Abstract
OBJECTIVE
Over 30 loci have been associated with risk of type 2 diabetes at genome-wide statistical significance. Genetic risk scores (GRSs) developed from these loci predict diabetes in the general population. We tested if a GRS based on an updated list of 34 type 2 diabetes–associated loci predicted progression to diabetes or regression toward normal glucose regulation (NGR) in the Diabetes Prevention Program (DPP).
RESEARCH DESIGN AND METHODS
We genotyped 34 type 2 diabetes–associated variants in 2,843 DPP participants at high risk of type 2 diabetes from five ethnic groups representative of the U.S. population, who had been randomized to placebo, metformin, or lifestyle intervention. We built a GRS by weighting each risk allele by its reported effect size on type 2 diabetes risk and summing these values. We tested its ability to predict diabetes incidence or regression to NGR in models adjusted for age, sex, ethnicity, waist circumference, and treatment assignment.
RESULTS
In multivariate-adjusted models, the GRS was significantly associated with increased risk of progression to diabetes (hazard ratio [HR] = 1.02 per risk allele [95% CI 1.00–1.05]; P = 0.03) and a lower probability of regression to NGR (HR = 0.95 per risk allele [95% CI 0.93–0.98]; P < 0.0001). At baseline, a higher GRS was associated with a lower insulinogenic index (P < 0.001), confirming an impairment in β-cell function. We detected no significant interaction between GRS and treatment, but the lifestyle intervention was effective in the highest quartile of GRS (P < 0.0001).
CONCLUSIONS
A high GRS is associated with increased risk of developing diabetes and lower probability of returning to NGR in high-risk individuals, but a lifestyle intervention attenuates this risk.
Widespread collaboration and recent advances in genetic knowledge and technology have permitted discovery of many new loci associated with risk of type 2 diabetes (1,2). The Diabetes Genetics Replication And Meta-analysis (DIAGRAM) consortium has carried out genome-wide meta-analyses of type 2 diabetes as a categorical trait in populations of European descent (3,4) and the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) has done likewise for glycemic quantitative traits (5–7). Both efforts have revealed many genetic variants associated with type 2 diabetes at genome-wide significance levels (P < 5 × 10−8). The most recent report from DIAGRAM (including 42,542 type 2 diabetes case subjects and 98,912 control subjects of European descent) has added 12 new loci (4), producing a total of over 30 single nucleotide polymorphisms (SNPs) now accepted as associated with type 2 diabetes.
As fine-mapping and functional studies proceed, this new genetic knowledge has already revealed unsuspected biological pathways that help increase our understanding of pathophysiological mechanisms leading to the disease. Other investigators have tested the ability of genetic information to predict who is likely to develop type 2 diabetes in prospective general population–based cohorts (8–10); however similar analyses in a population already at high risk for type 2 diabetes are lacking.
The Diabetes Prevention Program (DPP) was designed to test the preventive effects of a lifestyle intervention or medication on progression to diabetes in high-risk individuals. We have previously shown that participants who carry the risk allele at TCF7L2 (the common type 2 diabetes locus with the strongest effect yet reported) are at increased risk of developing diabetes (11), but most of the other individual variants examined were not statistically associated with diabetes incidence (12,13). It is unknown if a genetic risk score (GRS) using all currently type 2 diabetes–associated loci is associated with progression to type 2 diabetes in a multiethnic population such as the DPP cohort, whose participants are already at a very high baseline risk based on clinical characteristics.
We therefore tested the hypothesis that a higher GRS, which includes 34 type 2 diabetes–associated loci, would be associated with a greater risk of developing type 2 diabetes in DPP participants after considering treatment arms (lifestyle intervention and metformin) and other risk factors for progression toward the disease. Because all participants had impaired glycemic regulation at baseline, we conducted similar analyses to test the association between the GRS and regression to normal glucose regulation (NGR). Finally, we also tested if the GRS was associated with physiologic traits (insulin sensitivity and insulin secretion indices), and whether the preventive interventions maintained their effectiveness in those with the highest genetic risk.
RESEARCH DESIGN AND METHODS
Description of DPP study design and participants.
Details of the DPP study design and characteristics of the participants at baseline have been described previously (14,15). In brief, the DPP was a multicenter trial that was designed to test whether intensive lifestyle modification or pharmacologic intervention prevents progression to diabetes in individuals at high risk of developing type 2 diabetes. The active intervention phase was conducted from 1996 through 2001 in 27 U.S.-based medical centers.
Participants were included if they had a fasting plasma glucose between 95 and 125 mg/dL (5.3–6.9 mmol/L) and 2-h plasma glucose between 140 and 199 mg/dL (7.8–11.0 mmol/L) on oral glucose tolerance testing (OGTT). A total of 3,234 participants were randomized to intensive lifestyle modification (goal >7% weight loss and >150 min/week of physical activity), metformin (850-mg twice daily), or placebo. A fourth troglitazone arm was stopped early because of the risk of hepatotoxicity. The participants included in this report represent both sexes (66.8% women) and ethnically diverse backgrounds (56.4% were of European descent, 20.2% African American, 16.8% Hispanic, 4.3% Asian, and 2.4% American Indian) as planned in the study design. At baseline, mean (± SD) age was 50.6 ± 10.7 years and mean (± SD) BMI was 34.0 ± 6.7 kg/m2. The primary end point of the DPP was reduction of diabetes incidence: after 2.8 years of mean follow-up, there was a 58% (95% CI 48–66%) reduction of diabetes incidence in the lifestyle intervention group and a 31% (95% CI 17–43%) reduction in the metformin group compared with placebo (16). Institutional review board approval was obtained by each participating medical center; the 2,843 (947 placebo, 955 lifestyle, and 941 metformin) participants included in this report provided written informed consent for the main study and for subsequent genetic investigations.
Definitions of diabetes incidence and regression to NGR.
Follow-up of glycemic regulation was performed with fasting glucose measurement every 6 months and by OGTT every 12 months. Diagnosis of diabetes was made based on American Diabetes Association (ADA) guidelines (fasting glucose above ≥126 mg/dL or 2-h glucose levels ≥200 mg/dL on an OGTT, confirmed by a second test within 6 weeks). Regression to NGR was defined as normalization of both fasting and 2-h glucose (fasting glucose <100 mg/dL and 2-h glucose <140 mg/dL).
Quantitative glycemic physiologic traits.
