Abstract
OBJECTIVE
Nominally resistant mouse strains such as C57BL/6 (B6) harbor latent type 1 diabetes susceptibility genes uncovered in outcross to disease-susceptible NOD mice. However, identification of possible recessively acting B6-derived susceptibility genes is limited because very few F2 progeny derived from outcrossing this strain with NOD develop spontaneous autoimmune diabetes. Thus, we assessed whether a transgenic T-cell receptor (TCR) disease transfer model allowed the mapping of recessively acting B6 genetic loci that in the proper context contribute to diabetes.
RESEARCH DESIGN AND METHODS
CD8 T-cells transgenically expressing the diabetogenic AI4 TCR were transferred into 91 (NODxB6.H2g7)F1xB6.H2g7 first-backcross (BC1) females. A genome-wide scan was performed for loci affecting clinical diabetes and insulitis severity.
RESULTS
A major locus on chromosome 11 in tight linkage with the marker D11Mit48 (logarithm of odds score = 13.2) strongly determined whether BC1 progeny were susceptible to AI4 T-cell–mediated diabetes. Mice homozygous versus heterozygous for B6 markers of this chromosome 11 genetic locus were, respectively, highly susceptible or resistant to AI4-induced insulitis and diabetes. The genetic effect is manifest by host CD4 T-cells. Microarray analyses of mRNA transcript expression identified a limited number of candidate genes.
CONCLUSIONS
The distal region of chromosome 11 in B6 mice harbors a previously unrecognized recessively acting gene(s) that can promote autoreactive diabetogenic CD8 T-cell responses. Future identification of this gene(s) may further aid the screening of heterogeneous humans at future risk for diabetes, and might also provide a target for possible disease interventions.
Although particular major histocompatibility complex (MHC) haplotypes are the strongest contributor to T-cell–mediated autoimmune type 1 diabetes development in both humans and NOD mice, disease pathogenesis requires interactions with multiple other susceptibility (Idd) genes (1). This is illustrated by the fact that C57BL/6 background mice congenic for the NOD-derived H2g7 MHC haplotype (B6.H2g7) are normally diabetes-resistant (2). However, certain T-cell receptor (TCR) molecules contributing to diabetes development in NOD mice paradoxically exert even greater pathogenic activity when transgenically expressed in the B6.H2g7 strain (3,4). This finding, along with previous linkage analyses and congenic approach studies (5,6), indicate normally diabetes-resistant strains harbor some genes that in the proper combination can actually contribute to aggressive disease development.
Analyses of F2 rather than first backcross (BC1) progeny is the preferable approach for mapping diabetic genes, because this allows for the identification of both susceptibility or resistance variants from NOD mice or the outcross partner strain. However, identifying possible recessively acting B6-derived susceptibility genes after the outcross to NOD has been hampered because very few F2 progeny develop spontaneous diabetes even when the H2g7 MHC is fixed in all segregants (7). Nevertheless, analyses of progeny from a first backcross to B6.H2g7 mice revealed recessive alleles from this strain on chromosomes 1, 2, 7, and 15 promoting the diabetogenic activity of CD4 T-cells transgenically expressing the BDC2.5 TCR (3). We now report a similar BC1 strategy using another transgenic TCR that reveals at least one additional B6 origin recessive Idd susceptibility gene on chromosome 11 contributing to the peripheral activation of pathogenic CD8 T-cells.
RESEARCH DESIGN AND METHODS
Mice.
NOD/ShiLtDvs mice are maintained in a specific pathogen-free research colony. B6.H2g7 mice are maintained at the N8 backcross generation (4). NOD mice transgenically expressing the TCR from the diabetogenic CD8+ T-cell clone AI4 plus a functionally inactivated Rag1 gene (NOD.Rag1null.AI4) have been described (8).
Adoptive transfer of diabetes.
Indicated female mice were sublethally irradiated (600 R) and injected intravenously with 1 × 107 NOD.Rag1null.AI4 splenocytes to induce diabetes. In one experiment, recipients were injected intraperitoneally with 250 μg/mouse of the CD25-depleting PC61.5 antibody 1 day before AI4 T-cell transfer. In another experiment, donor cells were prelabeled with 2.5 μmol/L carboxy fluorescein succinimidyl ester (CFSE). After 4 days, viable AI4 T-cells from collagenase d-digested spleen and pancreatic lymph nodes were identified by flow cytometry using CFSE and a CD8-specific antibody (53–6.7). Expression of various T-cell surface markers was assessed using antibodies specific for CD44 (IM7.8.1), CD25 (PC61.5), and CD62 L (MEL-14). Another experiment used female mice injected with AI4 T-cells preactivated in culture for 3 days with 100 nmol/L antigenic mimotope peptide YFIENYLEL and 50 units/mL interleukin 2.
