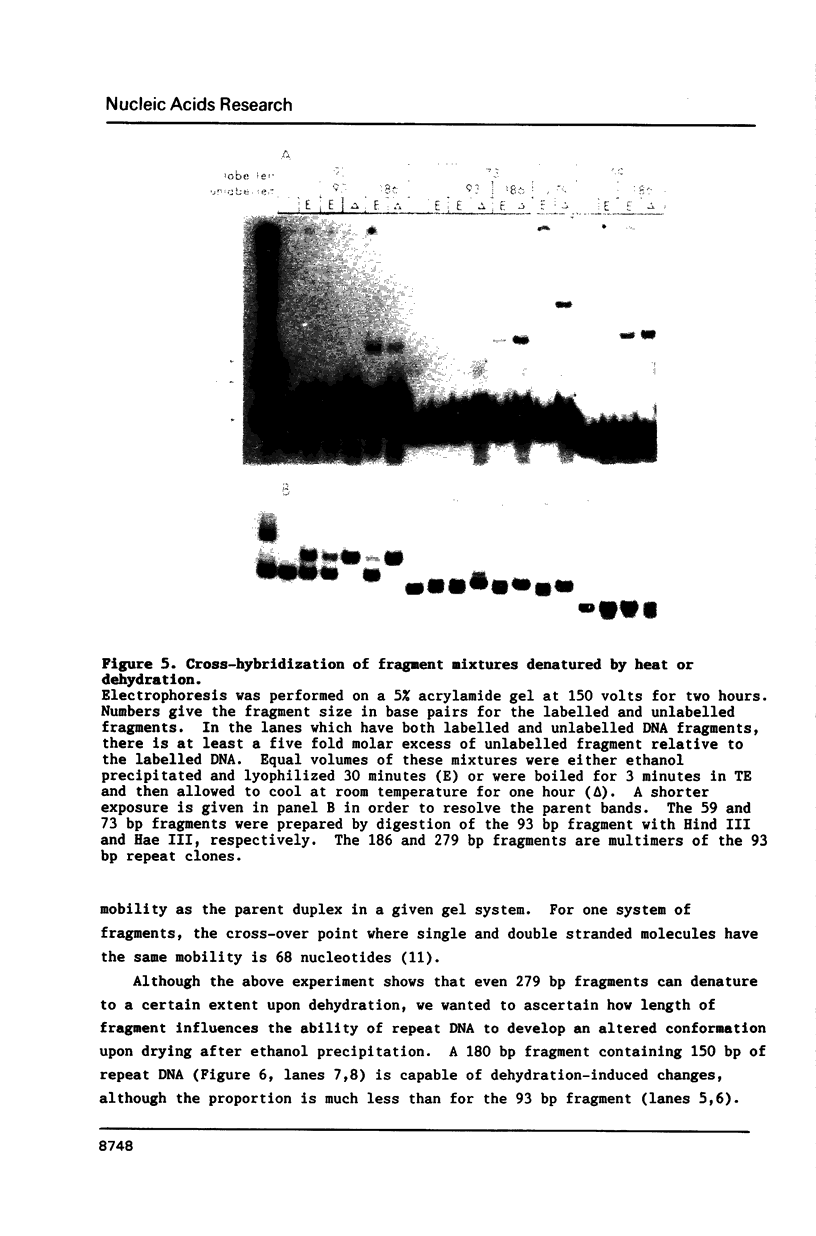






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BRAHMS J., MOMMAERTS W. F. A STUDY OF CONFORMATION OF NUCLEIC ACIDS IN SOLUTION BY MEANS OF CIRCULAR DICHROISM. J Mol Biol. 1964 Oct;10:73–88. doi: 10.1016/s0022-2836(64)80029-2. [DOI] [PubMed] [Google Scholar]
- Blakesley R. W., Dodgson J. B., Nes I. F., Wells R. D. Duplex regions in "single-stranded" phiX174 DNA are cleaved by a restriction endonuclease from Haemophilus aegyptius. J Biol Chem. 1977 Oct 25;252(20):7300–7306. [PubMed] [Google Scholar]
- Courey A. J., Wang J. C. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983 Jul;33(3):817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Hofer B., Ruhe G., Koch A., Köster H. Primary and secondary structure specificity of the cleavage of 'single-stranded' DNA by endonuclease Hinf I. Nucleic Acids Res. 1982 May 11;10(9):2763–2773. doi: 10.1093/nar/10.9.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEVINE L., GORDON J. A., JENCKS W. P. The relationship of structure to the effectiveness of denaturing agents for deoxyribonucleic acid. Biochemistry. 1963 Jan-Feb;2:168–175. doi: 10.1021/bi00901a030. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Mizusawa H., Kakefuda T. Unwinding of double-stranded DNA helix by dehydration. Proc Natl Acad Sci U S A. 1981 May;78(5):2838–2842. doi: 10.1073/pnas.78.5.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pandolfo M., Valentini O., Biamonti G., Morandi C., Riva S. Single stranded DNA binding proteins derive from hnRNP proteins by proteolysis in mammalian cells. Nucleic Acids Res. 1985 Sep 25;13(18):6577–6590. doi: 10.1093/nar/13.18.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M., Igo-Kemenes T., Zachau H. G. Nucleotide sequence of a highly repetitive component of rat DNA. Nucleic Acids Res. 1979 Sep 25;7(2):417–432. doi: 10.1093/nar/7.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sealey L., Chalkley R. At least two nuclear proteins bind specifically to the Rous sarcoma virus long terminal repeat enhancer. Mol Cell Biol. 1987 Feb;7(2):787–798. doi: 10.1128/mcb.7.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sealy L., Hartley J., Donelson J., Chalkley R., Hutchison N., Hamkalo B. Characterization of a highly repetitive sequence DNA family in rat. J Mol Biol. 1981 Jan 15;145(2):291–318. doi: 10.1016/0022-2836(81)90207-2. [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Broyles S. S., Pettijohn D. E. Perfect palindromic lac operator DNA sequence exists as a stable cruciform structure in supercoiled DNA in vitro but not in vivo. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1797–1801. doi: 10.1073/pnas.80.7.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. R., Reddigari S., Patel G. L. Identification of a nucleic acid helix-destabilizing protein from rat liver as lactate dehydrogenase-5. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5260–5264. doi: 10.1073/pnas.82.16.5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]