Abstract
Intestinal stem cells (ISCs) have been studied for more than three decades; however, their isolation has remained a challenge. We hypothesized that, just as for stem cells of other tissues, one or more membrane markers would allow positive selection of ISCs by antibody-based sorting. To explore this hypothesis, microarray data of putative ISC fractions generated by side population sorting and laser capture microdissection were subjected to bioinformatic analysis to identify common membrane antigens. The microarray comparison suggested CD24 as a candidate surface marker, and immunohistochemistry showed expression of CD24 in epithelial cells of crypt bases. Flow cytometry of jejunal epithelial preparations revealed a CD24+ CD45− fraction comprising ∼1% of the cells. Analysis with epithelial cell adhesion molecule and CD31 confirmed that the cell preparations were epithelial and without endothelial contamination. Cycling cells identified by prior injection with 5-ethynyl-2′-deoxyuridine were found predominantly in the CD24lo subfraction. Transcript analysis by real-time RT-PCR showed this subfraction to be enriched in the ISC markers leucine-rich-repeat-containing G-protein-coupled receptor 5 (40-fold) and Bmi1 (5-fold), but also enriched in lysozyme (10-fold). Flow cytometry with anti-lysozyme antibodies demonstrated that Paneth cells comprise ∼30% of the CD24lo subfraction. Additional flow analyses with leucine-rich-repeat-containing G-protein-coupled receptor 5-enhanced green fluorescent protein (EGFP) epithelium demonstrated colocalization of EGFPhi and CD24lo. In contrast, CD24 cells were negative for the quiescent ISC marker doublecortin and CaM kinase-like-1. Culture of CD24lo cells in Matrigel generated organoid structures, which included all four epithelial lineages, thus giving functional evidence for the presence of ISCs. We conclude that the CD24lo fraction of jejunal epithelium is highly enriched with cycling ISCs. This isolation method should be useful to many investigators in the field to advance both the basic understanding of ISC biology and the therapeutic applications of ISCs.
Keywords: small intestine, cycling cells, 5-ethynyl-2′-deoxyuridine, fluorescence-activated cell sorting, leucine-rich-repeat-containing G-protein-coupled receptor 5
the existence of multipotent intestinal stem cells (ISCs) has been recognized since the pioneering studies by Cheng and Leblond (5) in 1974. These early studies showed that undifferentiated cells, termed “crypt-base columnar cells,” (CBCs), located in the intestinal crypts just above and between the Paneth cells, are responsible for the generation of all differentiated lineages of the small intestinal epithelium (5). In the intervening years, there was extensive focus on the supra-Paneth CBCs (now often called “+4” cells) due to their critical role in radiation damage and to the fact that these cells demonstrate long-term label retention of thymidine analogs (30). The cells in the +4 zone are now known to be characterized by a variety of markers, including p-β-catenin (Ser552), Bmi1 polycomb ring finger oncogene (Bmi1), telomerase, and doublecortin and CaM kinase-like-1 (DCAMKL1) (25, 30, 38). They also appear to be resistant to differentiation in the face of loss of proliferation (19). Lineage tracing experiments with Bmi1 (35) and telomerase (32) have confirmed that these cells can self-renew and give rise to all four differentiated cell lineages of the epithelium, although there is debate as to whether these +4 ISCs are slowly cycling or quiescent (25, 30, 38). In contrast, the inter-Paneth CBCs, which have recently been demonstrated to be marked by the leucine-rich-repeat-containing G-protein-coupled receptor 5 (Lgr5) (2), appear to be an actively cycling population of ISCs. The extent to which the supra-Paneth “+4” CBCs and inter-Paneth CBCs represent distinct populations remains controversial (3, 25, 38); however, it is clear that, in agreement with the original studies of Cheng and Leblond (5), cells in both locations demonstrate the properties of self-renewal and multilineage differentiation. Some commonality in their biology is also indicated by the fact that they share several markers, including Musashi-1 (18, 29), CD133/Prominin 1 (40, 43), and SRY-box containing gene 9 (Sox9) (11).
Compared with stem cells of other tissues, ISCs have proven remarkably difficult to isolate. The classical approach of antibody-based fluorescence-activated cell sorting (FACS) has been hampered by the lack of appropriate membrane markers. In an effort to overcome this problem, in 2005, our laboratory (8) reported the use of side population (SP) sorting to isolate ISCs in an antibody-independent fashion. These studies (8) demonstrated that mouse intestinal epithelium contained a fraction of cells displaying an SP phenotype, and that intestinal SP cells are enriched for Musashi-1 expression (at that time the only known marker of ISC). Subsequent microarray, in situ hybridization, resection, and damage studies (6, 7, 15) have provided further evidence that the SP from mouse small intestine includes the ISCs. While it was clear from the outset that the intestinal SP is heterogeneous, until 2009 there had been no reports of alternate approaches to the isolation of ISCs. During the past year or so, there have been significant advances in the isolation of ISCs via use of genetically engineered mice with ISC promoters driving fluorescent reporters: specifically telomerase-green fluorescent protein (GFP) (4, 32), Sox9-GFP (11), and Lgr5-GFP (36). For the latter, the combination of FACS with a novel in vitro culture system has allowed the demonstration that single Lgr5 positive epithelial cells are capable of both self-renewal and multilineage differentiation (36). Recently, Sox9-GFP cells have also been reported to display these stemness qualities in vitro (14). While the reporter-based isolation methods are of unquestionable value for studying the biology of ISCs, their use is limited to these specifically engineered mice.
In 2009, May et al. (24) reported the first membrane marker for isolation of ISCs, namely DCAMKL1. While this represented an important contribution, it appears to select for a quiescent ISC population and thus is not useful for isolation of actively cycling ISCs. As the latter are essential for maintenance of the epithelium under homeostatic conditions, the goal of the present study was to develop the technology needed to isolate cycling ISCs from wild-type mice in a manner that would be readily assimilated and applicable to other species, including human.
We hypothesized that comparison of microarray data from the intestinal SP with similar data from a putative ISC fraction generated by laser capture microdissection (LCM) would identify potential membrane antigens suitable for sorting. Among the candidates identified by this approach, the adhesion molecule CD24 appeared most promising, based on the availability of well-documented antibodies that have been used to isolate CD24+ stem cells from other tissues (28, 31, 33, 39, 42).
We report here that immunostaining of mouse jejunal tissue with these antibodies revealed CD24 expression as being restricted to cells at the crypt base. Flow cytometric analysis showed a CD24+ fraction that includes cells of bone marrow origin. The latter could be excluded by either negative selection with anti-CD45 or by appropriate gating. Within the CD24+ fraction, cycling cells [identified by in vivo labeling with the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU)] were found primarily in the CD24lo subfraction. Gene expression studies showed this subfraction to be markedly enriched in the ISC markers Lgr5 and Bmi1, but also enriched in Paneth cell transcripts. Flow cytometric analyses of jejunal epithelial cells stained with anti-lysozyme antibodies demonstrated that Paneth cells comprise ∼30% of the CD24lo subfraction. Finally, culture of the CD24lo fraction with a Matrigel-based protocol led to the generation of crypt-like structures within 14 days, giving functional evidence for the presence of ISC.
