Abstract
The p53 tumor suppressor is principally regulated by post-translational modifications and proteasome-dependent degradation. Various kinases have been shown to phosphorylate p53, but little is known about the counteracting phosphatases. We demonstrate here that the newly identified complex GAS41-PP2Cβ, and not PP2Cβ alone, is specifically required for dephosphorylation of serine 366 on p53. Ectopic expression of GAS41 and PP2Cβ reduces UV radiation-induced p53 up-regulation, thereby increasing the cell survival upon genotoxic DNA damage. To our knowledge, the GAS41-PP2Cβ complex is the first example in which substrate specificity of a PP2C family member is controlled by an associated regulatory subunit. Because GAS41 is frequently amplified in human gliomas, our finding illustrates a novel oncogenic mechanism of GAS41 by p53 dephosphorylation.
Keywords: DNA Damage, p53, PP2C, Protein Phosphatase, Protein-Protein Interactions
Introduction
The p53 tumor suppressor is known as a genome guardian because of its pivotal role in maintaining genome integrity and preventing cells from cancer development (1). The major mechanism that regulates the level of p53 involves Mdm2-dependent negative feedback regulation in which Mdm2 targets p53 for ubiquitination and subsequent proteasomal degradation (2, 3). In response to DNA damage, the levels of p53 dramatically increase as a consequence of disruption of the p53-Mdm2 interaction. The relevance of p53 stabilization by post-translational modifications, especially phosphorylation, has been extensively studied in relation to the p53-Mdm2 interaction (4–6). In general, phosphorylation of p53 blocks the p53-Mdm2 interaction and thus stabilizes p53 in stressed cells (7, 8). However, phosphorylation at specific p53 sites by TAF1 (threonine 55), the COP9 signalosome kinase (threonine 155), Aurora kinase A (serine 315), and IκB kinase 2 (IKK2) (serine 366) is known to induce p53 destabilization (9–12).
Reversible protein phosphorylation by a kinase/phosphatase pair constitutes a fundamental mechanism to regulate signal transduction pathways in response to various environmental and metabolic stimuli. The protein phosphatase magnesium-dependent phosphatase PP2C2 family includes at least 18 different human members, all of which share high sequence homology within the catalytic domain (13). Despite the functional importance of PP2C proteins in embryonic development, apoptosis, stress response, and tumor transformation, the regulatory mechanisms that govern activation, targeting, and deactivation of PP2C have not been elucidated (14). Because no known regulatory proteins have been shown to be required for modulating PP2C activity and specificity, it has been generally assumed that phosphatase activity is controlled by passive mechanisms such as subcellular compartmentalization, cell type-specific expression, and degradation (15).
GAS41 is a highly conserved nuclear protein whose gene is frequently amplified in certain brain tumors (16). GAS41 is a member of a protein family that is characterized by the presence of an N-terminal YEATS domain (Yaf9, ENL, AF9, Taf14, Sas5) (17). Because YEATS domain-containing proteins are found in chromatin-modifying and transcription regulatory complexes, this domain is proposed to function in histone and chromatin interactions (18). We previously reported that GAS41 plays a role in repressing the p53 tumor suppressor and appears to reside in a different complex that is independent of the GAS41-containing TIP60 and SRCAP chromatin-remodeling/modifying complexes (19). Here we report the identification of a novel GAS41-PP2Cβ complex that shows phosphatase specificity for phosphorylated serine 366 of p53. Our results indicate that substrate specificity of PP2Cβ and its effect on p53 stability are dependent on the associated GAS41 subunit.
EXPERIMENTAL PROCEDURES
Expression Plasmids
The cDNAs for FLAG-PP2Cβ and phosphatase-defective PP2Cβ (Arg-179 → Gly) were obtained from Dr. Gaynor (32). The CbF expression plasmids for FLAG-GAS41, FLAG-GAS41 (Δ193–225), and FLAG-GST-Coil (193–225) were described previously (19). HA-tagged cDNAs for GAS41 (1–225), GAS41ΔCoil (1–192), and GST-Coil (193–225) were constructed in CbS-3HA mammalian expression vector. GST fusion and His-tagged constructs for recombinant protein preparations were made in pGEX4T-1 and 6HpET11d vectors, respectively.