In a separate experiment, female B6.H2g7 mice were lethally irradiated (1,200 R) and injected intravenously with 5 × 106 bone marrow cells from BC1 progeny heterozygous or homozygous for the microsatellite marker D11Mit48. Fourteen weeks later, recipient mice were sublethally irradiated and injected intravenously with 1 × 107 NOD.Rag1null.AI4 splenocytes to induce diabetes. In another study, B6.H2g7, NOD and F1 mice all homozygous for the Rag1null mutation were injected with 1 × 107 NOD.Rag1null.AI4 splenocytes. Finally, magnetic bead purified CD4 T-cells from D11Mit48B6/B6 or D11Mit48NOD/B6 BC1 progeny were cotransferred with 1 × 107 NOD.Rag1null.AI4 splenocytes into B6.H2g7.Rag1null recipients. Recipients were either killed at the indicated time or monitored for diabetes development.
Assessment of diabetes and insulitis.
Diabetes was assessed by daily monitoring of glycosuria with Ames Diastix (Bayer, Diagnostics Division, Elkhart, IN), with disease onset defined by two consecutive values of ≥3. Previously described criteria (9) were used to establish insulitis scores ranging from 0 (individual islet with no leukocytic infiltration, normal β-cell mass) to 4 (complete destruction) for the indicated mice.
Genotyping and linkage analyses.
BC1 progeny were genotyped as previously described (10) for 131 single nucleotide polymorphisms (SNP) at approximately 20-Mb intervals across the genome (National Center for Biotechnology Information [NCBI] build 37 marker positions). Linkage markers for genes controlling insulitis severity and diabetes development in response to AI4 T-cell transfer were identified as previously described (11). PCR typing of the polymorphic D11Mit48 microsatellite marker was also done.
Microarray analyses.
CD4 T-cells were purified from BC1 progeny either heterozygous or homozygous for D11Mit48 and cotransferred into female B6.H2g7.Rag1null recipients with 1 × 107 NOD.Rag1null.AI4 splenocytes. RNA was isolated from CD4 T-cells sorted from spleens of these recipients by flow cytometry ∼3 weeks after adoptive transfer. Three biologic replicates were generated for both genotypes, with each sample emanating from purified CD4 T-cells pooled from two donor mice. Microarray analysis of comparative CD4 T-cell gene expression was conducted using the Affymetrix 430v2 GeneChip array (Santa Clara, CA). R/Bioconductor software summarized the probe intensities for each gene using the robust multiaverage method (12,13). R/MAANOVA software (Churchill Group, Bar Harbor, ME) was used to generate lists of differentially expressed genes between the tested samples (14). Differentially expressed genes were identified by using ts, a modified t statistic incorporating shrinkage estimates of variance components from within the R/MAANOVA (15).
RESULTS
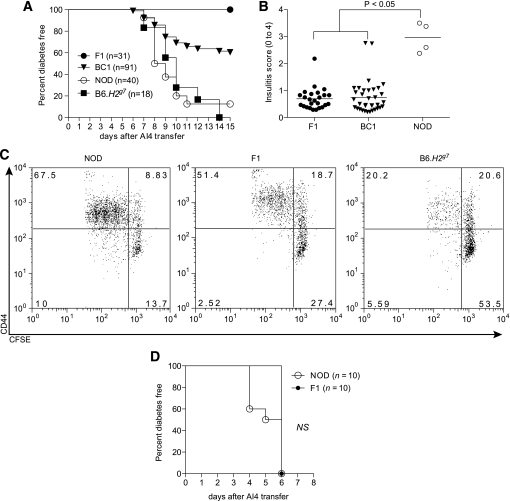
We previously developed NOD mice transgenically expressing the TCR from the diabetogenic AI4 CD8 T-cell clone and also homozygous for the Rag1null mutation (NOD.Rag1null.AI4). Adoptively transferred AI4 T-cells from such donors rapidly induce diabetes in both sublethally irradiated NOD and B6.H2g7 mice (16). Thus, we were surprised AI4 T-cells failed to transfer diabetes or significant levels of insulitis to sublethally irradiated (NODxB6.H2g7)F1 hybrids (Fig. 1A and B). This difference was not due to varying post-transfer stimulation of AI4 T-cells within pancreatic lymph nodes, because the proliferation of such effectors at this site in F1 hybrids was similar to that of NOD mice and even greater than in B6.H2g7 recipients (Fig. 1C). Furthermore, no differences in expression of CD44, CD25, and CD62 L activation markers were detected between AI4 T-cells isolated from the parental or F1 mice (Fig. 1C and data not shown). However, AI4 T-cells preactivated in culture rapidly transferred diabetes to F1 recipients (Fig. 1D). Hence, the F1 genetic environment allows for less efficient initial pathogenic activation of diabetogenic CD8 T-cells than in either parental strain.