We calculated the insulin sensitivity index (ISI) as the reciprocal of homeostasis model assessment of insulin resistance, determined as 22.5/[(fasting insulin × fasting glucose)/18.01] (17). We estimated insulin secretion by the insulinogenic index using the formula [(insulin at 30 min) − (insulin at 0 min)]/[(glucose at 30 min) − (glucose at 0 min)] (18). The oral disposition index was calculated using the formula [insulinogenic index/fasting insulin] (19). The ISI and insulinogenic index were also calculated at 1 year to estimate the change in insulin sensitivity or secretion over time: we used the change in each index in subsidiary analyses ([ISI at 1 year – ISI at baseline] and [insulinogenic index at 1 year – insulinogenic index at baseline]). We chose 1 year because changes in weight were most pronounced at that time point, and it contained the highest number of measures for analysis within the DPP population, as individuals who had developed diabetes no longer had an OGTT.
SNP selection and genotyping.
SNPs associated with type 2 diabetes were selected based on published reports from the literature (3,5,20–23) as well as a personal communication from the DIAGRAM+ investigators (4). SNPs associated with type 2 diabetes at genome-wide statistical significance (P < 5 × 10−8) were included. Where the index SNP was not available, a suitable proxy was selected (r2 >0.8 in HapMap CEU). The mean genotyping success was 96.8%, and the minimum call rate was 92.1%. No SNP had to be excluded for deviation from Hardy-Weinberg equilibrium (ethnic-specific P < 0.001).
DNA was extracted from peripheral blood leukocytes in a standard fashion. Genotyping was carried out by allele-specific primer extension of multiplex-amplified products and detection using matrix-assisted laser desorption ionization time-of-flight mass spectrometry on a Sequenom iPLEX platform (24).
Construction of GRS.
Using previously described methods to construct genetic scores for type 2 diabetes prediction (9,25), we created a weighted GRS per participant by multiplying the number of risk alleles present per SNP by the β-estimate reported for that SNP in the MAGIC and DIAGRAM studies and summing the results over the 34 SNPs (resulting in a possible score ranging from 0 to 68). We reported the effect of the GRS “per risk allele,” representing an “average level” per risk allele after weighting. The β-estimates are the natural log of the odds ratios listed in Table 1. One hundred and fifty-five individuals missing more than 3 SNPs were excluded from the analysis. The genotype was imputed for those missing 1–3 SNPs by using the genotype occurring in the highest frequency within each ethnicity subgroup (n = 313 individuals with imputed genotypes).
TABLE 1.
Type 2 diabetes risk loci genotyped in DPP participants
| Genetic locus | SNP typed | Alleles (risk/other) | Strand | OR* | Study | Overall study (all ethnic groups) |
||
|---|---|---|---|---|---|---|---|---|
| A = risk allele/a = other allele | ||||||||
| AA |
Aa |
aa |
||||||
| N (%) | N (%) | N (%) | ||||||
| TCF7L2 | rs7903146 | T/C | + | 1.37 | DIAGRAM | 287 (9.6) | 1,227 (41.1) | 1,469 (49.3) |
| CDKN2A/2B | rs10811661 | T/C | + | 1.26 | DIAGRAM | 2,245 (75.3) | 664 (22.3) | 71 (2.4) |
| CDKAL1 | rs7754840 | C/G | + | 1.25 | DIAGRAM | 453 (15.2) | 1,348 (45.2) | 1,180 (39.6) |
| PPARG | rs1801282 | C/G | + | 1.18 | DIAGRAM | 2,501 (83.6) | 476 (15.9) | 16 (0.5) |
| HHEX | rs1111875 | C/T | – | 1.17 | DIAGRAM | 1,218 (40.8) | 1,337 (44.8) | 432 (14.5) |
| IGF2BP2 | rs1470579 | C/A | + | 1.17 | DIAGRAM | 598 (20.2) | 1,185 (40.1) | 1,175 (39.7) |
| KCNJ11 | rs5219 | T/C | + | 1.16 | DIAGRAM | 318 (10.6) | 1,242 (41.6) | 1,426 (47.8) |
| SLC30A8 | rs13266634 | C/T | + | 1.15 | DIAGRAM | 1,723 (57.7) | 1,072 (35.9) | 191 (6.4) |
| THADA | rs7578597 | A/G | – | 1.15 | DIAGRAM | 2,259 (77.7) | 587 (20.2) | 63 (2.2) |
| CENTD2 | rs1552224 | A/C | – | 1.14 | DIAGRAM+ | 2,363 (81.9) | 494 (17.1) | 29 (1.0) |
| NOTCH2 | rs10923931 | T/G | + | 1.13 | DIAGRAM | 95 (3.3) | 697 (24.0) | 2,118 (72.8) |
| ADCY5 | rs11708067 | A/G | + | 1.12 | MAGIC | 1,891 (65.7) | 863 (30.0) | 125 (4.3) |
| WFS1 | rs10010131 | G/A | + | 1.11 | DIAGRAM | 1,315 (44.0) | 1,302 (43.6) | 371 (12.4) |
| CDC123 | rs4747969 | C/T | + | 1.11 | DIAGRAM | 136 (4.7) | 959 (33.0) | 1,814 (62.4) |
| IRS1 | rs7578326 | A/G | + | 1.11 | DIAGRAM+ | 1,299 (47.1) | 1,119 (40.6) | 340 (12.3) |
| CHCHD9 | rs13292136 | C/T | + | 1.11 | DIAGRAM+ | 2,431 (85.5) | 389 (13.7) | 25 (0.9) |
| JAZF1 | rs864745 | G/A | + | 1.10 | DIAGRAM | 493 (17.0) | 1,348 (46.5) | 1,059 (36.5) |
| HNF1B | rs757210 | T/C | – | 1.10 | DIAGRAM | 517 (17.9) | 1,318 (45.7) | 1,049 (36.4) |
| HMGA2 | rs1531343 | C/G | + | 1.10 | DIAGRAM+ | 114 (4.1) | 692 (24.8) | 1,987 (71.1) |
| ADAMTS9 | rs4607103 | C/T | + | 1.09 | DIAGRAM | 1,478 (50.8) | 1,201 (41.3) | 230 (7.9) |
| TSPAN8 | rs7961581 | C/T | + | 1.09 | DIAGRAM | 217 (7.5) | 1,089 (37.4) | 1,603 (55.1) |
| MTNR1B | rs10830963 | G/C | + | 1.09 | MAGIC | 200 (6.9) | 998 (34.5) | 1,692 (58.6) |
| BCL11A | rs243021 | A/G | – | 1.08 | DIAGRAM+ | 704 (24.4) | 1,392 (48.3) | 787 (27.3) |
| SLC22A18AS | rs231362 | G/A | – | 1.08 | DIAGRAM+ | 1,150 (40.3) | 1,274 (44.7) | 427 (15.0) |
| ZBED3 | rs4457053 | G/A | + | 1.08 | DIAGRAM+ | 251 (8.8) | 1,148 (40.3) | 1,447 (50.8) |
| PROX1 | rs340874 | C/T | – | 1.07 | MAGIC | 630 (22.1) | 1,305 (45.7) | 921 (32.3) |
| GCK | rs917793 | T/A | – | 1.07 | MAGIC | 168 (5.8) | 1,015 (35.1) | 1,707 (59.1) |
| TSGA13 | rs972283 | G/A | + | 1.07 | DIAGRAM+ | 1,099 (38.8) | 1,274 (45.0) | 460 (16.2) |