GST Pulldown Assay
2 μg of GST fusion proteins were bound to glutathione-Sepharose 4B beads, and 1 μg of purified proteins was mixed in binding buffer (20 mm Tris-Cl (pH 7.5), 150 mm KCl, 0.2 mm EDTA, 20% glycerol, 0.05% Nonidet P-40, 1 mg/ml BSA, and 0.5 mm PMSF). After a 3-h incubation at 4 °C, the beads were washed with binding buffer, and bound proteins were resolved by SDS-PAGE and immunoblot analysis.
Protein Purification
For purification of GAS41-PP2Cβ complexes from 293T cells that stably express FLAG-GAS41, nuclear extracts were prepared by a modified Dignam procedure and directly applied to a Sepharose CL6B gel filtration column to remove GAS41-containing nuclear chromatin-remodeling complexes (19). The fractions corresponding to the GAS41 protein peak around 158 kDa of apparent molecular mass were combined and subjected to M2 agarose affinity purification. After being serially washed with BC300 (20 mm Tris-Cl (pH 7.9), 0.2 mm EDTA, 20% glycerol, 300 mm KCl, 0.5 mm PMSF) and BC500 (20 mm Tris-Cl (pH 7.9), 0.2 mm EDTA, 20% glycerol, 500 mm KCl, 0.5 mm PMSF) buffer containing 0.1% Nonidet P-40, captured FLAG-GAS41 was eluted by the addition of 250 μg/ml FLAG peptide at 4 °C.
Protein Phosphatase Assay
The Promega non-radioactive serine/threonine phosphatase assay system was used for measuring the protein phosphatase activity of PP2Cβ1. For enzyme preparation, His-PP2Cβ1 was expressed in Escherichia coli and purified on Ni-NTA resin. For the heterodimeric PP2Cβ1 complex, plasmids expressing His-PP2Cβ1 and GST-GAS41 were co-introduced in E. coli, and the complexes were purified using double affinity chromatography of glutathione-Sepharose 4B and Ni-NTA resin. After normalizing the protein amount for the His-PP2Cβ1-associated and the GAS41-associated complex, protein phosphatase assays were conducted using phosphopeptide (RRA(phospho-T)VA) as a substrate. A detailed procedure and the composition of buffers were described in the manufacturer's protocol. For preparation of phosphorylated p53 as a substrate, FLAG-p53 was transiently overexpressed in 293T cells and purified on M2 agarose from cells after 3 h of UV irradiation. Prepared FLAG-p53 was subjected to phosphatase assay in the reaction buffer (10 mm Tris-HCl (pH 7.9), 10 mm MgCl2, 50 mm NaCl, 1 mm dithiothreitol, 50 μm okadaic acid, protease inhibitor mixture) at 37 °C for 30 min. Reactions were divided into two equal volumes and analyzed for phosphorylation status and protein amount by immunoblotting.