MATERIALS AND METHODS
Animals and Reagents
Adult male C57BL/6J mice and heterozygote breeder pairs of Lgr5-EGFP (enhanced green fluorescent protein)-IRES-creERT2 (Lgr5-EGFP) mice were procured from Jackson Laboratories and maintained on a 12:12-h light-dark cycle in American Association for Accreditation of Laboratory Animal Care-approved facilities. All animals were used within the age range of 6–10 wk of age, and all animal usage had Institutional Animal Care and Use Committee approval. Antibodies used for immunohistochemistry were purified anti-CD24 (BD Pharmingen catalog no. 557436) and anti-rat IgG-horseradish peroxidase (HRP) (Serotec catalog no. star72). Antibodies used for flow cytometric analysis included phycoerythrin (PE)-conjugated anti-CD24 (BD Pharmingen catalog no. 553262), fluorescein isothiocyanate (FITC)-conjugated anti-CD45 (BD Pharmingen catalog no. 553080), Pacific Blue (PB)-conjugated anti-CD24 (Biolegend catalog no. 101819), FITC-conjugated anti-epithelial cell adhesion molecule (EpCAM) (Biolegend catalog no. 118207), Alexa Fluor 647 (Alexa 647)-conjugated anti-CD31 (Biolegend catalog no. 102415), FITC-conjugated anti-lysozyme (Dako catalog no. F0372), and DCAMKL1 (Abcam catalog no. 37994) conjugated to DyLight 633 (Thermo Scientific catalog no. 53047).
Microarray Analyses
The generation [using dChip(20) software] of a list of transcripts enriched twofold or greater in mouse jejunal SP cells, compared with intact jejunum, has already been published (15). For the present study, an analogous list of enriched transcripts was generated for LCM-procured cells from the small intestinal crypt bases of germ-free CR2-tox176 mice (that are depleted of Paneth cells) (13, 41) compared with whole jejunum from wild-type, germ-free mice (17). This LCM fraction has previously been shown to bioinformatically fit into a stem cell-like grouping (9). Transcripts common to the SP-enriched list and the LCM-enriched list were subjected to gene ontology analysis, as described by Gulati et al. (15).
Immunostaining
For tissue preparation, small intestines were flushed with PBS. A 4-mm segment from midjejunum was then filled with optimal cutting temperature (OCT) medium, embedded in OCT, and frozen on dry ice. Sections (7 μm) were fixed in 100% acetone at 4°C for 10 min, washed with 1× PBS for 10 min at room temperature (RT), and blocked with normal serum diluted in PBS for 1 h at RT. The primary antibody, anti-mouse CD24, was applied at 1:50 overnight. Secondary labeling was performed using anti-rat IgG-HRP at 1:200 for 1 h at RT. The secondary HRP was then visualized using a diaminobenzidine reaction.
Organoids were prepared and immunostained with lysozyme, substance P, sucrase isomaltase, and mucin 2 antibodies, as described by Gracz et al. (14).
Flow Cytometry and FACS Analysis
Five different approaches were used for the cell sorting reported in this paper. Although the differences are subtle (e.g.. live cells vs. permeabilized cells), they are very important in allowing reproduction of the data and thus are described separately below.
Approach 1: Isolation of CD24+ CD45− cells and epithelial validation.
Jejunal epithelial cells were isolated using the EDTA/dispase method described by Formeister et al. (11). Cells were incubated with both CD24-PE and CD45-FITC antibodies, at 0.25 μg per 1 × 106 cells in 100 μl PBS/1% BSA, for 15-min RT, washed with PBS/1% BSA, and analyzed in a MoFlo FACS machine (Dako/Cytomation). For epithelial validation, the aforementioned method was used, and the cells were incubated with CD24-PB at 0.25 μg per 1 × 106 cells and EpCAM-FITC antibodies at 0.5 μg per 1 × 106 cells. To check for endothelial contamination, cells were also incubated with CD24-PB at 0.25 μg per 1 × 106 cells and CD31-Alexa 647 antibodies at 0.5 μg per 1 × 106 cells. Because the CD24+ fraction was negative for CD31, a control experiment was performed using digested lamina propria after removal of the epithelium. This showed a distinct CD31-positive fraction, thus validating the CD31 antibody. To assess colocalization of CD24 and DCAMKL1, cells were incubated with CD24-PB antibodies at 0.25/μg per 1 × 106 cells and DCAMKL1-DyLight 633 antibodies, applied at a 1:50 dilution. In these and all subsequent experiments, doublets were excluded using a bivariate plot of pulse width vs. forward scatter (FSC). Additionally the CD45+ population was identified and back-gated onto a FSC vs. side scatter (SSC) plot to exclude it for this and the following four approaches.
Approach 2: Labeling with CD24 and EdU.
Animals were injected intraperitoneally with 100 μg EdU (Invitrogen catalog no. A10202) and killed 3 h later. Epithelial cells were prepared from the jejunum as in approach 1, incubated with CD24-PB antibody at the concentration used for CD24-PE above, and then washed. Detection of the EdU via a coupling reaction with Alexa647 azide was performed after fixation and permeabilization using the aforementioned kit from Invitrogen, following the provided protocol. Cells were analyzed by flow cytometry on a CyAn (Dako/Cytomation) machine.
Approach 3: RNA isolation and quantitative RT-PCR.
In the same manner as approach 1, live cells were isolated, labeled with CD24-PB antibody, and subjected to FACS. The lower half of the CD24 positive, designated CD24lo, was then gated for collection. RNA was isolated using the RNAqueous Micro Kit from Ambion (Austin, TX), and cDNA was generated from this fraction as well as intact jejunum using the High Capacity cDNA Reverse Transcription Kit from Applied Biosystems (Foster City, CA). Quantitative PCR was conducted for each sample in triplicate. Taqman probes (Actb, Mm00607939_s1; Lgr5, Mm00438890_m1; Lyz, Mm00727183; SI, Mm01210305_m1, Acta2, Mm01546133_m1) were obtained from Applied Biosystems (Pleasanton, CA) and used according to the manufacturer's protocol. β-Actin RNA was used as an internal control gene, and ΔΔCT (cycle threshold) values were calculated to obtain fold changes vs. intact jejunum (37).
Approach 4: Coexpression of CD24 and Lgr5-EGFP.
For the analysis of jejunal epithelial cells from the Lgr5-EGFP mice, labeling methods described for CD24-PB in approach 1 were used, and the appropriate instrument channel was opened for endogenous Lgr5-EGFP fluorescence detection. The EGFPhi fraction, which has been shown to represent Lgr5+ ISCs, was gated as described by Sato et al. (36).
Approach 5: Analysis of CD24 and lysozyme.
Cells were isolated and labeled with CD24-PB as in approach 1, then fixed for 15 min in 4% paraformaldehyde. After washing with PBS, cells were resuspended in saponin permeabilization buffer (Invitrogen PB001) with lysozyme-FITC antibody (1:10) and incubated at RT for 30 min. The cells were again washed and then analyzed by flow cytometry.
