Abstract
Cryptococcus neoformans STE12α, a homologue of Saccharomyces cerevisiae STE12, exists only in MATα strains. We identified another STE12 homologue, STE12a, which is MATa specific. As in the case with Δste12α, the mating efficiency for Δste12a was reduced significantly. The Δste12a strains surprisingly still mated with Δste12α strains. In MATα strains, STE12a functionally complemented STE12α for mating efficacy, haploid fruiting, and regulation of capsule size in the mouse brain. Furthermore, when STE12a was replaced with two copies of STE12α, the resulting MATa strain produced hyphae on filament agar. STE12a regulates mRNA levels of several genes that are important for virulence including CNLAC1 and CAP genes. STE12a also modulates enzyme activities of phospholipase and superoxide dismutase. Importantly, deletion of STE12a markedly reduced the virulence in mice, as is the case with STE12α. Brain smears of mice infected with the Δste12a strain showed yeast cells with a considerable reduction in capsule size compared with those infected with STE12a strains. When the disrupted locus of ste12a was replaced with a wild-type STE12a gene, both in vivo and in vitro mutant phenotypes were reversed. These results suggest that STE12a and STE12α have similar functions, and that the mating type of the cells influences the alleles to exert their biological effects. C. neoformans, thus, is the first fungal species that contains a mating-type-specific STE12 homologue in each mating type. Our results demonstrate that mating-type-specific genes are not only important for saprobic reproduction but also play an important role for survival of the organism in host tissue.
Cryptococcus neoformans is a pathogenic fungus that primarily infects patients with impaired immune systems, but people with no known underlying immunodeficiencies are affected also (1). C. neoformans is a bipolar heterothallic species in which mating is controlled by two alleles: MATα and MATa (2, 3). In the laboratory, the sexual reproduction cycle begins when two strains of the opposite mating type are crossed under nutrient-deprivation conditions. This cycle is characterized by the formation of dikaryotic hyphae with typical basidiomycetous clamp connections and by the production of basidia. Basidiospores germinate to produce yeast cells that multiply by polar budding without hyphal formation. Previous studies indicate that MATα strains are found far more frequently than MATa strains among clinical as well as environmental isolates of serotype D (4). For serotype A strains, MATα is thus far the only mating type recovered. In a mouse systemic-infection model, MATα strains of serotype D are significantly more virulent than MATa strains, which suggests an important role of the MATα locus for fungal pathogenicity (5). In the absence of MATa strains, MATα strains also undergo haploid fruiting in response to nitrogen starvation, which results in hyphal production and sporulation (6).
The signal-transduction pathways regulating morphogenesis and pathogenicity of C. neoformans have been studied, and homologues of several Saccharomyces cerevisiae mitogen-activated protein (MAP) kinase cascade genes involved in the pheromone-response pathway have been identified (7–9). STE12α, which encodes a protein similar to the ascomycetous Ste12p, was isolated during molecular analysis of hyphal production in a MATα strain of C. neoformans (7). The transcriptional factor Ste12p is one of the well conserved elements of the pheromone-response pathway studied in several fungi. In S. cerevisiae, STE12 is an integral part of the conserved MAP kinase signal-transduction pathway involved in mating, pseudohyphal development, and haploid invasive growth (for reviews see refs. 10–13). Unlike the STE12 genes in other fungi, STE12α of C. neoformans exists only in MATα strains (7). Although STE12α influences mating efficiency in C. neoformans, the Δste12α strains of C. neoformans are still fertile (14, 15). Deletion of STE12α from a wild-type strain of C. neoformans had no effect on yeast cell growth but abolished its ability to undergo haploid fruiting on filament agar. In serotype D strains, STE12α was found to modulate the expression of several genes associated with virulence. As a consequence, deletion of STE12α significantly reduced virulence in a serotype D strain (14). However, disruption of STE12α in a serotype A strain showed no effect on virulence (14, 15).
Because STE12α is dispensable for mating, it is possible that C. neoformans contains another STE12 homologue that could substitute for the function of Ste12αp during the mating process. To investigate this possibility, we attempted to isolate a putative STE12 homologue from the cDNA of a culture during mating. Here, we report the identification of another STE12 homologue, STE12a, which only exists in the MATa strain of C. neoformans. After deleting the STE12a gene from a MATa strain, we examined the phenotypes and the virulence of the ste12a deletants. Deletion of STE12a affected the expression of several virulence-associated genes and showed reduced virulence. When the disrupted ste12a locus was reconstituted with a wild-type STE12a, the resulting strain expressed phenotypes and virulence similar to wild-type levels. These results provide molecular evidence for the important role of the mating type a-specific gene, STE12a, in the virulence of C. neoformans MATa strains.
Materials and Methods
Strains, Media, and General Methods.