| VPS33B | rs8042680 | A/C | + | 1.07 | DIAGRAM+ | 978 (34.4) | 1,086 (38.2) | 783 (27.5) |
| HNF1A | rs7957197 | T/A | + | 1.07 | DIAGRAM+ | 2,009 (70.3) | 756 (26.5) | 92 (3.2) |
| DGKB | rs2191349 | T/G | + | 1.06 | MAGIC | 870 (30.1) | 1,401 (48.5) | 618 (21.4) |
| GCKR | rs780094 | C/T | – | 1.06 | MAGIC | 1,258 (43.5) | 1,281 (44.3) | 351 (12.2) |
| PLEKHF2 | rs896854 | T/C | – | 1.06 | DIAGRAM+ | 826 (29.0) | 1,379 (48.4) | 644 (22.6) |
| BCL2A1 | rs11634397 | G/A | + | 1.06 | DIAGRAM+ | 988 (34.3) | 1,348 (46.7) | 548 (19.0) |
OR, odds ratio.
*OR reported by previous literature (column study).
Statistical analyses.
We assessed baseline characteristics in each quartile of the weighted GRS: parametric trend tests were constructed using linear contrasts for continuous variables, and the Jonckheere-Terpstra trend test was used for qualitative variables. The GRS was analyzed in general linear models predicting baseline (and year 1 change from baseline) for ISI (1/homeostasis model assessment of insulin resistance), the insulinogenic index, proinsulin-to-insulin ratio, and the oral disposition index. Insulin sensitivity/secretion indices models were adjusted for age, sex, self-reported ethnicity, and waist circumference (the GRS was more strongly associated with waist circumference than BMI at baseline). The year 1 analysis included a test for treatment × GRS interaction. Model fit was assessed by residual analysis including quintile-quintile plots for residual normal distribution.
For diabetes incidence and regression to NGR, we analyzed proportional hazards models (GRS modeled per risk allele) adjusted for treatment group, sex, age at randomization, self-reported ethnicity, and waist circumference. We analyzed treatment × GRS interactions by including interaction treatment terms in models only if significant; if not significant, models included the full cohort to test the impact of GRS on diabetes incidence and regression to NGR. To test the stability of our models, we performed sensitivity analyses first by excluding the top 5% (n = 142) participants with GRS ≥44 (n = 2,701), then excluding the bottom 5% (n = 142) participants with GRS ≤30 (n = 2,701). Deleting the tails introduced only minor changes in the estimates of the hazard ratios (HRs), which suggests that the β-estimates are robust and that the models are stable. Calibration tests of the Cox models were performed using methods described by Grønnesby and Borgan (26) and Parzen and Lipsitz (27); there was no evidence of potential problems in the fit of the Cox models. To assess the additional predicting value of the weighted GRS, we calculated the C-statistic (28) and the integrated discrimination improvement (29) of the main multivariable models (adjusted for age, sex, ethnic background, treatment arm, and waist circumference) with and without the GRS.
Furthermore, we analyzed proportional hazards models for association with diabetes incidence with two sets of covariates. The first set, termed a clinical model, was intended to ascertain the extent to which the GRS provided information not contained in easily obtained clinical measurements: it included the covariates treatment group, sex, age at randomization, self-reported ethnicity, family history of diabetes, BMI, fasting plasma glucose, history of hypertension, HDL cholesterol, and log triglycerides (similar to a previous model used in the general population [9,30]). The second set of covariates represented a physiological model, designed to examine the functional mechanisms that have generally been hypothesized in pathways leading to diabetes: in addition to treatment group, sex, age at randomization, it included the covariates waist circumference, fasting plasma glucose, 2-h plasma glucose, log insulinogenic index, log fasting insulin, log alanine aminotransferase (a surrogate for hepatic fat), and log C-reactive protein (a surrogate for inflammation). We assessed collinearity by analyzing correlations, variance-inflation factors, and the condition index; there was no evidence that collinearity was a problem in these data. The weighted GRS was added to each model and assessed for predicting diabetes. Madalla R-square (31) was used to describe the amount of variation explained by each variable in the models. To obtain adjusted incidence rates, we used a Poisson model through a general estimating equation. Rates were adjusted for age, sex, self-reported ethnicity, and waist circumference, and linear contrasts were used to test incidence rates between treatment groups in the 4th quartile of GRS. The Holm procedure was used to correct for multiple comparisons.
As exploratory analyses, each individual locus was tested for associations with progression to diabetes and regression to NGR taking into account treatment arms: interactions were tested and results are presented adjusted for treatment arms (if interaction tests were nonsignificant) and in each treatment arm. Finally, we performed main analyses in each ethnic subgroup to assess how the GRS (based on loci found in populations of European descent) performed in groups other than white; nevertheless, we are aware that sample size is limiting and consider these subgroup analyses as exploratory.
RESULTS
The type 2 diabetes–associated SNPs genotyped in DPP participants and the frequencies of the risk alleles in the overall group are presented in Table 1; frequencies for each ethnic group are presented in Supplementary Table 1. Table 1 also shows the reported odds ratios we used when constructing the weighted GRS based on reported effects in recent type 2 diabetes meta-analyses (3–5,20–23).