RESULTS
Microarray Cross-Comparison Identifies Potential Membrane Markers of ISC
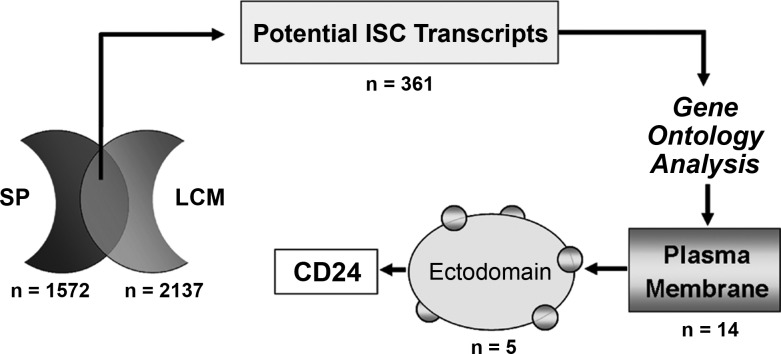
Given our prior demonstration by in situ hybridization that SP cells from mouse small intestine map to the stem cell zone (15), we used our SP microarray data as a starting point to search for potential membrane antigens suitable for sorting. As can be seen in Fig. 1, a total of 1,572 transcripts were found to be enriched in the SP. To determine which of these transcripts was most likely to represent ISC markers, we cross-referenced this list with the 2,137 transcripts enriched within a putative ISC population, which was harvested by LCM from crypt bases of mice lacking Paneth cells (13, 41). The rationale for the cross-comparison was that populations of cells isolated by these different methods are likely to be enriched for ISCs, but also contain other distinctly different cell types. Thus comparison of the two data sets should facilitate identification of common ISC-enriched transcripts. As shown in Fig. 1, this cross-comparison yielded a total of 361 potential ISC transcripts (listed in Supplemental Table S1; The online version of this article contains supplemental data). Gene ontology analysis showed that 14 of these transcripts encode plasma membrane proteins (asterisked in Supplemental Table S1), 5 of which have ecto domains that appear suitable for antigen-based cell sorting (double asterisked in Supplemental Table S1). Of these five, CD24 appeared the most promising because it is known to be a stem cell marker in other tissues (28, 31, 33, 39, 42) and in tumors (10).
Fig. 1.
Cross-comparison strategy using side population (SP) and laser capture microdissection (LCM) populations to identify possible intestinal stem cell (ISC) membrane markers. Numbers of transcripts (n) are shown for each stage of the analysis.
Localization of CD24 Expression in Jejunal Tissue
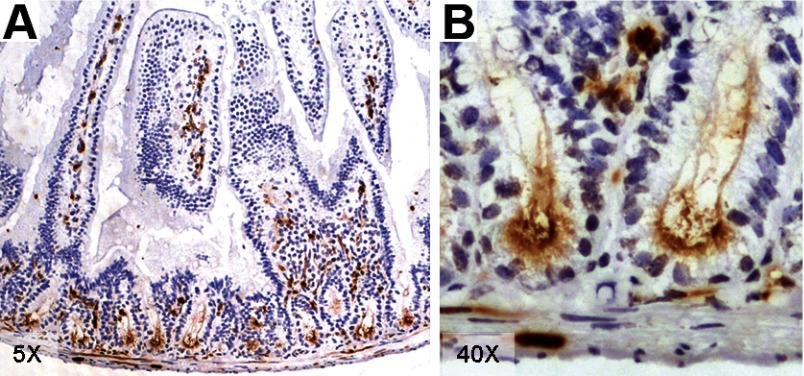
To verify that CD24 is expressed in the stem cell zone of the mouse small intestine, jejunal sections were subjected to immunohistochemical staining with CD24 antibody. As can be seen in the low power image in Fig. 2A, CD24 is restricted to epithelial cells at the crypt base and to cells within the lamina propria. Based on the literature, the latter are likely to be a mix of blood cells and endothelial cells (10, 22). At higher power (Fig. 2B), it appears that the epithelial staining is primarily apical and probably includes both the supra- and inter-Paneth CBC, as well as the Paneth cells themselves. Based on this staining pattern, together with the original microarray data, we concluded that sorting epithelial preparations with anti-CD24 should isolate a small fraction that is enriched in both ISCs and Paneth cells.
Fig. 2.
Mouse jejunum stained with CD24 antibody at low (A) and high power (B).
Flow Cytometric Identification of a CD24+ CD45− Epithelial Fraction
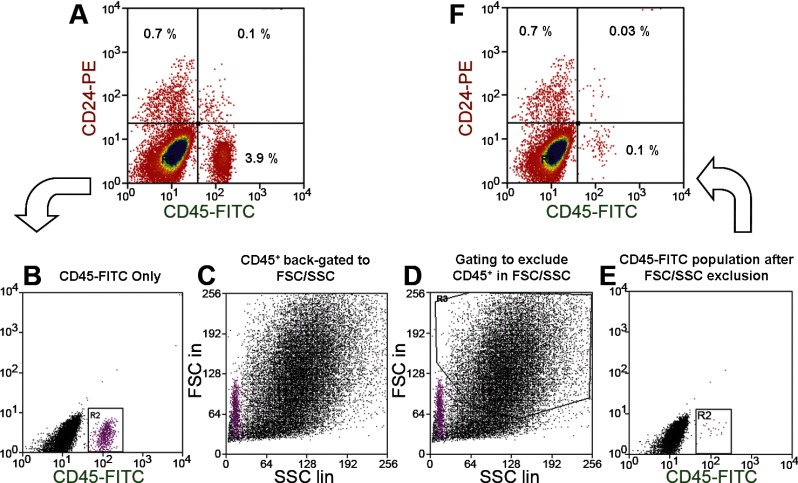
Using an EDTA-based isolation procedure, which should yield epithelial cells relatively free of lamina propria, in our first approach, we stained cells with anti-CD24 for positive selection, as well as anti-CD45 to identify and eliminate cells of hematopoietic origin. Figure 3A shows that a majority of the CD24+ cells were CD45−. In the representative figure, CD24+ CD45− cells comprised 0.7% of the total, and replicate experiments gave the mean ± SE (n = 5) for this fraction to be 0.94 ± 0.13 (Supplemental Table S2). In the initial sorting (Fig. 3A), the double-positive CD24+ CD45+ cells made up 0.1% of the total. To exclude the double-positive fraction in future experiments without the need for the anti-CD45, this CD45+ fraction was identified (Fig. 3B), back-gated to the FSC/SSC plot (Fig. 3C), and then gated out (Fig. 3, D and E). Figure 3F shows that, with this exclusion gating, the CD24+ CD45+ fraction was reduced to 0.03%, which is considered an acceptable level for future studies.
Fig. 3.
Flow cytometric identification of a CD24+ CD45− fraction from jejunal epithelium. A: cells labeled with both CD24-PE (phycoerythrin) and CD45-FITC antibodies. B: selection of the entire CD45+ population for back-gating. C: CD45+ population as it relates to forward scatter (FSC)/side scatter (SSC) properties. D: exclusion of the CD45+ cells in FSC/SSC. E: sorting of gated cells with anti-CD45. F: validation of reduction in CD45+ cells.