All strains used in this study were serotype D and are listed in Table 1. YEPD, minimal media, and filament agar have been described (14). The method used for the quantitative assay of mating frequency has been described (14). In brief, two auxotrophic MATα strains (JEC31 and JEC33) were used as tester strains to determine the mating frequency of any given MATa strain carrying different auxotrophic markers. The relative mating frequency was expressed as a percentage of the mating frequency of the reference strain (LP2). The data represent the average of results derived from matings with JEC31 and JEC33. The experiments were repeated at least twice to confirm reproducibility. All crosses were performed on V-8 juice agar (16). Phospholipase activity was determined on egg-yolk agar as described (17). Cultures were incubated at 30°C for 72 h, and the diameter of colony plus the zone of precipitate around the colony was measured. The index of phospholipase activity was determined by dividing the colony diameter by the total diameter of the colony plus hue. There is an inverse relationship between the index and enzyme activity; the smaller the index, the higher the enzyme activity of the strain. To determine the superoxide dismutase (SOD) activity, the yeast cells were grown in Sabouraud broth for 20 h, harvested, and resuspended in 50 mM Tris (pH 7.5)/10 mM MgSO4/100 mM KCl/1 mM EDTA. The resuspended cells were disrupted with glass beads in a minibead beater (Biospec Products, Bartlesville, OK). The suspension was centrifuged at 13,000 × g for 10 min at 4°C, and the crude cell extract was obtained. SOD activity in the protein extract was detected as described (18). A calibration curve was obtained by using bovine erythrocyte SOD (Sigma 2515) as a standard, and the sample SOD activity was determined from the regression line for standard. The method for virulence studies and the preparations of brain smears have been described (14).
Table 1.
List of strains relevant to this study
| Strain | Genotype/Comment | Source |
|---|---|---|
| B4500 | MATα; congenic strain of B-4476 | Ref. 5 |
| B4476 | MATa; congenic strain of B-4500 | Ref. 5 |
| LP1 | MATα ura5 ade2 | Ref. 24 |
| LP2 | MATa ura5 ade2 | Ref. 24 |
| LP8 | MATa ura5 ade2 | Ref. 24 |
| JE31 | MATα lys1 | Gift of J. C. Edmen |
| JE33 | MATα lys2 | Gift of J. C. Edmen |
| TYCC245 | MATα ura5 ade2 Δste12α∷ADE2 | Ref. 14 |
| TYCC245F1 | MATα Δste12α∷ADE2 | Ref. 14 |
| TYCC409AF1 | MATα Δste12α∷STE12α | Ref. 14 |
| TYCC384 | MATa ura5 Δste12a∷ADE2 | This study |
| C429 | MATa Δste12a∷ADE2; F2 of TYCC384 | This study |
| C437 | MATa ura5 ade2 Δste12a∷STE12a | This study |
| C446 | MATα ura5 ade2 Δste12α∷STE12a | This study |
| C448 | MATa ura5 ade2 GAL7(p)∷STE12a∷ADE2 | This study |
| C449 | MATa Δste12a∷STE12a; F2 of C437 | This study |
| C460 | MATα ura5 ade2 GAL7(p)∷STE12a∷ADE2 | This study |
| C487 | MATa ura5 ade2 Δste12a∷2×STE12α | This study |
| C488 | MATa ura5 ade2 Δste12a∷STE12α | This study |
| C489 | MATα Δste12α∷STE12a; F1 of C446 | This study |
| C490 | MATa Δste12a∷2×STE12α; F1 of C487 | This study |
Identification of the STE12a Gene.
Degenerate primers were synthesized based on the conserved homeodomain regions among the STE12 gene of S. cerevisiae, the steA gene of Aspergillus nidulans, and the STE12α gene of C. neoformans (AARAARTTYGARGARGGIRTITT and CCARWARAAIACYTTYGYTTYTT; R = A or G; Y = C or T; W = A or T; I = inosine). Reverse transcription–PCR was performed by using RNAs isolated from 6-h mating cultures of MATa and MATα strains by 30 cycles of 94°C for 30 sec, 48°C for 30 sec, and 72°C for 1 min. The ≈150-bp PCR product obtained was cloned and sequenced. One of the 46 PCR clones contained a sequence clearly different from the STE12α sequence, whereas the rest showed sequences identical to the STE12α. To isolate the longer cDNA clone, pYCC360, the rapid amplification of cDNA ends was performed in accordance with the protocol of the Marathon cDNA amplification kit (CLONTECH). The genomic clone containing the entire STE12a, pYCC367 (Fig. 2B), was obtained by screening the genomic library of B-3502 (a gift from J. C. Edman) with pYCC360 as a probe.
Figure 2.
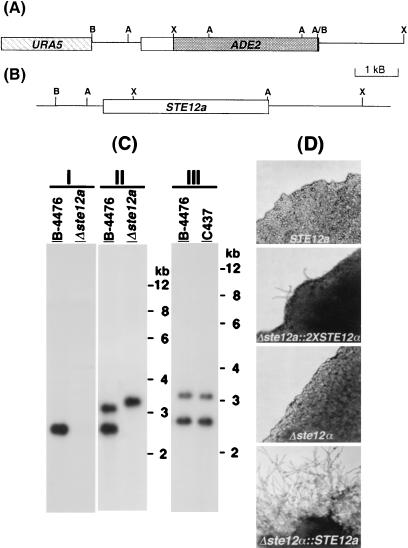
Deletion and reconstitution of STE12a. (A) Map of the deletion plasmid construct, pYCC384. (B) Map of STE12a reconstitution construct, pYCC367. A, AseI; B, BamHI; X, XbaI. (C) Southern blot analysis of the Δste12a strain (I and II) and STE12a reconstituted strain (III). DNA was digested with AseI, fractionated on an agarose gel, and transferred to a nylon membrane. The resulting blot was hybridized with 2.1-kb XbaI–AseI probe of STE12a (I) or 3.6-kb XbaI–XbaI probe of STE12a (II and III). Wild type, B-4476; Δste12a, TYCC384; Δste12∷STE12a, C437. (D) Hyphae formation on filament agar. Strains B-4476, C490, TYCC245F1, and C489 were inoculated on filament agar. Photographs were taken after 3 days of incubation at room temperature.