After imputation, the median-weighted GRS score was 37.0 (ranging from 23.7 to 51.5). The baseline characteristics of participants in each quartile of the weighted GRS are presented in Table 2. There was a higher proportion of individuals from European descent than of other ethnicities in the lowest risk quartile. As hypothesized, more participants in the highest GRS quartile reported a family history of type 2 diabetes. Also as hypothesized, a higher GRS was associated with indices of diminished β-cell function (as determined by the insulinogenic index, r = −0.04; P = 0.04) and of impaired insulin processing (proinsulin-to-insulin ratio, r = 0.06; P = 0.003). On the other hand, participants in the highest GRS quartile showed a better metabolic profile mainly in terms of central obesity (lower waist circumference) and insulin resistance-related traits (lower fasting insulin and triglycerides, higher HDL levels) or estimated insulin sensitivity (ISI, r = 0.05; P = 0.009).
TABLE 2.
Baseline characteristics of DPP participants in each quartile of GRS
| Quartile of weighted GRS |
|||||
|---|---|---|---|---|---|
| Baseline characteristic | 1st | 2nd | 3rd | 4th | P* |
| Weighted genetic score | 32 (24–34) N = 710 | 36 (34–37) N = 711 | 38 (37–40) N = 711 | 42 (40–51) N = 711 | |
| Female sex (%) | 460 (64.8) | 484 (68.1) | 486 (68.4) | 482 (67.8) | 0.049 |
| Self-reported ethnicity | |||||
| White (%) | 481 (67.7) | 444 (62.4) | 375 (52.7) | 295 (41.5) | <0.001 |
| African American (%) | 41 (5.8) | 77 (10.8) | 166 (23.3) | 295 (41.5) | |
| Hispanic (%) | 121 (17.0) | 121 (17.0) | 130 (18.3) | 94 (13.2) | |
| Asian/Pacific Islander (%) | 41 (5.8) | 47 (6.6) | 19 (2.7) | 16 (2.3) | |
| American Indian (%) | 26 (3.7) | 22 (3.1) | 21 (3.0) | 11 (1.5) | |
| Self-reported family history of DM (%) | 477 (67.4) | 483 (67.9) | 491 (69.1) | 529 (74.4) | <0.001 |
| Age (years) | 50 (28–85) | 50 (26–84) | 50 (25–84) | 49 (26–83) | <0.001 |
| BMI (kg/m2) | 33 (23–66) | 33 (23–58) | 33 (23–65) | 32 (22–71) | 0.06 |
| Waist (cm) | 105 (69–182) | 104 (70–190) | 104 (73–159) | 102 (74–155) | <0.001 |
| Fasting plasma glucose (mg/dL) | 105 (89–136) | 105 (85–139) | 106 (82–139) | 106 (90–138) | <0.001 |
| 2-h plasma glucose (mg/dL) | 162 (140–199) | 165 (140–199) | 162 (140–199) | 162 (140–199) | 0.38 |
| Hypertension (%) | 165 (23.2) | 161 (22.6) | 167 (23.5) | 154 (21.7) | 0.90 |
| HDL (mg/dL) | 43 (21–91) | 43 (22–105) | 44 (20–103) | 45 (19–101) | 0.002 |
| Fasting triglycerides (mg/dL) | 152 (37–823) | 151 (30–695) | 136 (39–835) | 125 (36–920) | <0.001 |
| ALT | 18 (3–106) | 17 (1–112) | 17 (2–114) | 17 (1–114) | <0.001 |
| CRP (mg/L) | 0.38 (0.01–12.60) | 0.38 (0.01–7.36) | 0.38 (0.01–11.80) | 0.36 (0.01–11.11) | 0.83 |
Data are n (%) or median (minimum to maximum) unless otherwise indicated. ALT, alanine transaminase; CRP, C-reactive protein; DM, diabetes.
*P value for F tests from general linear models.
Adjustment for age, sex, ethnic background, and waist circumference strengthened the association of higher GRS with lower insulin secretion (insulinogenic index β = −0.004 [SE 0.001], P < 0.001) and impaired insulin processing (proinsulin-to-insulin ratio β = 0.008 [SE 0.002], P = 0.001) (Table 3). The apparently paradoxical association between a high GRS and insulin sensitivity was attenuated but remained nominally statistically significant after multivariable adjustment (ISI β = 0.006 [SE 0.002], P = 0.02). The oral disposition index tended to be lower in individuals with a higher GRS (β = −0.004 [SE 0.002]; P = 0.06), driven by the significantly lower insulin secretion but attenuated by slightly better insulin sensitivity after adjustment for potential confounders.
TABLE 3.
Associations between GRS and insulin secretion or insulin sensitivity indices adjusted for age, sex, ethnic background, and waist circumference in DPP participants
| Quartile of weighted GRS |
β* (SE) | P | ||||
|---|---|---|---|---|---|---|
| 1st |
2nd | 3rd | 4th | |||
| N | 710 | 711 | 711 | 711 | ||
| Weighted genetic score | 32 (24–34) | 36 (34–37) | 38 (37–40) | 42 (40–51) | ||
| Insulinogenic index | 1.24 (1.17–1.32) | 1.25 (1.18–1.33) | 1.25 (1.17–1.32) | 1.12 (1.05–1.20) | −0.004 (0.001) | <0.001 |
| ISI (= 1/HOMA-IR) | 0.157 (0.150–0.164) | 0.157 (0.150–0.164) | 0.159 (0.152–0.166) | 0.161 (0.154–0.169) | 0.006 (0.002) | 0.02 |
| Proinsulin-to-insulin ratio | 0.178 (0.170–0.186) | 0.181 (0.173–0.189) | 0.184 (0.176–0.192) | 0.191 (0.182–0.200) | 0.008 (0.002) | 0.001 |
| Disposition index (oral) | 0.048 (0.046–0.051) | 0.050 (0.047–0.053) | 0.050 (0.047–0.053) | 0.046 (0.043–0.049) | −0.004 (0.002) | 0.06 |
Data are least squares mean (95% CI) unless otherwise indicated. P values for β in the regression model. HOMA-IR, homeostasis model assessment of insulin resistance.
*Regression model, per unit increase in GRS.
Over the first year, a higher GRS tended to be associated with worsening of insulin secretion, but this was not significant (change in insulinogenic index β = −0.008 [SE 0.005], P = 0.13 after adjustment for age, sex, ethnic background, waist circumference, and treatment arms). Having a high GRS did not seem to influence the 1-year change in other insulin sensitivity/secretion indices.