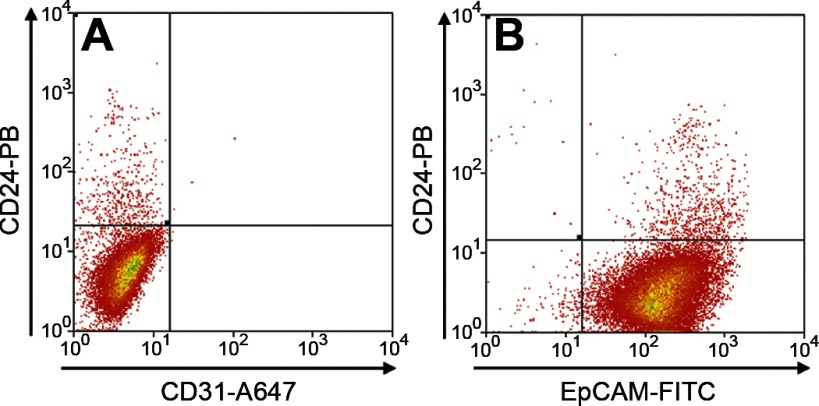
In view of the expression of CD24 on endothelial cells within the lamina propria (Fig. 2), the CD24+ fraction from an epithelial preparation was subjected to further flow cytometry using the pan-endothelial marker CD31 (16). As can be seen from the representative analysis in Fig. 4A, essentially no CD31+ cells were detected. In contrast, as shown in Fig. 4B, when the CD24+ fraction was examined for presence of the pan-epithelial marker EpCAM (1), it was found to be almost entirely epithelial. Replicate analyses (n = 3) of the CD24+ fraction showed it to be 97.4 ± 0.3% EpCAM+ and only 1.7 ± 0.5% CD31+.
Fig. 4.
Flow cytometric identification of CD24+ CD31− and CD24+ EpCAM+ (epithelial cell adhesion molecule) fractions from jejunal epithelium. A: cells labeled with both CD24-PB (Pacific Blue) and CD31-Alexa 647 antibodies. B: CD24-PB and EpCAM-FITC antibodies.
Labeling with EdU Identifies a Cycling CD24+ Fraction
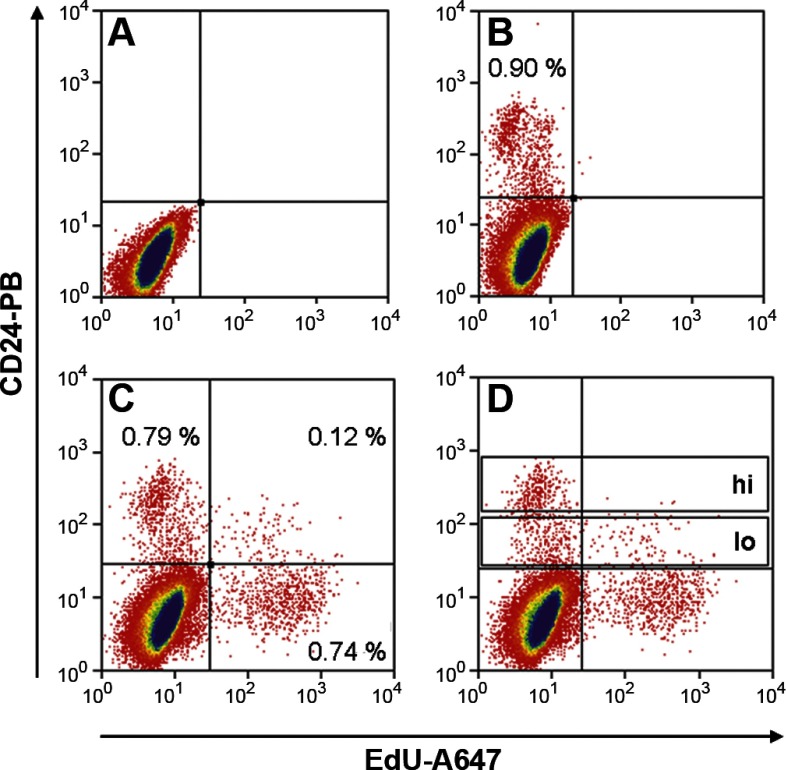
Although there may be a quiescent population of ISCs responsible for repair of epithelial damage (21, 24, 38), normal homeostasis of the intestinal epithelium is maintained by an actively cycling population of ISCs (3, 30, 38). We hypothesized that the latter may represent a subfraction within the relatively broad range of fluorescence intensities exhibited by the CD24+ cells in Fig. 3. To identify the actively cycling cells in this fraction, in approach 2, the thymidine analog EdU was administered to mice 3 h before tissue collection. Epithelial cells that incorporated EdU were then detected by flow cytometry using a fluorescent azide dye following fixation and permeabilization. Figure 5, A and B, shows that sorting with anti-CD24 alone under these conditions yielded a positive fraction much like that shown in Fig. 3. When the azide dye was applied (Fig. 5C), cycling cells were readily identified as an EdU+ fraction on the x-axis. The majority of these were CD24−. Based on the lack of staining in the midcrypts (Fig. 2), we assume these CD24− EdU+ cells are the cycling transit amplifying cells. Within the CD24+ fraction, there was obvious accumulation of EdU+ cells at the lower fluorescence intensities (Fig. 5C). Subdividing the CD24+ fraction into CD24hi and CD24lo subfractions (Fig. 5D) showed that EdU+ cells represented 2.7 ± 1.0% of the CD24hi subfraction (Supplemental Table S3) compared with 19.5 ± 2.2% (n = 4) of the CD24lo subfraction (Supplemental Table S4). Thus the latter is proposed to contain a majority of the actively cycling ISCs. Unfortunately, because cells are killed by the permeabilization and fixation steps required for FACS with EdU, the CD24lo EdU+ fraction cannot be used for RNA analyses or for culture. However, since the actively cycling EdU+ cells fall primarily in the CD24lo subfraction, we focused on this subfraction for subsequent analyses.
Fig. 5.
Flow cytometric analysis of isolated jejunal epithelial cells 3 h post-EdU (5-ethynyl-2′-deoxyuridine) injection. A: no stain. B: CD24-PB antibody only. C: CD24-PB antibody and Edu-Alexa 647. D: CD24-PB antibody and Edu-Alexa 647 with CD24hi and CD24lo gates labeled.
The CD24lo Population Is Highly Enriched for the ISC Marker Lgr5
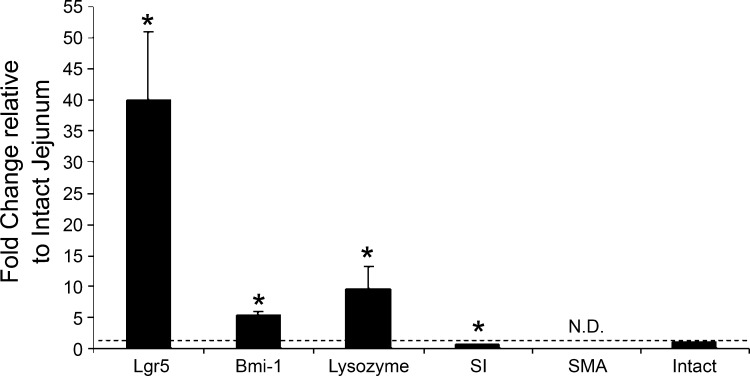
To assess the presence of ISCs in the CD24lo population, in approach 3 we returned to the use of live cells and collected the CD24lo subfraction for mRNA analyses. Figure 6 shows that this subfraction is highly enriched (40-fold) for the ISC marker Lgr5 and modestly enriched (5-fold) for the ISC marker Bmi1. Not surprisingly, the CD24lo cells are also enriched for lysozyme mRNA (10-fold), indicating the presence of Paneth cells. In contrast, transcripts of sucrase-isomaltase, a marker for absorptive cells of the villus, were deenriched as predicted by the CD24 staining pattern shown in Fig. 2. Transcripts for smooth muscle actin, a myofibroblast marker, were deenriched to the point of being nondetectable, further indicating that the EDTA protocol is indeed epithelial specific and excludes underlying mesenchymal cells. Overall, these mRNA data confirm the predictions from the staining pattern shown in Fig. 2. When taken together with the EdU analyses (Fig. 5), we conclude that the CD24lo population includes ISCs as well as some Paneth cells.