Deletion and Reconstruction of STE12a.
For the ste12a deletion construct, the 1.3-kb BamHI–XbaI fragment of STE12a was subcloned into the BamHI–XbaI site of pCIP3 to yield pYCC382, and the 1.5-kb AseI–XbaI fragment of STE12a was cloned into the BamHI–XbaI site of pYCC123 to yield pYCC383. The 3.3 kb XbaI–ApaI fragment of pYCC382 was cloned into the EcoRI–ApaI site of pYCC383 to yield the final deletion construct, pYCC384 (Fig. 2A). The electroporation-based positive-negative selection method (19) was used to delete the STE12a gene, and a PCR protocol was used to screen the putative deletants as described (14). The deleted ste12a locus was reconstituted back to the wild type by using plasmids pYCC367 and pYCC331 (20) by a biolistic-based cotransformation method (14). All putative reconstituted adenine auxotrophs were transferred on YEPD three consecutive times to cure the cotransformed telomere-based plasmid, pYCC331. Uracil and adenine auxotrophs were isolated, and their DNAs were analyzed by Southern blot. To replace the deleted ste12α with STE12a, plasmid pYCC411 was constructed. Briefly, the coding region of STE12α in p18-S1 was replaced with the coding region of STE12a from pYCC367 to yield pYCC411. Both pYCC411 and pYCC331 were cotransformed into the ste12α deletant, TYCC245. C446 is the resulting strain in which the deleted ste12α locus was replaced with STE12a and devoid of the cotransformed plasmid pYCC331 (Table 1). Similarly, plasmid pYCC511 was constructed to replace the deleted ste12a with STE12α. Briefly, the region containing the STE12a in pYCC367 was replaced with the region containing the entire STE12α from p18-S1 to yield pYCC511. Both pYCC511 and pYCC331 were cotransformed into the ste12a deletant, TYCC384. C487 and C488 were the resulting strains devoid of the cotransformed plasmid pYCC331 and contained one or two copies of STE12α at the deleted ste12a locus, respectively (Table 1).
Overexpression of STE12a.
An NdeI site was generated at the first ATG of STE12a by PCR, and the resulting construct containing the entire coding and 3′ flanking region of STE12a was subcloned into the XbaI–NdeI site of pYCC246 to yield pYCC405. The plasmid pYCC405 was linearized with ApaI to transform LP1 and LP8. Stable transformants were selected from both mating types by transferring the cultures four times on nonselective media (YEPD). The existence of the intact GAL7∷STE12a construct was confirmed by PCR with primers flanking the construct. C448 and C460 were the resulting strains (Table 1).
Preparation and Analysis of Nucleic Acids.
Genomic DNA isolation and analysis were performed as described (19, 21). For low stringency hybridization, the blot was hybridized at 50°C with 6× SSC, 5× Dehnhardt's, and 0.5% SDS and was washed at 50°C with 0.5× SSC/0.1% SDS. To isolate RNA, cells were harvested from 45-h culture as described (14). Total RNA was isolated by using the FastRNA kit (Bio 101), and poly(A)+ RNA was isolated by using the oligotex mRNA kit (Qiagen, Chatsworth, CA). Northern blot analysis was performed as described (22). After hybridization, the blot was exposed to a PhosphorImager Screen and quantified with IMAGEQUANT 1.1 (Molecular Dynamics). Each gene-specific signal was normalized to that of the actin gene. The relative expression levels of each gene were compared between the deletant and its congenic wild-type strain and expressed as a percentage of the wild-type levels.
Results
Cloning a STE12 Homologue.