FIG. 1.
NOD and B6.H2g7 but not (NODxB6.H2g7)F1 mice succumb to AI4 T-cell–induced diabetes. A: Incidence of diabetes in 6- to 8-week-old female NOD, B6.H2g7, F1, and (F1xB6.H2g7)BC1 recipient mice that were sublethally irradiated (600 R) and injected intravenously with 1 × 107 NOD.Rag1null.AI4 splenocytes. Incidence of diabetes in BC1 recipients was significantly different (P < 0.0001) from NOD, B6.H2g7, and F1 mice. B: Insulitis scores (0 = no insulitis to 4 = no remaining islet cell mass) for surviving nondiabetic NOD.Rag1null.AI4 splenocyte recipients. Insulitis severity was significantly greater in surviving NOD mice compared with F1 and BC1 recipients according to the Mann-Whitney test. C: In vivo proliferation and activation of CFSE-labeled NOD.Rag1null.AI4 T-cells at 4 days after transfer in pancreatic lymph nodes of NOD, B6.H2g7, and F1 mice. D: Incidence of diabetes in 6- to 8-week-old sublethally irradiated female NOD and F1 recipients of in vitro activated AI4 T-cells.
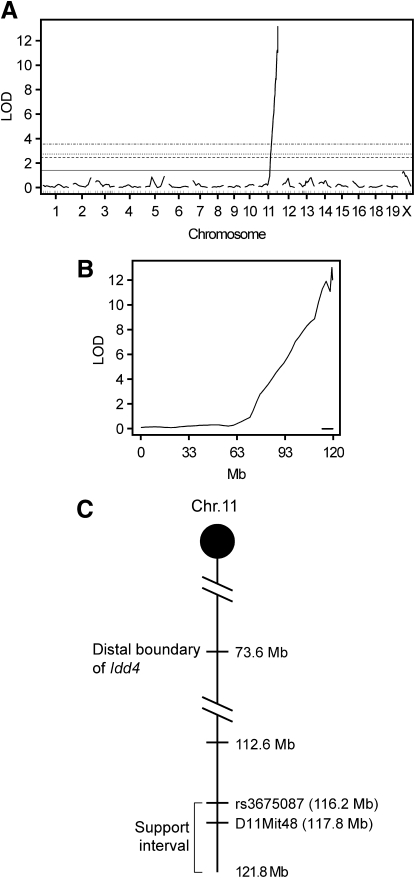
F1 hybrid resistance to AI4-induced diabetes indicates the NOD and B6.H2g7 genomes harbor separate recessively acting alleles supporting pathogenic CD8 T-cell activation. To map such unrecognized recessive B6 alleles, NOD.Rag1null.AI4 splenocytes were transferred into 91 sublethally irradiated female (NODxB6.H2g7)F1xB6.H2g7 BC1 progeny that were then monitored for diabetes development and also genotyped. Diabetes developed in 40% of BC1 segregants at a rate similar to both NOD and B6.H2g7 mice (Fig. 1A). Nondiabetic BC1 mice were examined for insulitis levels. Similar to F1 hybrids, insulitis levels were low in nondiabetic BC1 mice (Fig. 1B). This sharp dichotomy among BC1 progeny suggested that rather than contributions from multiple loci, which would result in a broad spectrum of insulitis scores, a limited number of genes rendered B6.H2g7 mice susceptible to AI4 T-cell–induced diabetes. Indeed, a SNP-based genome-wide one-dimensional scan of BC1 progeny revealed only one B6 genomic region on chromosome 11 that was highly linked to diabetes susceptibility and insulitis (logarithm of odds [LOD] score = 13.2; Fig. 2A).