Fig. 6.
Quantitative RT-PCR of mRNA isolated from the CD24lo CD45− fraction of jejunal epithelium compared with intact jejunum. Transcripts shown are markers for the following: ISCs [leucine-rich-repeat-containing G-protein-coupled receptor 5 (Lgr5) and Bmi1 polycomb ring finger oncogene (Bmi1)]; Paneth cells (lysozyme); absorptive cells [sucrase-isomaltase (SI)]; and myofibroblasts [smooth muscle actin (SMA)]. Bars show means + SE (n = 7). Dashed line indicates intact jejunum. N.D., nondetectable. *P < 0.05.
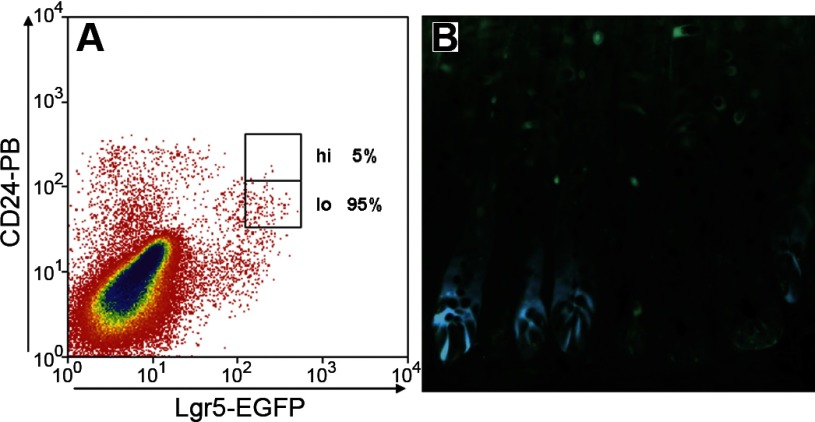
To verify the presence of actively cycling ISCs, we used Lgr5-EGFP mice (36) to assess the distribution of Lgr5-EGFP cells within the CD24 fraction. As can be seen in Fig. 7A, the Lgr5 EGFPhi fraction, which has been demonstrated to represent actively cycling ISCs (36), includes CD24-expressing cells. Most importantly, the majority of the Lgr5-EGFPhi cells fall within the CD24lo subfraction (95% in the representative analysis shown in Fig. 7 and 85 ± 5% in 3 replicate analyses). While it would be of significant interest to also know what proportion of CD24lo cells express Lgr5, this is not possible with the current Lgr5-EGFP mice because, as shown in Fig. 7B, the EGFP expression is highly mosaic in the jejunum of these mice. Thus the large number of cells in Fig. 7A that are CD24lo but EGFP-negative presumably reflects crypts that do not express EGFP.
Fig. 7.
Use of Lgr5-EGFP (enhanced green fluorescent protein) mice to assess colocalization of CD24 and Lgr5. A: flow cytometric analysis of jejunal epithelial cells displaying Lgr5-EGFPhi distribution with respect to the CD24hi and CD24lo fractions. B: histological section of Lgr5-EGFP mouse jejunum, showing mosaicism of EGFP-expressing crypts.
DCAMKL1 Sorted Cells Are Not Present in the CD24 Population
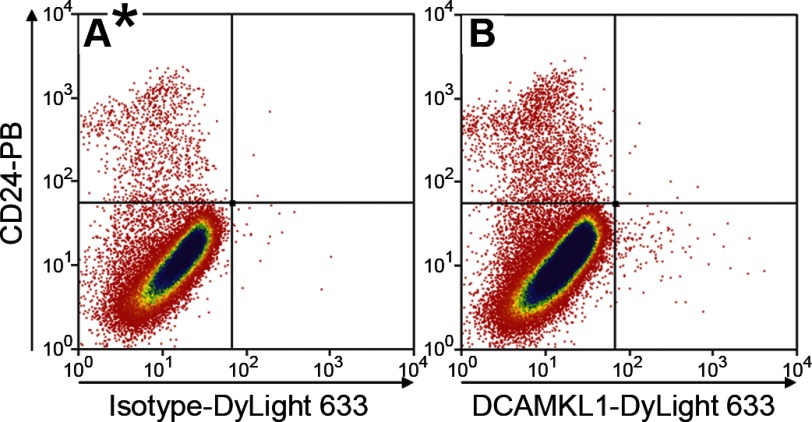
Since antibodies to DCAMKL1 have been reported to isolate a fraction that represents quiescent ISCs (24), we used flow cytometry of live cells to assess coexpression of DCAMKL1 and CD24. Although a distinct DCAMKL1-positive fraction was observed (Fig. 8), essentially all of these cells fell within the CD24-negative quadrant. Thus we conclude that the CD24lo fraction is not enriched with the putative quiescent ISCs marked by DCAMKL1.
Fig. 8.
Flow cytometric analysis of jejunal epithelial cells labeled with CD24-PB and isotype-DyLight 633 antibodies (A) and CD24-PB and doublecortin and CaM kinase-like-1 (DCAMKL1)-DyLight 633 antibodies (B).
Paneth Cells Comprise Approximately 30% of the CD24lo Population
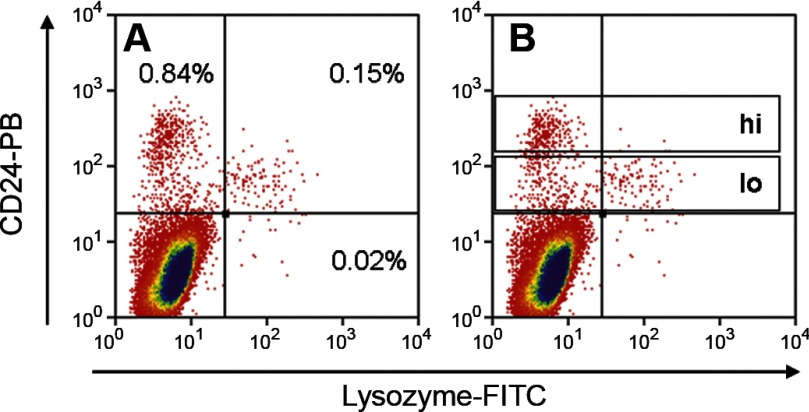
Although the RT-PCR data for lysozyme shown in Fig. 6 clearly indicated the presence of Paneth cells, since transcript analyses do not allow inference regarding cell numbers, we used approach 4 to determine what proportion of the CD24lo subfraction actually represents Paneth cells. For this purpose, epithelial preparations were stained with anti-CD24, then fixed, permeabilized, and stained with anti-lysozyme. As expected from the CD24 immunohistochemistry shown in Fig. 2, most of the lysozyme+ cells were CD24+ (Fig. 9A). When the CD24lo subfraction was analyzed (Fig. 9B), 32.5% was found to be lysozyme+. In replicates (Supplemental Table S4), the mean ± SE for lysozyme+ cells in the CD24lo subfraction was 29.8 ± 1.4% (n = 3). Thus it appears that ∼30% of the cells in the CD24lo fraction are Paneth cells. As these are postmitotic, their presence should not preclude functional studies of the ISCs that are also predicted to be in the CD24lo subfraction.