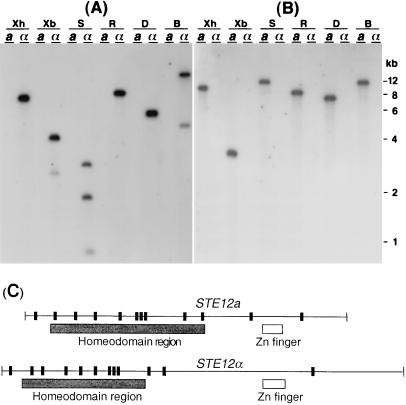
Previous studies showed that the Δste12α strain of C. neoformans remains fertile after mating with MATa strains albeit with reduced mating efficiency (14, 15). This observation is in contrast to S. cerevisiae, in which deletion of the STE12 gene causes sterility. It is possible that a different STE12 homologue exists in C. neoformans, which may augment the function of STE12α in the mating process. Existence of another STE12 homologue in MATα or MATa strains, however, was not detectable by Southern blot analysis using a probe of STE12α cDNA under low stringency conditions (Fig. 1A). Because all the STE12 homologues identified thus far contain a conserved homeodomain region, it was assumed that the new proposed STE12 homologue would also contain this conserved homeodomain region. Degenerated PCR primers were designed according to a conserved sequence of the STE12 homeodomain region. Reverse transcription–PCR was performed then by using RNAs that were isolated from a mating culture containing a mixture of MATa and MATα cells. A new STE12 homologue was identified, and then the cDNA and genomic clones containing this STE12 homologue were isolated. We found that this homologue existed only in the MATa strain B-4476 (Fig. 1B). This STE12 homolog is present also in the three other genetically unrelated MATa strains reported thus far but was absent in the five randomly chosen genetically unrelated MATα strains (data not shown). Because of the sequence similarity and a mating-type specificity, we designated this STE12 homologue as STE12a. Sequence analysis suggested that, as in STE12α, STE12a contains both the homeodomain and the zinc finger regions (7). Further comparison of the sequences between STE12a and STE12α indicated that the number and distribution pattern of introns in these two genes are similar (Fig. 1C), but the overall DNA sequence similarity between STE12a and STE12α is only 45%. Furthermore, the predicted size of the proteins for Ste12ap and Ste12αp are 69 and 94 kDa, respectively. A higher sequence similarity at the amino acid level was observed between STE12a and STE12α within the homeodomain (74%) and the zinc finger regions (62%).
Figure 1.
Detection and structure of STE12a and STE12α. (A) Southern blot analysis of STE12α. (B) Southern blot analysis of STE12a. DNAs of B-4500 (MATα) and B-4476 (MATa) were digested with different restriction enzymes, fractionated on an agarose gel, and transferred to a nylon membrane. The resulting blot was hybridized with a probe of STE12α cDNA (A) or STE12a cDNA (B) and washed at low stringency. B, BamHI; D, HinDIII; R, EcoRI; S, SalI; Xb, XbaI; Xh, XhoI. (C) Structure of STE12a- and STE12α-coding region. Vertical bars represent the intron positions.
Deletion and Reconstitution of the STE12a Gene.
To study the function of STE12a, we deleted STE12a from the MATa strain, LP2, by using a positive-negative selection method (19). Putative transformants were identified first by PCR and then confirmed by Southern blot analysis. DNA blot was hybridized with a probe of the 2.1-kb XbaI–AseI DNA fragment that was deleted in pYCC384 (Fig. 2C I). The wild-type strain (B-4476) showed a hybridization signal corresponding to the STE12a gene, whereas no signal was detected in the putative deletant. When the 3.6-kb XbaI–XbaI DNA fragment was used as a probe to hybridize to the same blot, the signals detected in the putative deletant corresponded to fragments of the predicted size (Fig. 2C II). These results indicated that we had disrupted the STE12a gene. To obtain a relevant control strain for further analysis of STE12a function, it was important to complement the deletant. Because complementation by an ectopic copy of a wild-type gene could generate undesirable effects (14), we designed a cotransformation method that enabled us to reintroduce the wild-type gene into the deletant at the homologous site (14). Adenine auxotrophic transformants, which were derived from a gene-replacement event at the disrupted Δste12a locus with the wild-type copy of STE12a, were isolated and analyzed by Southern blot. Fig. 2C III demonstrates that the putative STE12a reconstituted strain exhibits the same hybridization pattern as B-4476. This result indicated that we had obtained a STE12a reconstituted strain. The auxotrophic markers in the resulting strain were removed subsequently by mating with a wild-type strain.
Phenotype of Δste12a.
Because one of the presumed functions for STE12a was its involvement in the mating process, we first examined the influence of the ste12a deletion on mating. Like the Δste12α strain, deletion of STE12a affected the mating efficiency, but the Δste12a strain was still able to mate with MATα strains. When the mating frequency was measured quantitatively (14), the Δste12a strain (TYCC384) and STE12a reconstituted strain (C437) had a mating frequency of 18.4 and 106%, respectively, compared with the parent strain, LP2. Although the 5.4-fold reduction in mating efficiency of ste12a mutants seems substantial, the degree of reduction is negligible when compared with S. cerevisiae ste12 mutants (23). Therefore, although STE12a is not essential for mating, STE12a plays an important role in the regulation of mating proficiency of C. neoformans. In addition, when the coding region of STE12α was swapped with the coding region of STE12a at the disrupted ste12α locus, the mating frequency of such a reconstituted strain (C446) was 7.2-fold higher than that of the original Δste12α strain. This result suggested that STE12a could partially substitute the function of STE12α in mating. Most surprisingly, however, C. neoformans remained fertile even when the STE12 homologues were removed from both of its mating types. During mating between a Δste12a and a Δste12α strain, typical chains of basidiospores were produced, and the spores were viable. Although the mating efficacy of Δste12a × Δste12α could not be quantitated by the method described above, we observed an additive effect of the mutation in this cross. The amount of hyphae produced in the Δste12a × Δste12α culture was even less than that observed in the cross of Δste12a × STE12α or Δste12α × STE12a (data not shown).