Progression to diabetes.
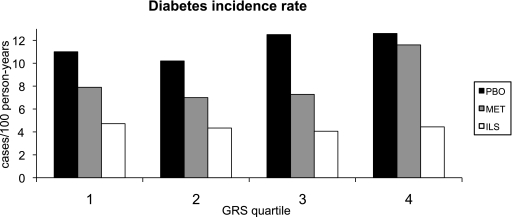
We did not find significant interactions between treatment arms and the weighted GRS (GRS × metformin interaction, P = 0.67; GRS × lifestyle interaction, P = 0.13); therefore, we pursued our multivariable models in the full cohort adjusting for treatment arms. The GRS tended to be associated with higher risk of progression to diabetes when adjusting for treatment arms (HR = 1.02 per risk allele [95% CI 1.00–1.04]; P = 0.08) (Table 4). When further adjusting for major risk factors for type 2 diabetes (age, sex, ethnic background, and waist circumference), the weighted GRS was nominally associated with higher risk of type 2 diabetes over the average 3.2-year follow-up (HR = 1.02 per risk allele [95% CI 1.00–1.05]; P = 0.03). Adding BMI to the model did not influence the associations, and the weighted GRS remained nominally associated with progression to diabetes (HR = 1.02; P = 0.03). Figure 1 illustrates the incidence rate of diabetes (case subjects/100 person-years) by quartile of GRS per treatment arm. The C-statistic value for the main multivariable model was 0.628 without the weighted GRS and 0.631 when adding the weighted GRS; this improvement was not statistically significant (P = 0.34). The integrated discrimination improvement (IDI) measure for the GRS was also not statistically significant (IDI = −0.001; P = 0.38) in the multivariable model taking diabetes incidence as the main outcome. When examining individuals at the highest genetic risk of type 2 diabetes (4th quartile of GRS), there was no statistical difference in diabetes incidence between the placebo and metformin arms (P = 0.32); however, diabetes incidence was significantly higher in the placebo arm than in the lifestyle intervention (P < 0.0001) (Fig. 1).
TABLE 4.
Multivariable models to predict progression to diabetes in DPP participants
| Minimally adjusted for treatment arms |
Multiadjusted model |
|||
|---|---|---|---|---|
| Variables | HR (95% CI) | P | HR (95% CI) | P |
| Weighted GRS (per risk allele) | 1.02 (1.00–1.04) | 0.08 | 1.02 (1.00–1.05) | 0.03 |
| Metformin (vs. placebo) | 0.71 (0.59–0.86) | 0.0005 | 0.70 (0.58–0.85) | 0.0003 |
| Lifestyle (vs. placebo) | 0.46 (0.37–0.57) | <0.0001 | 0.45 (0.36–0.56) | <0.0001 |
| Female (vs. male) | 0.99 (0.83–1.19) | 0.94 | ||
| African American (vs. white) | 1.03 (0.82–1.29) | 0.81 | ||
| Hispanic (vs. white) | 1.12 (0.88–1.42) | 0.37 | ||
| Asian/Pacific Islander (vs. white) | 1.56 (1.04–2.33) | 0.03 | ||
| American Indian (vs. white) | 0.90 (0.53–1.54) | 0.70 | ||
| Age at randomization (per year) | 1.00 (1.00–1.01) | 0.45 | ||
| Waist circumference (per cm) | 1.02 (1.02–1.03) | <0.0001 | ||
FIG. 1.
Diabetes incidence rate (case subjects/100 person-year) in each GRS quartile per treatment arm adjusted for age, sex, ethnic background, and waist circumference. Although the GRS was associated with diabetes incidence in the full cohort, not all P values reached nominal statistical significance in the stratified treatment arms (PBO, P = 0.152; MET, P = 0.039; ILS, P = 0.877). In the 4th quartile of GRS: PBO vs. MET, P = 0.315; PBO vs. ILS, P < 0.0001. PBO, placebo group; MET, metformin treatment arm; ILS, intensive lifestyle treatment arm.
We conducted sensitivity analyses in the largest ethnic group within the DPP (1,595 participants of European descent) to reduce the likelihood that population stratification was influencing our results. This subgroup is essentially free of non-European ethnic admixture (32). In the DPP, diabetes incidence did not differ by self-reported ethnicity, further minimizing concerns for such an effect (16). As hypothesized, a higher GRS was associated with an increased risk of progression to diabetes in white DPP participants (HR = 1.03 per risk allele [95% CI 1.00–1.06]; P = 0.04 in multivariable model including treatment, age, sex, and waist circumference). We performed the same multivariable models in each ethnic subgroup, but the results should be interpreted with caution because of the small numbers in those subgroups (Supplementary Table 2).
We derived two separate prediction models composed of nongenetic variables. The clinical prediction model contained variables deemed to be easily available in routine clinical care and was used to estimate the extent to which the available genetic information contributes to existing prediction tools. The physiological prediction model contains variables derived through metabolic investigations and was used to examine which area of diabetes physiology was best captured by the genetic data. The HRs for the weighted GRS and each variable included in the models are presented in Supplementary Table 3. The effect of the weighted GRS remained in the same direction but was not significantly associated with progression to diabetes when added to clinical or physiological models (HR = 1.01 per risk allele [0.99–1.03], P = 0.35 and HR = 1.01 per risk allele [0.99–1.04], P = 0.29, respectively). Using Madalla R2 to assess the amount of variation explained by each variable, glucose concentrations explained the largest R2 in both models (R2 = 8.58% for fasting glucose in the clinical model; R2 = 5.87% for fasting glucose; and R2 = 2.78% for 2-h glucose in the physiological model). Once glucose variables were excluded from the models, the weighted GRS was nominally associated with diabetes incidence (P = 0.04 in the clinical model; P = 0.02 in the physiological model).
We explored associations between each locus and progression to diabetes (Supplementary Table 4). Risk alleles located in or near HNF1A, PLEKHF2, and KCNJ11 showed nominally significant interactions with treatment arms to predict progression to diabetes.
Regression to NGR.