Fig. 9.
Flow cytometric analysis of jejunal epithelial cells labeled with CD24-PB and lysozyme-FITC antibodies. A: representative numbers in the positive quadrants. B: gating of the CD24hi and CD24lo subfractions.
The CD24lo Population Generates Intestinal Organoids In Vitro
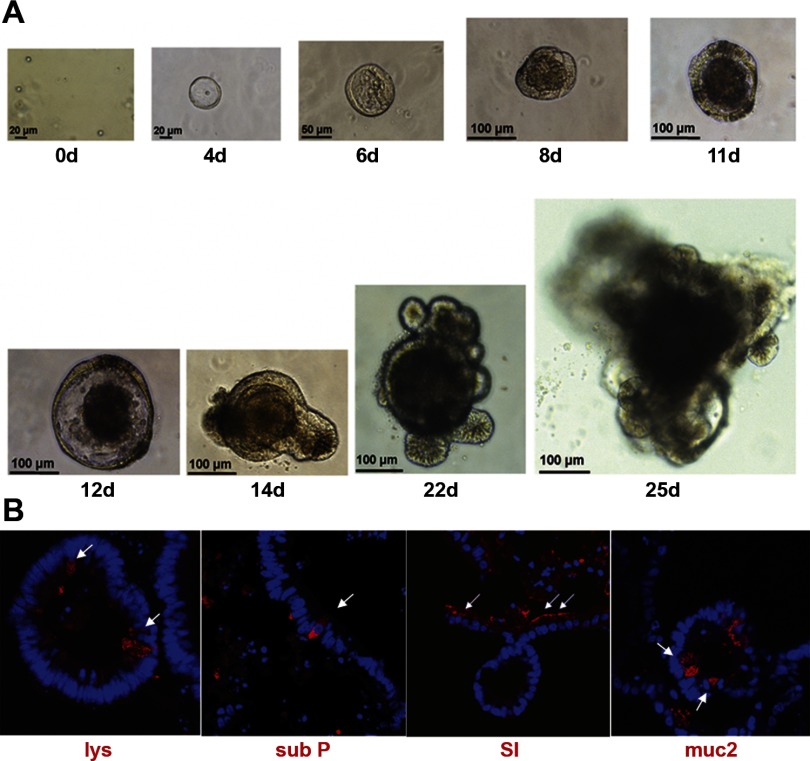
The Matrigel-based culture system developed by Sato et al. (36) for Lgr5-GFP cells and subsequently employed by Gracz et al. (14) for Sox9-GFP cells provides a useful way to assess the functional behavior of preparations of putative ISCs. As can be seen in Fig. 10A, when our CD24lo population was subjected to the Sato protocol, multicellular organoids were observed to develop. The time course of formation of these intestinal organoids appeared a little slower than that shown by Sato et al. for Lgr5-GFP cells (36) and by Gracz et al. for Sox9-GFP cells (14). However, the morphology is very similar to these previous reports; with an early phase (days 4–12) of growth as a simple spheroid and subsequent budding of multiple cryptlike structures (beginning on day 14 and becoming extensive by day 22) to generate complex organoids. In contrast to the results with the CD24lo population, CD24− cells showed zero in vitro growth. Figure 10B shows that, when 25-day organoids from our CD24lo cells were collected and subjected to immunofluorescent staining, cells of all four differentiated epithelial lineages were detected. Although passaging has not been demonstrated with C57BL/6J mice, CD24lo organoids from CD1 mice have been passaged up to three times with successful formation of new organoids (A. D. Gracz and S. T. Magness, unpublished observations).
Fig. 10.
Growth of CD24lo CD45− cells in 3D Matrigel culture as per Sato et al. (36). A: phase-contrast images depict progression of organoid growth over 25 days (0d to 25d) with scale as shown. B: immunofluourescent staining of a CD24lo CD45− organoids, harvested at 25 days, with markers of Paneth cells (lysozyme), enteroendocrine cells [substance P (sub P)], enterocytes (SI), and goblet cells [mucin 2 (muc2)].
DISCUSSION
This paper reports the successful identification of a membrane marker, CD24, that can be used for antibody-based sorting of ISCs. Multiple lines of evidence have been presented to demonstrate that the CD24+ fraction from murine jejunal epithelium is enriched with ISCs. First is the fact that CD24 transcripts were found in the putative ISC transcriptome profile generated by two independent approaches, namely SP sorting and LCM. Second, immunohistochemistry revealed CD24 protein expression within the jejunal epithelium to be restricted to cells at the crypt base. Third, sorting epithelial preparations with antibodies to CD24 demonstrated a positive fraction (CD24lo), which included actively cycling cells, and is highly enriched for the ISC marker Lgr5. Finally, this fraction is capable of in vitro growth to yield organoid-like units, which include all four epithelial lineages.
Since CD24 is known to be expressed on white blood cells, in our initial sorting with anti-CD24, we assessed the leukocytic component by labeling with anti-CD45, a pan-leukocyte marker. A distinct CD24+ CD45− fraction was observed (Fig. 3), comprising 0.94 ± 0.13% of the jejunal epithelium, a range consistent with the pattern of expression of CD24 protein, as assessed by immunohistochemistry (Fig. 2). Subsequent manipulations demonstrated that the leukocytic CD45+ fraction can be effectively gated out, thus simplifying the sorting procedure. Figure 4 shows that CD24-expressing cells in the lamina propria are effectively excluded by our EDTA-based cell preparation, which is selective for the epithelial cells.
The presence of Paneth cells in the CD24+ fraction was predicted by the immunohistochemistry and confirmed by RT-PCR for lysozyme. As there is currently no published membrane marker for Paneth cells, their elimination is not straightforward. Since our interest lay primarily in isolating actively cycling ISCs, we hypothesized that this quality might effectively distinguish the ISCs from the Paneth cells within the CD24+ population. However, when the thymidine analog EdU was used to identify proliferating cells, the EdU+ subfraction (Fig. 5) was found to be intermixed with the lysozyme+ subfraction (Fig. 9) in the CD24lo region of the CD24+ population.