Because STE12α is required for haploid fruiting (14, 15), we tested whether STE12a could substitute for the function of STE12α in this process. Like the wild-type strain B-4500, Δste12α∷STE12a strain (C489) produced hyphae on filament agar, whereas the ste12α deletant (TYCC245F1) produced only yeast cells (Fig. 2D, Δste12α∷STE12a vs. Δste12α). Levels of filamentation in strain C489 were similar to that of the wild-type MATα strain B-4500 (data not shown). These data suggested that STE12a could substitute for the function of STE12α in the production of hyphae on filament agar. It has been shown that MATa cells do not produce hyphae on filament agar (6). When the deleted ste12a gene was replaced with two copies of the STE12α gene, however, the resulting MATa strain (C490) produced hyphae sparsely on filament agar (Fig. 2D, STE12a vs. Δste12a∷2 × STE12α). Conversely, when only one copy of the STE12α gene was inserted in the ste12a locus, the resulting MATa strain produced only yeast cells on filament agar (data not shown).
It is known also that when STE12α is overexpressed, the MATα cells produce hyphal protrusions, and the morphology of MATα cells become abnormal (7). To test whether similar morphological changes occur when STE12a is overexpressed, we placed STE12a under the control of a GAL7 promoter, and the resulting construct was transformed into both mating-type strains. The resulting stable transformants of both mating types produced protrusions resembling the short hyphal tubes on galactose medium, whereas control strains retained yeast morphology. These structures never extended to form true hyphae, indicating that overexpression of STE12a causes abnormal cell morphology regardless of mating type. As in the case of growth on minimal medium with galactose, we observed no clear filamentation on filament agar with galactose when STE12a was overexpressed in both mating types (data not shown).
STE12a and Virulence.
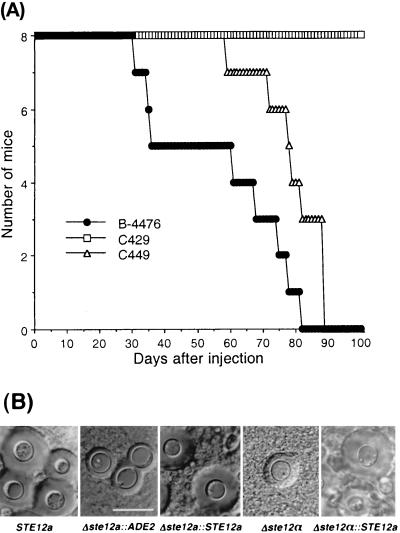
Like STE12α, deletion of STE12a did not affect the growth rate at 37°C, capsule size or melanin production in vitro (data not shown). Because deletion of STE12α dramatically affected the virulence of C. neoformans, we also tested the importance of STE12a in virulence. Groups of mice were infected with F2 prototrophs of the Δste12a strain (C429), STE12a reconstituted strain (C449), or wild-type congenic strain (B-4476). Although health of the mice infected with these strains was affected, the Δste12a strain produced no mortality during the 100-day experimental period, whereas all of the mice infected with B-4476 died within 82 days after infection (Fig. 3A). In addition, virulence was restored nearly to the wild-type level when the ste12a gene was replaced with STE12a. Therefore, it was clear that deletion of the STE12a gene severely reduced the ability of C. neoformans to cause fatal infection. To examine the size of the capsule of yeast cells microscopically, brain smears were prepared from mice infected with each strain of C. neoformans. The capsule size of yeast cells in brain smears from mice infected with C429 was considerably smaller (ranging from 1–3 μm) than that seen in mice infected with B-4476 (ranging from 2–7 μm; Fig. 3B, Δste12a∷ADE2 vs. STE12a). Furthermore, the capsule size of yeast cells in brain smears from mice infected with the STE12a reconstituted strain was the same as that of mice infected with B-4476 (Fig. 3B, Δste12a∷STE12a vs. STE12a).
Figure 3.
Effect of STE12a on virulence. (A) Virulence study. Female BALB/c mice (6–8 weeks old) were injected via the lateral tail vein with 0.2 ml (1 × 106 cells) of a suspension of wild type (B-4476), Δste12a (C429), and Δste12a∷ STE12a (C449). Mortality was monitored for 100 days. (B) Brain smear of mice challenged with different yeast strains examined under a microscope with a Normalski interference condenser. (Bar = 10 μm.)
Because we have demonstrated that STE12a could replace several functions of STE12α, we also examined the in vivo capsule size of the strain in which the ste12α gene was substituted with STE12a. The yeast cells of brain smear from the mice infected with strain C489 showed a capsule size similar to the wild-type MATα strain and was markedly larger than that of the ste12α deletant, TYCC245F1 (Fig. 3, Δste12α∷STE12a vs. Δste12α).
STE12a and Virulence-Associated Genes.