FIG. 2.
A single genetic locus primarily contributes to B6.H2g7 susceptibility to AI4 T-cell–induced diabetes. Whole genome (A) and chromosome 11–specific (B) LOD score analysis of SNP markers linked to diabetes susceptibility and insulitis severity in sublethally irradiated BC1 recipients of 1 × 107 NOD.Rag1null.AI4 splenocytes. Horizontal lines depict LOD scores indicative of 1, 5, 10, and 63% linkage support thresholds. The 1 and 63% thresholds, respectively, indicate significant and suggestive linkage. The lower bold line indicates the 95% CI for linkage. C: Schematic representation of the distal region of chromosome 11. The positions of the markers (Mb) are based on NCBI build 37. The distance between markers is not drawn to scale.
On the basis of original typing of SNP markers only, the 95% CI for the region of interest was originally narrowed to a 3.6-Mb segment at the distal end of chromosome 11 (112.6–116.2 Mb), ∼40 Mb below the previously identified Idd4 locus (Fig. 2B and C). Susceptibility to AI4 T-cell–induced diabetes/insulitis was associated with homozygosity for B6 markers within this region of chromosome 11, with LOD scores increasing up to and including the most distal NOD/B6 distinguishing SNP (rs3675087 at 116.2 Mb) that was typed. However, it remained possible that a gene(s) controlling susceptibility to AI4 transfer resided distally to the 116.2-Mb position. Thus, BC1 progeny were regenotyped for the polymorphic microsatellite marker D11Mit48 located at 117.76 Mb. Linkage between B6 homozygosity at D11Mit48 and diabetes/insulitis was very similar to that for rs3675087 (Supplementary Table 1). Sanger sequence analysis (www.sanger.ac.uk) indicated there are NOD/B6 polymorphisms in protein-encoding regions of two genes distal to D11Mit48, but these were not covered by the SNP typing panel available to us. For this reason, we redefined the support interval controlling differential sensitivity to type 1 diabetes induced by transferred AI4 T-cells to between 112.6 Mb and the end of chromosome 11 (121.8 Mb; Fig. 2C).
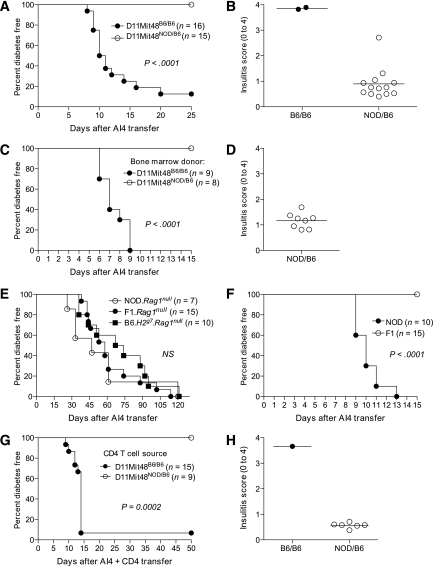
We tested whether genotyping BC1 mice for D11Mit48 alone could predict susceptibility to AI4 T-cell–induced diabetes. An additional cohort of 31 BC1 mice was genotyped for the D11Mit48 polymorphism before receiving NOD.Rag1null.AI4 splenocytes. None of the 15 heterozygous (D11Mit48NOD/B6) animals developed diabetes and usually no more than mild peri-insulitis (Fig. 3A and B). Conversely, 14 of 16 homozygous (D11Mit48B6/B6) mice developed diabetes, and the remaining two mice were severely insulitic (Fig. 3A and B). This simple segregation pattern strongly indicates that a recessively acting B6 origin gene(s) tightly linked to D11Mit48 is a primary contributor to the pathogenic activation of diabetogenic CD8 T-cells, whereas the NOD allelic variant actually dominantly suppresses this process. However, the protective effect of this NOD chromosome 11 allelic variant must normally be masked by the large number of other diabetes susceptibility genes characterizing this strain.
FIG. 3.