RNA analyses of the CD24lo cells (Fig. 6) suggested that this fraction is highly enriched in cycling ISCs, as judged by Lgr5 transcripts being 40-fold more abundant than intact jejunum. Given that transcripts for the +4 marker Bmi1 were enriched only fivefold, it would appear that the CD24lo ISC fraction represents primarily the intra-Paneth CBCs, which are marked by Lgr5. Expression of Lgr5 in the CD24lo fraction was confirmed by flow cytometric analyses of jejunal epithelium from Lgr5-EGFP mice (Fig. 7). Finally, we used the culture technique of Sato et al. (36) to confirm that the CD24lo cells, just like the Lgr5-EGFP cells (36) and the Sox9-GFP cells (14), are capable of extensive growth in vitro and give rise to organoid-like structures, which include all four epithelial lineages (Fig. 10).
While this work was in progress, Gracz et al. (14) independently generated evidence that CD24 antibodies could be used to sort murine ISCs. Their approach was based on the use of Sox9-GFP mice, which yielded a GFPlo fraction enriched in ISCs, as evidenced by the presence of ISC markers (including Lgr5) and by the ability to both proliferate and differentiate in vitro. When RNA from Sox9-GFPlo cells was analyzed for the presence of surface markers known to be expressed in stem cells of other tissues, CD24 transcripts were found to be enriched. Moreover, when jejunal epithelial preparations were sorted with GFP and anti-CD24, the Sox9-GFPlo cells were found in the CD24lo region, consistent with our data indicating the presence of ISCs in that fraction. Just as in our studies with wild-type mice, Gracz et al. (14) were able to isolate CD24lo cells from Sox9-GFP mice and demonstrate stemlike behavior in vitro. These independent studies with two distinct strains of mice (wild-type C57BL/6J in the present study vs. Sox9-GFP on the CD1 background) point to the broad utility of CD24 as a membrane marker that facilitates isolation of ISCs. Furthermore, the extensive characterization of the CD24 fraction presented in the present study significantly extends our understanding of the composition of this fraction.
As noted in the Introduction, CD24 has previously been reported to be a useful marker for isolating stem cells from various tissues. The best studied of these are mammary epithelial stem cells, where sorting with anti-CD24 has been shown to yield a 10-fold increase in the clonogenic capacity of Lin−Sca-1l°CD49fhi cells (42), and a single Lin−CD29hi CD24+ cell can give rise to a functional mammary gland (39). Likewise, hepatic stem cells, which repopulate the liver after damage, are characterized by CD24 expression and can be isolated as a CD24+ fraction (28). Similar to our findings in the intestinal epithelium, studies with mouse brain have shown that neural stem cells are enriched in a CD24lo subfraction (33), and, encouragingly, work with human neural stem cells also points to CD24lo as defining the critical fraction (31).
Beyond its utility for stem cell isolation, CD24 is also likely to play one or more functional roles in stem cell biology. CD24 is a glycosyl phosphatidylinositol-anchored membrane protein, which functions in cell-cell and cell-matrix adhesion, as well as in extracellular-to-intracellular signal transduction (10, 22). Both of these properties could be important to ISCs. With respect to adhesion, CD24 may be one of the molecules responsible for anchoring ISCs in the crypt base, by partnering with a ligand on Paneth cells, or the basement membrane, or both. Likewise, CD24 could be involved in communication between ISCs and other cells of the stem cell niche, such as endothelial cells and neurons, which express known ligands for CD24, namely P-selectin and L1, respectively (22, 34). Basolateral interactions of CD24 with either bound or secreted ligands or apical interactions with luminal factors may elicit signaling in the ISCs. The most likely role of such signaling would be to provide homeostatic control of stem cell number and proliferation rate. This suggestion is based on the literature reports that signaling via CD24 can elicit apoptosis and inhibit proliferation (22). Most cogently, in CD24-deficient mice, increased proliferation has been observed in brain, skin, and cornea (27). Conversely, overexpression of CD24 leads to decreased clonogenic behavior (27). Although CD24-deficient mice have not been reported to have an overt intestinal phenotype (26, 27), no intestinal histology has been described. Moreover, a homeostatic role may be revealed only when ISCs are challenged in some way (e.g., by damage, resection, oncogenesis, or carcinogenesis). Clearly, there is much work needed to explore these intriguing possibilities.
Before the present study, the techniques available for isolation of ISCs from wild-type mice (or other nonengineered species) were extremely limited. As noted in the Introduction, initial studies used SP sorting (8, 15) and generated evidence that this fraction includes the ISC and numerically can be considered a surrogate for ISC number (6, 7). However, SP sorting has not come into wide use, presumably because it does not generate a highly enriched ISC fraction and because it requires the use of a UV laser, which is not available in many flow cytometry cores. As use of fluorophore-labeled antibodies to surface proteins overcomes the latter limitation, the search for membrane markers for ISCs has been intense. Beyond the CD24 reported here and by Gracz et al. (14), the only other membrane marker of ISC in the literature to date is DCAMKL1 (23, 24). However, this protein appears to identify postmitotic differentiated cells (12) and/or quiescent stem cells (24). In contrast, our CD24lo subfraction is essentially negative for DCAMKL1 (Fig. 8) and clearly includes actively cycling ISCs, as judged by in vitro labeling with EdU, enrichment in Lgr5, and in vitro growth into intestinal organoids. Finally, since CD24 antibodies are available for many species, including human, sorting with such antibodies (either using FACS or magnetic beads) should have wide application in both the basic understanding of ISC biology and the therapeutic applications of ISCs.
GRANTS
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grants R01-DK69585, T32-DK07664, P30-DK034987, and UO1-DK085547. The latter grant is part of the Intestinal Stem Cell Consortium, a collaborative research project funded by the NIDDK and the National Institute of Allergy and Infectious Diseases.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Kirk McNaughton and Kimberly Etzel for performing the immunohistochemistry; Doug von Allmen for the fluorescent image of intestine from Lgr5-EGFP mice; Dr. Larry Arnold and staff for FACS; and Drs. Chris Dekaney and Jennifer Uno for constructive input on the manuscript.