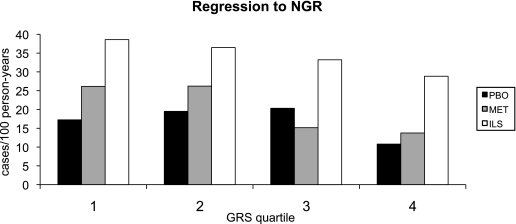
At baseline, all participants had impaired glucose tolerance (IGT) and elevated fasting glucose. A higher weighted GRS was strongly associated with a lower probability of regression toward NGR over the 3.2-year follow-up (HR = 0.95 per risk allele [0.93–0.98]; P < 0.0001) in the multivariable adjusted model (Table 5). Older age and larger waist circumference were also associated with a lower probability of regression to normoglycemia. Once again we did not detect significant interactions between the GRS and treatment arms (weighted GRS × metformin interaction, P = 0.98; weighted GRS × lifestyle interaction, P = 0.85). The incidence rate of regression to NGR (case subjects/100 person-years) in each quartile of GRS per treatment arm is illustrated in Fig. 2. The C-statistic value for prediction of regression to NGR was 0.646 for the multivariable model (adjusted for age, sex, waist, ethnic background, treatment arm); the C-statistic value increased to 0.658 when adding the weighted GRS in the model (P = 0.03). The IDI was nevertheless nonsignificant (IDI = −0.007; P = 0.10). When examining individuals at the highest genetic risk of type 2 diabetes (4th quartile of GRS), there was no statistical difference in regression to NGR between the placebo and metformin arms (P = 0.062); however, regression to NGR was significantly higher in the lifestyle intervention arm than in the placebo arm (P < 0.0001) (Fig. 2).
TABLE 5.
Multivariable models to predict regression to NGR in DPP participants
| Variables | HR (95% CI) | P |
|---|---|---|
| Weighted GRS (per risk allele) | 0.95 (0.93–0.98) | <0.0001 |
| Metformin (vs. placebo) | 1.26 (0.98–1.62) | 0.08 |
| Lifestyle (vs. placebo) | 2.38 (1.89–2.99) | <0.0001 |
| Female (vs. male) | 0.81 (0.66–0.99) | 0.04 |
| African American (vs. white) | 1.22 (0.95–1.58) | 0.12 |
| Hispanic (vs. white) | 0.96 (0.74–1.24) | 0.76 |
| Asian/Pacific Islander (vs. white) | 0.52 (0.31–0.84) | 0.009 |
| American Indian (vs. white) | 1.16 (0.63–2.14) | 0.64 |
| Age at randomization (per year) | 0.98 (0.97–0.99) | <0.0001 |
| Waist circumference (per cm) | 0.98 (0.98–0.99) | <0.0001 |
FIG. 2.
Regression rate (case subjects/100 person-year) to NGR in each GRS quartile per treatment arm adjusted for age, sex, ethnic background, and waist circumference. Although the GRS was associated with regression to NGR in the full cohort, not all P values reached nominal statistical significance in the stratified treatment arms (PBO, P = 0.123; MET, P = 0.001; ILS, P = 0.027). In the 4th quartile of GRS: PBO vs. MET, P = 0.062; PBO vs. ILS, P < 0.0001. PBO, placebo group; MET, metformin treatment arm; ILS, intensive lifestyle treatment arm.
Results of multivariable models in each ethnic subgroup are presented in Supplementary Table 2 and should be interpreted with caution because of the small numbers in some subgroups. In exploratory analyses testing associations between each single locus and regression to NGR, we observed significant interactions of treatment arms with SNPs located in or near PLEKHF2, MTNR1B, HHEX, and KCNJ11 (Supplementary Table 5).
DISCUSSION
We have shown that a weighted GRS based on 34 type 2 diabetes loci is associated with an increased risk of progression toward diabetes and a lower probability of regressing toward NGR over 3.2 years of follow-up in DPP participants, a population at high risk for type 2 diabetes. The association between the GRS and diabetes incidence was best revealed once we adjusted for major type 2 diabetes risk factors such as age, sex, ethnic background, and waist circumference. The effect size per risk allele was lower than that observed in the progression from normoglycemia to type 2 diabetes (8–10), reflecting the greater metabolic similarity at enrollment between DPP participants who went on to develop diabetes and those who did not. The GRS was also associated with lower insulinogenic index and higher proinsulin-to-insulin ratio at baseline, illustrating that most of the type 2 diabetes loci identified so far (and thus included in the weighted GRS) are related to β-cell function. The lifestyle intervention was effective in those with the highest genetic risk.
We observed an association between a higher GRS and greater estimated insulin sensitivity at baseline, which some might find paradoxical. It is likely that this paradoxical association is because of the narrow ascertainment criteria of the DPP at enrollment: to be included in the DPP at baseline, participants had to be glucose intolerant but not have type 2 diabetes. Therefore, those whose β-cell function was most impaired by their genetic burden should have had better insulin sensitivity to remain free of diabetes and fit into the DPP inclusion criteria. Along the same lines, we also observed an association between higher GRS and a better metabolic profile indicative of insulin sensitivity, in particular smaller waist circumference. Once again, this is likely because of the narrow DPP inclusion criteria where participants with decreased insulin secretory capacity at baseline had to have lower metabolic risk based on abdominal adiposity and its correlates (waist circumference, triglycerides, and HDL levels) to compensate for this deficiency and be included in the study. These observations further support our original conclusion that the observed association of the TCF7L2 diabetes risk genotype with a lower waist circumference was because of ascertainment artifact (11). Given that in the DPP a higher GRS is associated with a better metabolic profile and a different ethnic distribution at baseline, it was essential to adjust for those potential confounders in our investigations of diabetes incidence and regression to NGR. Indeed, the association between the GRS and diabetes incidence was strongest when adjusting for those important risk factors.
We also explored the association between genetic risk and diabetes incidence by constructing multivariable models using variables that are easily measured clinically or reflect diabetes pathophysiology. Covariates included in the clinical model were based on previous clinical models in a general population cohort (30). The GRS was not an independent predictor of diabetes in the clinical model, again showing that common known risk factors easily measurable in practice capture most of the predictive information. Moreover, the improvement in C-statistics and the IDI value after adding the GRS to our main multivariable model were not significant. This is in accordance with previous reports from general population cohorts showing that even if a GRS is associated with diabetes incidence, it adds little to the common known clinical risk factors (8,9). In both clinical and physiological models, once we removed glucose levels from the models, the GRS was significantly associated with diabetes incidence, suggesting that the association between genetic variants and diabetes risk is mainly explained through the influence of risk alleles on glucose levels. As shown previously, a GRS may perform better in younger populations or in cohorts with longer follow-up than was attained here; in both instances, genetic markers gain predictive ability in comparison with clinical characteristics (8–10).