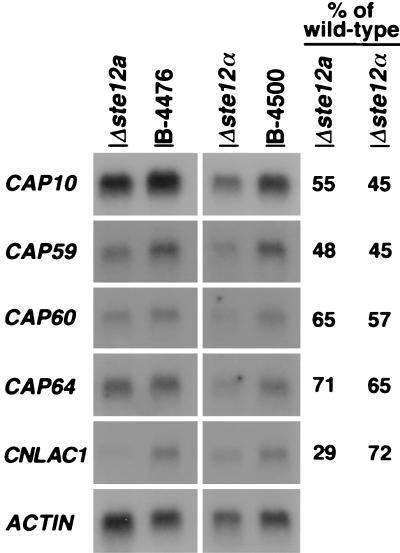
Because deletion of STE12a significantly affected virulence and the capsule size in vivo, it was of interest to study the influences of STE12a deletion on the expression of virulence-associated genes. Quantitative Northern blot analysis was used to measure the expression levels of four genes required for capsule formation (CAP10, CAP59, CAP60, and CAP64) and the laccase gene (CNLAC1). The CNLAC1 gene and the four capsule-associated genes have been shown to be important for the virulence of C. neoformans (19, 20, 22, 24, 25). Poly(A)+ RNAs were isolated from stationary phase cells (20) of wild-type MATa and MATα strains as well as deletants of ste12a and ste12α. The relative expression levels of these genes were compared between the deletant and its congenic wild-type strain. Fig. 4 demonstrated that the mRNA levels of CNLAC1 and all four capsule-associated genes were reduced in both Δste12a and Δste12α strains compared with their corresponding wild-type strains. The amount of reduction in the expression of these genes was similar between the Δste12a and Δste12α strains except that CNLAC1 mRNA levels were reduced more severely in the Δste12a than in the Δste12α strains. These data indicated that both STE12a and STE12α modulated expression of these virulence-associated genes.
Figure 4.
Northern blot analysis. Poly(A)+ RNA was isolated from yeast cells, fractionated on an agarose gel, and transferred to a nylon membrane. The resulting blot was hybridized with the indicated probe. The numbers in the last two columns represent the percentage of expression levels for each gene in the deletant compared with that of the congenic wild-type strain. Wild type, B-4476; Δste12a, C429; Δste12α, TYCC245F1.
During a subtraction library screening for genes whose expression levels were affected by deletion of STE12α, we found that the status of STE12α influenced mRNA levels of many genes including phospholipase and SOD (data not shown). Because both phospholipase and SOD are suggested to be putative virulence factors for C. neoformans, we measured the enzyme activity for these proteins in the background of Δste12a, Δste12α, and wild-type controls. The activity of phospholipase on egg-yolk agar was considerably lower in either Δste12a or Δste12α strains compared with that of the corresponding congenic wild-type strains (Table 2). The phospholipase activity was restored to nearly wild-type levels when the deleted gene was reconstituted. In contrast, we found that SOD enzyme activity increased in Δste12a and Δste12α strains compared with that of the corresponding congenic wild-type strains (Table 2). Reconstitution of STE12a and STE12α also restored SOD activity close to wild-type levels. These observations suggested that STE12a and STE12α modulate the enzyme activity of phospholipase and SOD in vitro.
Table 2.
Effect of STE12a and STE12α on the activity of phospholipase and SOD
| Strains† | Index of phospholipase activity‡ | SOD activity§ |
|---|---|---|
| STE12a | 0.49 ± 0.01** | 9.76 ± 0.05** |
| Δste12a | 0.63 ± 0.03 | 14.71 ± 0.06 |
| Δste12a∷STE12a | 0.53 ± 0.03* | 8.69 ± 0.04** |
| STE12α | 0.50 ± 0.03** | 7.05 ± 0.03** |
| Δste12α | 0.97 ± 0.03 | 12.39 ± 0.08 |
| Δste12α∷STE12α | 0.61 ± 0.03** | 7.57 ± 0.03** |
STE12a (B4476), Δste12a (C429), Δste12a∷STE12a (C449), STE12α (B4500), Δste12α (TYCC245F1), and Δste12α∷STE12α (TYCC409AF1).
There is an inverse relationship between index and enzyme activity, the smaller the index the higher the enzyme activity of the strain (see Materials and Methods). Data represent mean ± SD of three samples. *, P < 0.05 compared with the congenic deletant. **, P < 0.01 compared with the congenic deletant.
Bovine erythrocyte SOD was used as the standard for calibration of C. neoformans SOD activity. Data represent mean ± SD of three samples. **, P < 0.01 compared with the congenic deletant.
Discussion
C. neoformans is the first reported species containing a functional STE12 homologue in each of the opposite mating-type strains. Both STE12 homologues in C. neoformans, STE12a and STE12α, contain a conserved homeodomain and a zinc finger region. These two genes, however, do not cross-hybridize to each other, indicating that they are mating-type-specific genes. Unlike Ste12p of C. neoformans, Ste12 proteins of ascomycetous yeasts do not contain a zinc finger region. In this respect, C. neoformans Ste12p is similar to steAp of A. nidulans. C. neoformans Ste12p and steAp share another similarity; they lack a domain conserved among the homologous proteins from S. cerevisiae, Candida albicans, and Kluyveromyces lactis, which is essential for MAP kinase-mediated activation (26). Several transcription factors with two distinct DNA-binding domains have been described (27–33). Depending on the individual protein, homeodomain and zinc finger region can bind to DNA or function as regions of protein–protein interaction. Although Ste12αp binds DNA in vitro (unpublished data), the precise molecular function of the homeodomain and zinc finger region in Ste12ap and Ste12αp remains to be elucidated, and it is not clear that Ste12ap and Ste12αp directly interact with the virulence-associated genes analyzed in the present study.