A polymorphic gene(s) in close linkage with the D11Mit48 microsatellite marker controls susceptibility to AI4 T-cell–induced diabetes through effects on a CD4 T-cell population other than CD25+ Tregs. A: Mice homozygous for the B6 allele (B6/B6) vs. heterozygous (NOD/B6) for D11Mit48 were, respectively, highly susceptible and resistant to AI4 T-cell–induced diabetes. Results represent three independent experiments. B: Insulitis scores are shown for nondiabetic heterozygous and homozygous BC1 NOD.Rag1null.AI4 splenocyte recipients. C and D: B6.H2g7 mice previously reconstituted with bone marrow from D11Mit48B6/B6 or D11Mit48NOD/B6 BC1 segregants are, respectively, susceptible or resistant to diabetes and insulitis induced by subsequently infused AI4 T-cells. E: Incidence of diabetes in 6- to 8-week-old female B6.H2g7.Rag1null, NOD.Rag1null, and F1.Rag1null recipients of 1 × 107 NOD.Rag1null.AI4 splenocytes. F: Incidence of diabetes in 6- to 8-week-old sublethally irradiated NOD and F1 recipients of 1 × 107 NOD.Rag1null.AI4 splenocytes. Recipients were also injected intraperitoneally with a CD25-depleting antibody (PC61 250 μg/mouse) 1 day before AI4 T-cell transfer. G and H: B6.H2g7.Rag1null mice infused with purified CD4+ T-cells from D11Mit48B6/B6 or D11Mit48NOD/B6 BC1 segregants are, respectively, susceptible or resistant to diabetes and insulitis induced by subsequently infused AI4 T-cells.
To determine whether the chromosome 11 gene(s) controls diabetes susceptibility through effects on hematopoietic cells, NOD.Rag1null.AI4 splenocytes were transferred into B6.H2g7 mice previously reconstituted with bone marrow from D11Mit48B6/B6 or D11Mit48NOD/B6 BC1 progeny. Only recipients of D11Mit48B6/B6 but not D11Mit48NOD/B6 bone marrow developed diabetes (Fig. 3C) or significant levels of insulitis (Fig. 3D). Therefore, the D11Mit48-linked gene(s) controlling susceptibility to AI4 T-cell–mediated diabetic functions through a hematopoietic cell population(s).
Next, we established this gene(s) controls type 1 diabetes susceptibility through a lymphocyte population(s). This was determined by demonstrating NOD.Rag1null.AI4 splenocytes transferred diabetes with equal efficiency to B6.H2g7, NOD, and F1 mice all homozygous for the Rag1null mutation, eliminating all endogenous lymphocytes (Fig. 3E). We tested whether regulatory T-cells (Tregs) were the lymphocyte population rendering F1 mice resistant to AI4-mediated diabetes. NOD and F1 mice were treated with a CD25-depleting antibody 1 day before receiving NOD.Rag1null.AI4 splenocytes. This eliminated most CD4+CD25+ Tregs for the duration of the 15-day postadoptive transfer period during which AI4 T-cells normally induce diabetes (Supplementary Fig. 1). Anti-CD25–treated F1 mice remained resistant to AI4-induced diabetes (Fig. 3F).
We next tested whether the chromosome 11 gene(s) controls type 1 diabetes susceptibility through effects on a CD4 T-cell population other than Tregs. Total CD4 T-cells purified from D11Mit48B6/B6 or D11Mit48NOD/B6 BC1 progeny were cotransferred with NOD.Rag1null.AI4 splenocytes into B6.H2g7.Rag1null recipients. Only recipients of D11Mit48B6/B6 but not D11Mit48NOD/B6 CD4 T-cells developed AI4 T-cell–induced diabetes or high levels of insulitis (Fig. 3G and H). Therefore, the D11Mit48-linked gene(s) controlling susceptibility to AI4 T-cell–mediated diabetic functions through a non-Treg CD4 T-cell population(s).
We used microarray-based comparisons of mRNA transcript levels to identify candidates for a CD4 T-cell–expressed gene(s) within the chromosome 11 support interval regulating pathogenic activation of diabetogenic AI4 CD8 T-cells. This was accomplished by recovering CD4 T-cells purified from D11Mit48B6/B6 and D11Mit48NOD/B6 BC1 progeny that were previously cotransferred into B6.H2g7.Rag1null recipients with NOD.Rag1null.AI4 splenocytes. CD4 T-cells were repurified from spleens once all recipients of D11Mit48B6/B6 CD4 T-cells developed diabetes (all D11Mit48NOD/B6 CD4 T-cell recipients were disease-free). Table 1 describes 12 genes mapping within the chromosome 11 support interval (112.6–121.8 Mb) that were differentially expressed by these two classes of CD4 T-cells and also characterized by NOD/B6 polymorphisms (www.sanger.ac.uk). Interestingly, three of these genes (Rab37, Slc9a3r1, and CD300lf) map to the PSORS2 psoriasis susceptibility locus in humans (17–19).