With regard to preventive strategies, treatment with metformin or an intensive lifestyle intervention is effective at reducing the risk of diabetes incidence at any level of genetic risk. In fact, the data point toward the possibility that lifestyle could be even more effective in individuals with the highest GRS (Fig. 1). Nevertheless, the test for GRS by treatment interaction was not significant, and therefore we have no conclusive evidence that any treatment works better in a genetically defined category. In either case, genetic burden does not seem to undermine the DPP lifestyle intervention.
Perhaps as important as predicting progression to diabetes was the ability of the weighted GRS to predict regression to NGR. Clearly, the most desirable outcome is to return to a state of normal glucose homeostasis rather than remain in a high-risk state, such as IGT. We have previously reported both modifiable (lifestyle changes) and nonmodifiable (increasing age) factors that impact one’s ability to return to NGR (33). Most striking was the observation that diminished insulin secretion, not insulin sensitivity, impeded DPP participants from attaining NGR (33). In line with our previous report, we observed here that a higher GRS reflecting mostly impaired insulin secretory capacity at baseline was strongly associated with a lower chance to regress to NGR. This suggests that individuals with a high genetic risk of developing diabetes would need attention before they develop IGT if preventive interventions are to help them remain normoglycemic. On a positive note, we observed that even in individuals at high genetic risk, intensive lifestyle intervention was associated with a higher incidence of regression to NGR compared with metformin or placebo groups (Fig. 2).
Strengths and limitations.
The strengths of our study include that DPP is a randomized controlled trial with standardized glucose tolerance testing at regular intervals. SNP selection was based on the most recent findings from large meta-analyses (DIAGRAM+ and MAGIC), and genotyping was performed with high-quality control standards. However, our study also has limitations. All participants were classified as having IGT at baseline to be included in the study and could have regressed spontaneously to NGR; nevertheless, the rate of this spontaneous regression should not be different by treatment arm allocation. Imputation of missing genotypes slightly reduces the variance of the GRS, and thus results of multivariate models should be viewed with this in mind. The GRS was based on loci that were identified in populations of European descent (with the exception of KCNQ1); we tested our hypothesis in the overall DPP population (including participants of diverse ethnic backgrounds), which could raise the issue of population stratification. We addressed that issue by testing our hypothesis in the subgroup composed of white participants and found essentially the same level of association. Our analyses demonstrated that a GRS adds little to currently used phenotypic risk factors, and so clinical practice should continue to focus on well-established and easily measureable diabetes risk factors such as age, central adiposity, and glycemic levels (and/or other components of the metabolic syndrome). Analyses testing associations between each locus and progression to diabetes or regression to NGR were exploratory and should be interpreted with caution because P values were not corrected for multiple testing. Observations in each treatment arm also need to be interpreted with caution because we did not find a significant interaction between the GRS and treatment arms; in addition, the study might be underpowered to detect interactions by treatment. However, it is interesting to observe that DPP participants benefited from lifestyle intervention and showed lower progression to diabetes and a higher probability of regression to NGR, even in the highest quartile of GRS.
Conclusions.
In summary, we demonstrated that a higher type 2 diabetes genetic risk estimated with a score built from 34 known type 2 diabetes loci is associated with a greater likelihood of progressing toward diabetes and a lower likelihood of regressing to NGR in DPP participants. Currently, most of the loci identified as increasing the risk of type 2 diabetes are implicated in β-cell function. It is therefore not surprising that our GRS was associated with lower insulin secretion and impaired insulin processing at baseline. From a public health perspective, the knowledge that individuals with IGT who also have a high GRS have a greater impairment of β-cell function and a lower chance of regression toward NGR, suggesting that they may deserve medical attention before they reach this impaired status. Whether they would benefit from an earlier preventive intervention strategy remains to be tested.
ACKNOWLEDGMENTS
The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institutes of Health provided funding to the clinical centers and the Coordinating Center for the design and conduct of the study, and the collection, management, analysis, and interpretation of the data. The Southwestern American Indian Centers were supported directly by the NIDDK and the Indian Health Service. The General Clinical Research Center Program, National Center for Research Resources, and the Department of Veterans Affairs supported data collection at many of the clinical centers. Funding for data collection and participant support was also provided by the Office of Research on Minority Health, the National Institute of Child Health and Human Development, the National Institute on Aging, the Office of Research on Women’s Health, the Centers for Disease Control and Prevention, and the American Diabetes Association. This research was also supported, in part, by the intramural research program of the NIDDK.
This work was also funded by R01 DK072041 (to D.A., J.C.F., and K.A.J.). J.C.F. is also supported by Massachusetts General Hospital and a Clinical Scientist Development Award by the Doris Duke Charitable Foundation. This work was partially supported by a Doris Duke Charitable Foundation Distinguished Scientist Clinical Award to D.A. J.B.Me. is supported by NIDDK K24 DK080140. P.W.F. was supported by the Swedish Research Council, Novo Nordisk, the Swedish Diabetes Association, and the Swedish Heart-Lung Foundation. J.B.Me. currently has a research grant from GlaxoSmithKline and serves on a consultancy board for Interleukin Genetics. J.C.F. has received consulting honoraria from Daiichi-Sankyo and AstraZeneca.
Bristol-Myers Squibb and Parke-Davis provided medication. LifeScan Inc., Health O Meter, Hoechst Marion Roussel, Merck-Medco Managed Care, Merck and Co., Nike Sports Marketing, Slim-Fast Foods Co., and Quaker Oats Co. donated materials, equipment, or medicines for concomitant conditions. McKesson BioServices Corp., Matthews Media Group, and the Henry M. Jackson Foundation provided support services under subcontract with the Coordinating Center. No other potential conflicts of interest relevant to this article were reported.
SNPs were selected by J.C.F., D.A., J.B.Me., and R.S. based on previous meta-analyses, including unpublished data from the DIAGRAM Consortium. J.B.Mc. directed the genotyping with supervision from J.C.F. Recruitment and phenotyping were performed previously by the DPP Research Group. K.A.J. conducted statistical analyses with input from P.W.F. and W.C.K. M.-F.H., L.P., and J.C.F. wrote the manuscript. All authors (M.-F.H., K.A.J., L.P., R.S., J.B.Mc., P.W.F., R.F.H., S.E.K., S.H., J.B.Me., D.A., W.C.K., and J.C.F.) contributed to discussion and reviewed and edited the manuscript.