In C. neoformans, MATα strains predominate among clinical as well as natural isolates (4) contrary to other bipolar heterothallic fungi. The STE12α gene was isolated first on the basis of its ability to cause MATα cells to form hyphae (7). It has been proposed that the ability to undergo haploid fruiting by MATα strains but not by MATa strains may be a contributing factor for the predominance of MATα strain (6). Gene-disruption studies have shown that STE12α is required for haploid fruiting (14, 15). In this study, we found that not only could STE12a substitute for the function of STE12α in hyphae formation on filament agar, but also a replacement of STE12a with two copies of STE12α enabled the MATa strain to undergo the same morphogenesis process albeit with lower frequency. Our observations suggest that these two STE12 alleles of C. neoformans can complement each other functionally, conditional to interactions with other mating-type-related factors yet to be identified. The mating-type locus of C. neoformans harbors several other pheromone-response MAP-kinase cascade genes (9). It is reasonable, therefore, to assume that STE12 allele-specific interactions occur between the Ste12p and other proteins encoded by the mating-type locus including components of the MAP-kinase cascade. It is possible also that the MATa strains require stronger stimuli than just nitrogen starvation to undergo haploid fruiting, and STE12α is a stronger regulator compared with STE12a. The mechanism involved in regulating haploid fruiting is not clear.
It is known that STE12, a component of the MAP-kinase pathway that signals the mating pheromone response in S. cerevisiae, is involved also in filamentous morphogenesis in diploid as well as haploid cells (10–13). Unlike in C. neoformans, however, the components of the S. cerevisiae pheromone-response MAP-kinase pathway have no association with the mating-type locus. One of the intriguing findings in this study is that, in contrast to the sterile phenotype of S. cerevisiae ste12 mutants, the Δste12a strain was not only fertile after mating with STE12α strains, but also was able to mate with a ste12α-deleted strain. It has been shown in several species of basidiomycetes that interaction between different types of homeodomain-containing transcription factors are required for mating (34). It is possible, therefore, that C. neoformans contains other genes that could interact with or substitute for the function of Ste12ap or Ste12αp in the pheromone-response pathway. It is also possible that C. neoformans STE12α and STE12a belong to different pathways and the effects of both STE12a and STE12α on mating observed in our study could be an indirect result of cross-talk between the pathways. The involvement of STE12α in a different pathway is supported by the observations that STE12α is required for the haploid fruiting after nitrogen starvation (14, 15).
Like STE12α, STE12a seems to be required to express a wild-type level of virulence. Although mice infected with Δste12a or Δste12α strains survive significantly longer than those infected with wild-type or reconstituted strains, Δste12a and Δste12α cells persist in animal tissue and eventually cause fatal disease (data not shown). Disruption of STE12a altered the expression levels of many genes associated with virulence, and consequently the ste12a mutant was significantly less virulent in mice compared with the wild type. In fact, the decreased capsule size in vivo corroborated the reduced mRNA levels of capsule-associated genes tested in vitro. Conversely, when the Δste12a locus was reconstituted, virulence, capsule size in vivo, fertility, and phospholipase activity reverted close to that of wild type.
It is interesting that the enzyme activity of phospholipase was more significantly down-regulated in ste12α than in ste12a, whereas SOD activity was up-regulated in both Δste12a and Δste12α strains. The role of extracellular phospholipase as a potential virulence factor in pathogenic fungi, including C. albicans, C. neoformans, and Aspergillus, has gained acceptance recently (for review see ref. 35). Although SOD is an important housekeeping antioxidant and has been suggested to play a role in virulence, there has not been any experimental evidence supporting such a hypothesis (for review see ref. 36). Our observation with Δste12a and Δste12α strains seems to contradict the role of SOD on virulence of C. neoformans in mice. The elevated SOD level may benefit the fungus in a stressful environment with high concentration of oxygen radicals such as in animal tissue. The increased SOD activity in Δste12a or Δste12α strains, however, may not be enough to offset the negative effects caused by the STE12a or STE12α deficiency. It is unclear how STE12a and STE12α render different effects on the activity of phospholipase and SOD. Further studies on the relationship between Ste12p and affected proteins may reveal whether Ste12p directly affects the synthesis of these enzymes. We found additional genes either down-regulated or up-regulated in response to the deletion of STE12α (data not shown). These data implicate the multiple roles of STE12a and STE12α in regulating the expression of other genes and reveal the intricacy of how STE12a and STE12α modulate virulence.
Previous animal-model studies on virulence of isogenic MATa and MATα strains suggest that the mating-type locus is the main cause of the higher virulence of MATα cells (5). Our present study suggests that not only individual components of the MATα locus but also the MATa locus play important roles for virulence in the serotype D strains of C. neoformans. Therefore, these data support the notion that the fungal genes involved in mating have far more diverse roles than merely for reproduction.
Acknowledgments
We thank A. Varma for critical reading of the manuscript.
Abbreviations
- MAP
mitogen-activated protein
- SOD
superoxide dismutase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF242352).