RESULTS
GAS41 Associates with PP2Cβ
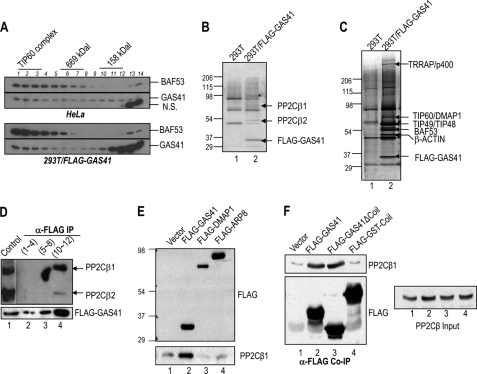
To test the hypothesis that GAS41 can form a nuclear complex distinct from GAS41-containing TIP60 or SRCAP complexes, we first analyzed the size distribution of GAS41 in HeLa nuclear extracts by Sepharose CL6B gel filtration and immunoblot (Fig. 1A, upper panel). As a control, the size distribution of BAF53, a common subunit of chromatin-remodeling complexes such as human SWI/SNF-related complexes, was also analyzed (20). Consistent with the apparent molecular mass of known BAF53-containing complexes, most BAF53 was found in molecular mass fractions greater than the 669-kDa marker. However, GAS41 showed distinct immunoblot signals around 158 kDa, suggesting that GAS41 is associated with a smaller sized protein complex distinct from known GAS41-containing chromatin-remodeling/modifying complexes. To identify GAS41-associated proteins in the ∼158-kDa fractions, we size-fractionated nuclear extracts derived from 293T HEK cells that stably express FLAG-GAS41 (Fig. 1A, bottom panel). The size distribution of FLAG-GAS41 was similar to that observed with HeLa nuclear extracts, except for the excess of monomeric FLAG-GAS41 (Fig. 1A, bottom panel, lanes 13 and 14). The FLAG-GAS41 complex in the ∼158-kDa fractions (Fig. 1A, bottom panel, lanes 10–12) was purified on M2 agarose and subsequently resolved by SDS-PAGE (Fig. 1B). This analysis revealed two unique bands, with apparent masses of 50 and 65 kDa, that were not evident in the control preparation of 293T HEK parental cells (Fig. 1B, lane 2 versus lane 1). Mass spectrometric analyses of these proteins identified two major human isoforms of PP2Cβ1 (65 kDa) and PP2Cβ2 (50 kDa). Because the purified protein preparation showed a polypeptide composition different from that of typical GAS41-containing TIP60 complex (Fig. 1C, lane 2), this GAS41-PP2Cβ complex appears to be a separable and independent nuclear complex. To confirm the association of GAS41 with PP2Cβ, three fractions from the gel filtration were pooled, immunoprecipitated with M2 agarose, and analyzed for enrichment of PP2Cβ by immunoblotting (Fig. 1D). Consistent with mass spectrometric analysis, two human isoforms of PP2Cβ were enriched in the ∼158-kDa fractions (Fig. 1D, lane 4) but were not detectable in the higher molecular mass fractions (Fig. 1D, lanes 2 and 3). The GAS41-PP2Cβ complex appears to exist as a stable complex in cells because the association between GAS41 and PP2Cβ1 was not dependent on DNA damage such as UV irradiation (supplemental Fig. 1A).
FIGURE 1.
A small GAS41 complex copurifies with two isoforms of human PP2Cβ. A, immunoblots of BAF53 and GAS41, which were resolved by Sepharose CL6B chromatography. Nuclear extracts from HeLa (upper panel) and 293T/FLAG-GAS41 (bottom panel) cells were analyzed with Sepharose CL6B gel filtration. Higher numbers in lanes represent lower molecular masses. N.S., not significant. B, SDS-PAGE/silver stain analysis of mock control and FLAG-GAS41 preparations from the Sepharose CL6B gel filtration analysis (A, lower panel, lanes 10–12) followed by affinity purification on M2 agarose. A nonspecific contaminant band is marked by the asterisk. C, SDS-PAGE/silver stain analysis of mock control and FLAG-GAS41 preparations from the Sepharose CL6B gel filtration analysis (A, lower panel, lanes 1–3) followed by affinity purification on M2 agarose. D, coimmunoprecipitation (IP) of PP2Cβ with FLAG-GAS41. Three pooled fractions (fractions 1–4, 5–8, and 10–12), originally resolved by Sepharose CL6B gel filtration (A, bottom panel), were subject to immunoprecipitation by M2 agarose. FLAG-GAS41 and enrichment of endogenous PP2Cβ1 and PP2Cβ2 were analyzed by immunoblot with anti-FLAG and anti-PP2Cβ antibodies. HeLa nuclear extract was used as a control (lane 1). E, selective association of PP2Cβ1 with GAS41 but not with a subunit of chromatin-remodeling complexes in 293T cells. FLAG-tagged GAS41, DMAP1, and ARP8 were ectopically expressed in 293T cells and subjected to immunoprecipitation using M2 agarose. F, interaction of PP2Cβ1 with the YEATS domain of GAS41. FLAG-tagged GAS41 (1–225), FLAG-GAS41ΔCoil (C-terminal-deleted GAS41 (1–192)), and FLAG-GST-Coil (GAS41 C-terminal domain fused with GST) were ectopically expressed in 293T cells and subjected to immunoprecipitation by M2 agarose.