A complete list of centers, investigators, and staff can be found in the Supplementary Data.
The investigators gratefully acknowledge the commitment and dedication of the participants of the DPP.
Footnotes
Clinical trial reg. no. NCT00004992, clinicaltrials.gov.
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db10-1119/-/DC1.
The opinions expressed are those of the investigators and do not necessarily reflect the views of the Indian Health Service or other funding agencies.
*Complete lists of the members of the Diabetes Prevention Program Research Group and the DIAGRAM investigators are provided in the Supplementary Data.
REFERENCES
- 1.McCarthy MI, Zeggini E. Genome-wide association studies in type 2 diabetes. Curr Diab Rep 2009;9:164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stolerman ES, Florez JC. Genomics of type 2 diabetes mellitus: implications for the clinician. Nat Rev Endocrinol 2009;5:429–436 [DOI] [PubMed] [Google Scholar]
- 3.Zeggini E, Scott LJ, Saxena R, et al. Wellcome Trust Case Control Consortium Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008;40:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voight BF, Scott LJ, Steinthorsdottir V, et al. MAGIC investigators. GIANT Consortium Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010;42:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupuis J, Langenberg C, Prokopenko I, et al. DIAGRAM Consortium. GIANT Consortium. Global BPgen Consortium. Anders Hamsten on behalf of Procardis Consortium. MAGIC investigators New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saxena R, Hivert MF, Langenberg C, et al. GIANT consortium. MAGIC investigators Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet 2010;42:142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soranzo N, Sanna S, Wheeler E, et al. Common variants at 10 genomic loci influence hemoglobin A1C levels via glycemic and nonglycemic pathways. Diabetes 2010;59:3229–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyssenko V, Jonsson A, Almgren P, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med 2008;359:2220–2232 [DOI] [PubMed] [Google Scholar]
- 9.Meigs JB, Shrader P, Sullivan LM, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med 2008;359:2208–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Miguel-Yanes JM, Shrader P, Pencina MJ, et al. Genetic risk reclassification for type 2 diabetes by age below or above 50 years using 40 type 2 diabetes risk single nucleotide polymorphisms. Diabetes Care 2011;34:121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Florez JC, Jablonski KA, Bayley N, et al. Diabetes Prevention Program Research Group TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 2006;355:241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore AF, Jablonski KA, McAteer JB, et al. Diabetes Prevention Program Research Group Extension of type 2 diabetes genome-wide association scan results in the Diabetes Prevention Program. Diabetes 2008;57:2503–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Florez JC, Jablonski KA, McAteer J, et al. Diabetes Prevention Program Research Group Testing of diabetes-associated WFS1 polymorphisms in the Diabetes Prevention Program. Diabetologia 2008;51:451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Diabetes Prevention Program Research Group The Diabetes Prevention Program: design and methods for a clinical trial in the prevention of type 2 diabetes. Diabetes Care 1999;22:623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Diabetes Prevention Program Research Group The Diabetes Prevention Program: baseline characteristics of the randomized cohort. Diabetes Care 2000;23:1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knowler WC, Barrett-Connor E, Fowler SE, et al. Diabetes Prevention Program Research Group Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 2002;346:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–419 [DOI] [PubMed] [Google Scholar]
- 18.Byrne CD, Wareham NJ, Brown DC, et al. Hypertriglyceridaemia in subjects with normal and abnormal glucose tolerance: relative contributions of insulin secretion, insulin resistance and suppression of plasma non-esterified fatty acids. Diabetologia 1994;37:889–896 [DOI] [PubMed] [Google Scholar]
- 19.Utzschneider KM, Prigeon RL, Faulenbach MV, et al. Oral disposition index predicts the development of future diabetes above and beyond fasting and 2-h glucose levels. Diabetes Care 2009;32:335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandhu MS, Weedon MN, Fawcett KA, et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet 2007;39:951–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet 2009;41:1110–1115 [DOI] [PubMed] [Google Scholar]
- 22.Yasuda K, Miyake K, Horikawa Y, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 2008;40:1092–1097 [DOI] [PubMed] [Google Scholar]
- 23.Unoki H, Takahashi A, Kawaguchi T, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 2008;40:1098–1102 [DOI] [PubMed] [Google Scholar]
- 24.Tang K, Fu DJ, Julien D, Braun A, Cantor CR, Köster H. Chip-based genotyping by mass spectrometry. Proc Natl Acad Sci USA 1999;96:10016–10020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cornelis MC, Qi L, Zhang C, et al. Joint effects of common genetic variants on the risk for type 2 diabetes in U.S. men and women of European ancestry. Ann Intern Med 2009;150:541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grønnesby JK, Borgan O. A method for checking regression models in survival analysis based on the risk score. Lifetime Data Anal 1996;2:315–328 [DOI] [PubMed] [Google Scholar]
- 27.Parzen M, Lipsitz SR. A global goodness-of-fit statistic for Cox regression models. Biometrics 1999;55:580–584 [DOI] [PubMed] [Google Scholar]
- 28.Pencina MJ, D’Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med 2004;23:2109–2123 [DOI] [PubMed] [Google Scholar]
- 29.Pencina MJ, D'Agostino RB Sr, D'Agostino RB Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 2008;27:157–172; discussion 207-112 [DOI] [PubMed]
- 30.Wilson PW, Meigs JB, Sullivan L, Fox CS, Nathan DM, D’Agostino RB., Sr Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch Intern Med 2007;167:1068–1074 [DOI] [PubMed] [Google Scholar]
- 31.Lachin JM. Biostatistical methods, the assessment of relative risks. New York, John Wiley & Sons, Inc., 2000 [Google Scholar]
- 32.Jablonski KA, McAteer JB, de Bakker PI, et al. ; Florez JC for the Diabetes Prevention Program Research Group. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle interventions in the Diabetes Prevention Program. Diabetes 2010;59:2672–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perreault L, Kahn SE, Christophi CA, Knowler WC, Hamman RF, Diabetes Prevention Program Research Group Regression from pre-diabetes to normal glucose regulation in the diabetes prevention program. Diabetes Care 2009;32:1583–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]