REFERENCES
- 1. Balzar M, Winter MJ, de Boer CJ, Litvinov SV. The biology of the 17–1A antigen (Ep-CAM). J Mol Med 77: 699–712, 1999. [DOI] [PubMed] [Google Scholar]
- 2. Barker N, van Es JH, Kuipers J, Kujala P, van den BM, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007, 2007. [DOI] [PubMed] [Google Scholar]
- 3. Bjerknes M, Cheng H. Intestinal epithelial stem cells and progenitors. Methods Enzymol 419: 337–383, 2006. [DOI] [PubMed] [Google Scholar]
- 4. Breault DT, Min IM, Carlone DL, Farilla LG, Ambruzs DM, Henderson DE, Algra S, Montgomery RK, Wagers AJ, Hole N. Generation of mTert-GFP mice as a model to identify and study tissue progenitor cells. Proc Natl Acad Sci U S A 105: 10420–10425, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian theory of the origin of the four epithelial cell types. Am J Anat 141: 537–562, 1974. [DOI] [PubMed] [Google Scholar]
- 6. Dekaney CM, Fong JJ, Rigby RJ, Lund PK, Henning SJ, Helmrath MA. Expansion of intestinal stem cells associated with long-term adaptation following ileocecal resection in mice. Am J Physiol Gastrointest Liver Physiol 293: G1013–G1022, 2007. [DOI] [PubMed] [Google Scholar]
- 7. Dekaney CM, Gulati AS, Garrison AP, Helmrath MA, Henning SJ. Regeneration of intestinal stem/progenitor cells following doxorubicin treatment of mice. Am J Physiol Gastrointest Liver Physiol 297: G461–G470, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dekaney CM, Rodriguez JM, Graul MC, Henning SJ. Isolation and characterization of a putative intestinal stem cell fraction from mouse jejunum. Gastroenterology 129: 1567–1580, 2005. [DOI] [PubMed] [Google Scholar]
- 9. Doherty JM, Geske MJ, Stappenbeck TS, Mills JC. Diverse adult stem cells share specific higher-order patterns of gene expression. Stem Cells 26: 2124–2130, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fang XF, Zheng P, Tang J, Liu Y. CD24: from A to Z. Cell Mol Immunol 7: 100–103, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol 296: G1108–G1118, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerbe F, Brulin B, Makrini L, Legraverend C, Jay P. DCAMKL-1 expression identifies tuft cells rather than stem cells in the adult mouse intestinal epithelium. Gastroenterology 137: 2179–2180, 2009. [DOI] [PubMed] [Google Scholar]
- 13. Giannakis M, Stappenbeck TS, Mills JC, Leip DG, Lovett M, Clifton SW, Ippolito JE, Glasscock JI, Arumugam M, Brent MR, Gordon JI. Molecular properties of adult mouse gastric and intestinal epithelial progenitors in their niches. J Biol Chem 281: 11292–11300, 2006. [DOI] [PubMed] [Google Scholar]
- 14. Gracz AD, Ramalingam S, Magness ST. Sox9-expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol Gastrointest Liver Physiol 298: G590–G600, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gulati AS, Ochsner SA, Henning SJ. Molecular properties of side population-sorted cells from mouse small intestine. Am J Physiol Gastrointest Liver Physiol 294: G286–G294, 2008. [DOI] [PubMed] [Google Scholar]
- 16. Haimovitz Friedman A, Cordon Cardo C, Bayoumy S, Garzotto M, McLoughlin M, Gallily R, Edwards CK, Schuchman EH, Fuks Z, Kolesnick R. Lipopolysaccharide induces disseminated endothelial apoptosis requiring ceramide generation. J Exp Med 186: 1831–1841, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hooper LV, Wong MH, Thelin A, Hansson L, Falk PC, Gordon JI. Molecular analysis of commensal host-microbial relations hips in the intestine. Science 291: 881–884, 2001. [DOI] [PubMed] [Google Scholar]
- 18. Kayahara T, Sawada M, Takaishi S, Fukui H, Seno H, Fukuzawa H, Suzuki K, Hiai H, Kageyama R, Okano H, Chiba T. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett 535: 131–135, 2003. [DOI] [PubMed] [Google Scholar]
- 19. Lee G, White LS, Hurov KE, Stappenbeck TS, Piwnica-Worms H. Response of small intestinal epithelial cells to acute disruption of cell division through CDC25 deletion. Proc Natl Acad Sci U S A 106: 4701–4706, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A 98: 31–36, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science 327: 542–545, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lim SC. CD24 and human carcinoma: tumor biological aspects. Biomed Pharmacother 59, Suppl 2: S351–S354, 2005. [DOI] [PubMed] [Google Scholar]
- 23. May R, Riehl TE, Hunt C, Sureban SM, Anant S, Houchen CW. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells 26: 630–637, 2008. [DOI] [PubMed] [Google Scholar]
- 24. May R, Sureban SM, Hoang N, Riehl TE, Lightfoot SA, Ramanujam R, Wyche JH, Anant S, Houchen CW. DCAMKL-1 and LGR5 mark quiescent and cycling intestinal stem cells respectively. Stem Cells, 2571–2579, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Montgomery RK, Breault DT. Small intestinal stem cell markers. J Anat 213: 52–58, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nielsen PJ, Lorenz B, Muller AM, Wenger RH, Brombacher F, Simon M, vonderWeid T, Langhorne J, Mossmann H, Kohler G. Altered erythrocytes and a leaky block in B-cell development in CD24/HSA-deficient mice. Blood 89: 1058–1067, 1997. [PubMed] [Google Scholar]
- 27. Nieoullon V, Belvindrah R, Rougon G, Chazal G. Mouse CD24 is required for homeostatic cell renewal. Cell Tissue Res 329: 457–467, 2007. [DOI] [PubMed] [Google Scholar]
- 28. Ochsner SA, Strick-Marchand H, Qiu Q, Venable S, Dean A, Wilde M, Weiss MC, Darlington GJ. Transcriptional profiling of bipotential embryonic liver cells to identify liver progenitor cell surface markers. Stem Cells 25: 2476–2487, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Potten CS, Booth C, Tudor GL, Booth D, Brady G, Hurley P, Ashton G, Clarke R, Sakakibara S, Okano H. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation 71: 28–41, 2003. [DOI] [PubMed] [Google Scholar]
- 30. Potten CS, Gandara R, Mahida YR, Loeffler M, Wright NA. The stem cells of small intestinal crypts: where are they? Cell Prolif 42: 731–750, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pruszak J, Ludwig W, Blak A, Alavian K, Isacson O. CD15, CD24, and CD29 define a surface biomarker code for neural lineage differentiation of stem cells. Stem Cells 27: 2928–2940, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richmond C, Montgomery R, Ambruzs D, Carlone D, Breault D. Characterization of slowly cycling telomerase-expressing intestinal stem cells. Gastroenterology 138: S–52, 2010. [Google Scholar]
- 33. Rietze RL, Valcanis H, Brooker GF, Thomas T, Voss AK, Bartlett PF. Purification of a pluripotent neural stem cell from the adult mouse brain. Nature 412: 736–739, 2001. [DOI] [PubMed] [Google Scholar]
- 34. Sammar M, Aigner S, Altevogt P. Heat-stable antigen (mouse CD24) in the brain: dual but distinct interaction with P-selectin and L1. Biochim Biophys Acta 1337: 287–294, 1997. [DOI] [PubMed] [Google Scholar]
- 35. Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40: 915–920, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sato T, Vries RG, Snippert HJ, van de WM, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265, 2009. [DOI] [PubMed] [Google Scholar]
- 37. Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem 285: 194–204, 2000. [DOI] [PubMed] [Google Scholar]
- 38. Scoville DH, Sato T, He XC, Li L. Current view: intestinal stem cells and signaling. Gastroenterology 134: 849–864, 2008. [DOI] [PubMed] [Google Scholar]
- 39. Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature 439: 84–88, 2006. [DOI] [PubMed] [Google Scholar]
- 40. Snippert HJ, van Es JH, van den BM, Begthel H, Stange DE, Barker N, Clevers H. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology 136: 2187–2194, 2009. [DOI] [PubMed] [Google Scholar]
- 41. Stappenbeck TS, Mills JC, Gordon JI. Molecular features of adult mouse small intestinal epithelial progenitors. Proc Natl Acad Sci U S A 100: 1004–1009, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature 439: 993–997, 2006. [DOI] [PubMed] [Google Scholar]
- 43. Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, Poppleton H, Zakharenko S, Ellison DW, Gilbertson RJ. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 457: 603–607, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.