The GAS41-PP2Cβ Complex Is Distinct from GAS41-containing Chromatin-remodeling Complexes
To exclude the possibility that a small amount of PP2Cβ is also associated with the GAS41-containing TIP60 or SRCAP complexes, the enrichment of endogenous PP2Cβ1 was analyzed by coimmunoprecipitating ectopically expressed FLAG-DMAP1, a common subunit of TIP60 and SRCAP complexes (21), and FLAG-ARP8, a subunit of the human INO80 complex (22) that lacks GAS41. The FLAG-DMAP1 and FLAG-ARP8 immunoprecipitates showed only low background levels of endogenous PP2Cβ1 on the immunoblot (Fig. 1E, lanes 1, 3, and 4). However, PP2Cβ1 clearly coimmunoprecipitated with FLAG-GAS41 (Fig. 1E, compare lane 2 versus the other lanes), suggesting that the GAS41-PP2Cβ complex is distinct from known nuclear chromatin-remodeling complexes. We have previously shown that the coiled-coil domain of GAS41 is critical for assembly into the TIP60 and SRCAP complexes (19). To investigate the role of the coiled-coil domain in formation of the GAS41-PP2Cβ complex, a FLAG-GAS41 that lacks the C-terminal coiled-coil domain (FLAG-GAS41ΔCoil) and a FLAG-GST fused to GAS41 C-terminal coil domain (FLAG-GST-Coil) were used in coimmunoprecipitation analysis with endogenous PP2Cβ1 (Fig. 1F). PP2Cβ1 was coimmunoprecipitated by both FLAG-GAS41 and the mutant lacking the C-terminal coil domain, suggesting that the coiled-coil domain of GAS41 is dispensable for the interaction with PP2Cβ (Fig. 1F, lanes 2 and 3). These results suggest that the GAS41-PP2Cβ complex is an independent complex, not related to previously reported GAS41-containing chromatin-remodeling complexes.
A Reconstituted GAS41-PP2Cβ Heterodimer Retains Protein Phosphatase Activity
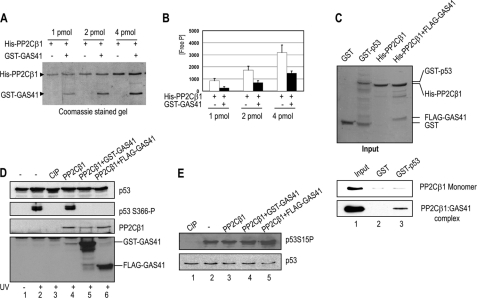
Diverse PP2C family members, including PP2Cβ, are known to exist as monomers and to interact transiently with their substrates (13). To investigate whether GAS41 plays a role in regulating PP2Cβ activity as a heterodimeric GAS41-PP2Cβ complex, we attempted to purify a recombinant heterodimer consisting of GAS41 and PP2Cβ1 from E. coli. Both GAS41 and PP2Cβ1 were coexpressed in E. coli and subjected to sequential affinity purifications on Ni-NTA and glutathione-Sepharose 4B (Fig. 2A). The individual components of the purified GAS41-PP2Cβ1 complex appear to exist in similar molar ratios based on the intensity of the Coomassie Brilliant Blue R-250-stained protein bands. This result indicates that the GAS41-PP2Cβ complex appears to be reasonably stable in vitro, given that it remains intact following two affinity purification steps. The GST tag is not likely to play a role in this assembly because we were able to purify a heterodimeric complex consisting of FLAG-GAS41 and His-PP2Cβ1 from E. coli (Fig. 2C, Input panel). The concentration of protein was normalized based on Coomassie Brilliant Blue R-250 staining of the PP2Cβ band and subjected to a malachite green-based phosphatase assay that measures inorganic phosphate release from a phosphopeptide substrate (Fig. 2B). The monomer and heterodimer of PP2Cβ1 showed a concentration-dependent increase in phosphatase activity. Despite the presence of similar amounts of PP2Cβ1 in each reaction, the GAS41-PP2Cβ1 complex showed a reduced phosphatase activity (∼50%) relative to that of the monomeric PP2Cβ1. These results indicate that the association of GAS41 with PP2Cβ1 reduces the activity of the latter for the synthetic peptide substrate in vitro. It remains to be determined whether the reduction of the PP2Cβ catalytic activity in vitro is relevant to the regulatory role of associated GAS41 in cells. However, based on the proposed role of GAS41 in substrate recognition in chromatin-remodeling complexes (17), we next explored a possible substrate for the GAS41-PP2Cβ complex that would be targeted specifically by the GAS41 subunit.