TABLE 1.
List of polymorphic genes within the chromosome 11 interval 112.6 to 121.8 Mb that are differentially expressed in CD4 T-cells purified from D11Mit48B6/B6 vs. D11Mit48NOD/B6 genotyped BC1 progeny
| Gene | Position (Mb)* | Description | Relative fold change: NOD/B6 over B6/B6 | Fs P | Function† |
|---|---|---|---|---|---|
| Socs3 | 117.8 | Suppressor of cytokine signaling 3 | −1.4 | 0.0529 | Inhibits activation and/or differentiation pathways in macrophages, dendritic cells, and T-cells |
| Mfsd11 | 116.7 | Major facilitator superfamily domain containing 11 | −1.4 | 0.0116 | Unknown |
| 1110017F19Rik | 115.8 | RIKEN cDNA | −1.3 | 0.0403 | Unknown |
| Slc9a3r1 | 115.0 | Solute carrier family 9 | 1.1 | 0.0387 | Multifunctional adaptor protein, recruiting cytoplasmic signaling proteins and membrane receptors/transporters into functional complexes. Defective regulation is linked with susceptibility to psoriasis |
| Cant1 | 118.3 | Calcium-activated nucleotidase 1 | 1.2 | 0.0088 | Calcium ion binding, nucleotide metabolism |
| Nt5c | 115.4 | 5′,3′-nucleotidase, cytosolic | 1.2 | 0.0210 | Unknown |
| Nploc4 | 120.2 | Nuclear protein localization 4 homolog | 1.2 | 0.0326 | Unknown |
| Rab37 | 115.0 | RAB37, member of RAS oncogene family | 2.1 | 0.0023 | GTPase expressed in mast cells |
| Fignl1 | 116.9 | Fidgetin-like 1 | 2.1 | 0.0072 | Unknown |
| BC018473 | 116.6 | cDNA sequence BC018473 | 2.4 | 0.0392 | Unknown |
| Cd300lf | 115.0 | CD300 antigen like family member F | 2.6 | 0.0089 | Member of an immunoglobulin superfamily gene cluster that may serve as an inhibitory receptor to regulate the maturation and differentiation of immune cells, helping to contain inflammation |
| St6galnac1 | 116.6 | ST6 (α-N-acetyl-neuraminyl-2,3-β-galactosyl-1,3)-N-acetylgalactosaminide α-2,6-sialyltransferase 1 | 2.6 | 0.0055 | Component of the transmembrane laminin glycoprotein receptor |
*Marker positions were taken from NCBI build 37.1 (www.ncbi.nlm.nih.gov).
DISCUSSION
Although the NOD genome contains a collection of genes supporting development of spontaneous diabetes, additional susceptibility alleles remain masked within normally disease-resistant mouse strains. Indeed, we found the distal region of chromosome 11 in B6.H2g7 mice harbors what is likely a single recessive gene acting through CD4 T-cells that more strongly promotes pathogenic activation of diabetogenic CD8 T-cells than the NOD allelic variant. Presumably, the normal efficient thymic deletion of pathogenic CD8 T-cells in B6.H2g7 mice (4,11) masks the activity of their prodiabetogenic gene(s) on chromosome 11. Determination of diabetes susceptibility genes hidden in normally resistant mouse strains might ultimately aid the identification of individuals among heterogeneous humans who are at high risk of future disease.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants DK-46266/DK-51090, Cancer Center Support Grant CA34196, and grants from the Juvenile Diabetes Research Foundation.
No potential conflicts of interest relevant to this article were reported.
J.P.D. researched data and wrote the manuscript. Y.-G.C. contributed to discussion. W.Z. carried out statistical analyses. S.A. researched data. D.V.S. directed research and wrote the manuscript.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db10-0885/-/DC1.