References
- 1.Kwon-Chung K J, Bennett J E. Medical Mycology. Philadelphia: Lea & Febiger; 1992. pp. 397–446. [Google Scholar]
- 2.Kwon-Chung K J. Mycologia. 1975;67:1197–1200. [PubMed] [Google Scholar]
- 3.Kwon-Chung K J. Mycologia. 1976;68:943–946. [PubMed] [Google Scholar]
- 4.Kwon-Chung K J, Bennett J E. Am J Epidemiol. 1978;108:337–340. doi: 10.1093/oxfordjournals.aje.a112628. [DOI] [PubMed] [Google Scholar]
- 5.Kwon-Chung K J, Edman J C, Wickes B L. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wickes B L, Mayorga M E, Edman U, Edman J C. Proc Natl Acad Sci USA. 1996;93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wickes B L, Edman U, Edman J C. Mol Microbiol. 1997;26:951–960. doi: 10.1046/j.1365-2958.1997.6322001.x. [DOI] [PubMed] [Google Scholar]
- 8.Wang P, Perfect J R, Heitman J. Mol Cell Biol. 2000;20:352–362. doi: 10.1128/mcb.20.1.352-362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karos M, Chang Y C, McClelland C M, Clarke D L, Fu J, Wickes B L, Kwon-Chung K J. J Bacteriol. 2000;182:6222–6227. doi: 10.1128/jb.182.21.6222-6227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 11.Roberts R L, Fink G R. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 12.Herskowitz I. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 13.Madhani H D, Fink G R. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- 14.Chang Y C, Wickes B L, Miller G F, Penoyer L A, Kwon-Chung K J. J Exp Med. 2000;191:871–882. doi: 10.1084/jem.191.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue C, Cavallo L M, Alspaugh J A, Wang P, Cox G M, Perfect J R, Heitman J. Genetics. 1999;153:1601–1615. doi: 10.1093/genetics/153.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon-Chung K J, Bennett J E, Rhodes J C. Antonie Leeuwenhoek. 1982;48:25–38. doi: 10.1007/BF00399484. [DOI] [PubMed] [Google Scholar]
- 17.Chen S C, Muller M, Zhou J Z, Wright L C, Sorrell T C. J Infect Dis. 1997;175:414–420. doi: 10.1093/infdis/175.2.414. [DOI] [PubMed] [Google Scholar]
- 18.Salin M L, McCord J M. J Clin Invest. 1974;54:1005–1009. doi: 10.1172/JCI107816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang Y C, Kwon-Chung K J. Mol Cell Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang Y C, Kwon-Chung K J. J Bacteriol. 1999;181:5636–5643. doi: 10.1128/jb.181.18.5636-5643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitkin J W, Panaccione D G, Walton J D. Microbiology. 1996;142:1557–1565. doi: 10.1099/13500872-142-6-1557. [DOI] [PubMed] [Google Scholar]
- 22.Chang Y C, Penoyer L A, Kwon-Chung K J. Infect Immun. 1996;64:1977–1983. doi: 10.1128/iai.64.6.1977-1983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malathi K, Ganesan K, Datta A. J Biol Chem. 1994;269:22945–22951. [PubMed] [Google Scholar]
- 24.Chang Y C, Kwon-Chung K J. Infect Immun. 1998;66:2230–2236. doi: 10.1128/iai.66.5.2230-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salas S D, Bennett J E, Kwon-Chung K J, Perfect J R, Williamson P R. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vallim M A, Miller K Y, Miller B L. Mol Microbiol. 2000;36:290–301. doi: 10.1046/j.1365-2958.2000.01874.x. [DOI] [PubMed] [Google Scholar]
- 27.Treisman J, Harris E, Desplan C. Genes Dev. 1991;5:594–604. doi: 10.1101/gad.5.4.594. [DOI] [PubMed] [Google Scholar]
- 28.Verrijzer C P, Alkema M J, van Weperen W W, Van Leeuwen H C, Strating M J, van der Vliet P C. EMBO J. 1992;11:4993–5003. doi: 10.1002/j.1460-2075.1992.tb05606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andres V, Chiara M D, Mahdavi V. Genes Dev. 1994;8:245–257. doi: 10.1101/gad.8.2.245. [DOI] [PubMed] [Google Scholar]
- 30.Darling D S, Gaur N K, Zhu B. Mol Cell Endocrinol. 1998;139:25–35. doi: 10.1016/s0303-7207(98)00076-8. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda K, Kawakami K. Eur J Biochem. 1995;233:73–82. doi: 10.1111/j.1432-1033.1995.073_1.x. [DOI] [PubMed] [Google Scholar]
- 32.Niimi T, Seimiya M, Kloter U, Flister S, Gehring W J. Development (Cambridge, UK) 1999;126:2253–2260. doi: 10.1242/dev.126.10.2253. [DOI] [PubMed] [Google Scholar]
- 33.Phelan S A, Loeken M R. J Biol Chem. 1998;273:19153–19159. doi: 10.1074/jbc.273.30.19153. [DOI] [PubMed] [Google Scholar]
- 34.Casselton L A, Olesnicky N S. Microbiol Mol Biol Rev. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghannoum M A. Clin Microbiol Rev. 2000;13:122–143. doi: 10.1128/cmr.13.1.122-143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamilton A J, Holdom M D. Med Mycol. 1999;37:375–389. doi: 10.1046/j.1365-280x.1999.00208.x. [DOI] [PubMed] [Google Scholar]