REFERENCES
- 1.Driver JP, Serreze DV, Chen YG. Mouse models for the study of autoimmune type-1 diabetes: a NOD to similarities and differences to human disease. Semin Immunopathol 2011;33:67–87 [DOI] [PubMed] [Google Scholar]
- 2.Yui MA, Muralidharan K, Moreno-Altamirano B, Perrin G, Chestnut K, Wakeland EK. Production of congenic mouse strains carrying NOD-derived diabetogenic genetic intervals: an approach for the genetic dissection of complex traits. Mamm Genome 1996;7:331–334 [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez A, Katz JD, Mattei M-G, Kikutani H, Benoist C, Mathis D. Genetic control of diabetes progression. Immunity 1997;7:873–883 [DOI] [PubMed] [Google Scholar]
- 4.Choisy-Rossi CM, Holl TM, Pierce MA, Chapman HD, Serreze DV. Enhanced pathogenicity of diabetogenic T cells escaping a non-MHC gene-controlled near death experience. J Immunol 2004;173:3791–3800 [DOI] [PubMed] [Google Scholar]
- 5.Hunter K, Rainbow D, Plagnol V, Todd JA, Peterson LB, Wicker LS. Interactions between Idd5.1/Ctla4 and other type 1 diabetes genes. J Immunol 2007;179:8341–8349 [DOI] [PubMed] [Google Scholar]
- 6.Ridgway WM, Peterson LB, Todd JA, et al. Gene-gene interactions in the NOD mouse model of type 1 diabetes. Adv Immunol 2008;100:151–175 [DOI] [PubMed] [Google Scholar]
- 7.Wicker LS, Miller BJ, Coker LZ, et al. Genetic control of diabetes and insulitis in the nonobese diabetic (NOD) mouse. J Exp Med 1987;165:1639–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiLorenzo TP, Lieberman SM, Takaki T, et al. During the early prediabetic period in NOD mice, the pathogenic CD8(+) T-cell population comprises multiple antigenic specificities. Clin Immunol 2002;105:332–341 [DOI] [PubMed] [Google Scholar]
- 9.Takaki T, Marron MP, Mathews CE, et al. HLA-A*0201-restricted T cells from humanized NOD mice recognize autoantigens of potential clinical relevance to type 1 diabetes. J Immunol 2006;176:3257–3265 [DOI] [PubMed] [Google Scholar]
- 10.Myakishev MV, Khripin Y, Hu S, Hamer DH. High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer-labeled primers. Genome Res 2001;11:163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serreze DV, Choisy-Rossi CM, Grier AE, et al. Through regulation of TCR expression levels, an Idd7 region gene(s) interactively contributes to the impaired thymic deletion of autoreactive diabetogenic CD8+ T cells in nonobese diabetic mice. J Immunol 2008;180:3250–3259 [DOI] [PubMed] [Google Scholar]
- 12.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249–264 [DOI] [PubMed] [Google Scholar]
- 13.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003;31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, Kerr MK, Cui J, Churchill G. MAANOVA: A Software Package for the Analysis Spotted cDNA Microarray Experiments. New York, Springer, 2003 [Google Scholar]
- 15.Cui X, Hwang JT, Qiu J, Blades NJ, Churchill GA. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics 2005;6:59–75 [DOI] [PubMed] [Google Scholar]
- 16.Driver JP, Scheuplein F, Chen YG, Grier AE, Wilson SB, Serreze DV. Invariant natural killer T-cell control of type 1 diabetes: a dendritic cell genetic decision of a silver bullet or Russian roulette. Diabetes 2010;59:423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danik JS, Paré G, Chasman DI, et al. Novel loci, including those related to Crohn disease, psoriasis, and inflammation, identified in a genome-wide association study of fibrinogen in 17,686 women: the Women’s Genome Health Study. Circ Cardiovasc Genet 2009;2:134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair RP, Henseler T, Jenisch S, et al. Evidence for two psoriasis susceptibility loci (HLA and 17q) and two novel candidate regions (16q and 20p) by genome-wide scan. Hum Mol Genet 1997;6:1349–1356 [DOI] [PubMed] [Google Scholar]
- 19.Tomfohrde J, Silverman A, Barnes R, et al. Gene for familial psoriasis susceptibility mapped to the distal end of human chromosome 17q. Science 1994;264:1141–1145 [DOI] [PubMed] [Google Scholar]