FIGURE 2.
A reconstituted GAS41-PP2Cβ heterodimer binds to and dephosphorylates p53 at serine 366 in vitro. A, double affinity purification of the heterodimeric GAS41-PP2Cβ complex. pET28b-His-PP2Cβ1 (KmR) and pGEX-GST-GAS41 (AmpR) were coexpressed in E. coli, and the protein complexes were purified by sequential affinity purifications on Ni-NTA and glutathione-Sepharose 4B resins. The preparations were resolved on an SDS-PAGE gel and stained with Coomassie Brilliant Blue R-250. B, in vitro serine/threonine phosphatase assay of PP2Cβ1 monomer and GAS41-PP2Cβ1 heterodimer. Three independent colorimetric measurements of phosphatase assays were carried out, and the average values are shown. Free P, free phosphate. Error bars indicate S.D. C, direct binding of GAS41-PP2Cβ1 to p53 in vitro. Recombinant His-PP2Cβ1 monomer and FLAG-GAS41-His-PP2Cβ1 heterodimer, purified from E. coli, were used in GST pulldown assays. The amounts of recombinant proteins used in the assays are shown in the Coomassie Brilliant Blue R-250-stained gel (upper Input panel). His-PP2Cβ1 binding to GST (bottom panel, lane 2) or GST-p53 (bottom panel, lane 3) proteins was scored by immunoblot using anti-PP2Cβ antibody. D, selective dephosphorylation of serine 366 on p53 by the GAS41-PP2Cβ1 complex. Equal amounts of purified FLAG-p53 derived from UV-treated 293T cells (lanes 2–6) were subjected to phosphatase assays using calf intestinal phosphatase (CIP, lane 3), recombinant His-PP2Cβ1 monomer (lane 4), or two preparations of PP2Cβ1 heterodimers containing GST-GAS41 (lane 5) and FLAG-GAS41 (lane 6), respectively. Dephosphorylation of serine 366 on the p53 was monitored by immunoblot with a phosphospecific p53 antibody. E, dephosphorylation assay of p53 at serine 15. Equal amounts of purified FLAG-p53 derived from UV-treated 293T cells (lanes 1–5) were subjected to phosphatase assays with calf intestinal phosphatase (lane 1), recombinant His-PP2Cβ1 monomer (lane 3), or two preparations of PP2Cβ1 heterodimers containing GST-GAS41 (lane 4) and FLAG-GAS41 (lane 5), respectively. Dephosphorylation of serine 15 on the p53 was monitored by immunoblot with a phosphospecific p53 antibody.
GAS41-PP2Cβ Interacts with p53 and Dephosphorylates Serine 366 of p53 in Vitro
Because both GAS41 and PP2C family members have been implicated in p53 regulation of human cancers (17, 23, 24), we first checked for a possible interaction between p53 and the GAS41-PP2Cβ1 complex. Monomeric His-PP2Cβ1 and heterodimeric FLAG-GAS41-His-PP2Cβ1 were purified from E. coli and tested by GST pulldown assays for interactions with p53 (Fig. 2C). In contrast to monomeric PP2Cβ1, which did not show selective binding to either GST or GST-p53 (Fig. 2C, PP2Cβ1 Monomer panel), the GAS41-PP2Cβ1 complex selectively bound GST-p53 (Fig. 2C, bottom panel, lane 3). The absence of an interaction between p53 and the PP2Cβ1 monomer in these assays suggests that the PP2Cβ interaction with p53 requires an associated GAS41 subunit. This is also supported by the fact that GAS41ΔCoil that is selectively associable with PP2Cβ was associated with p53 in cells (supplemental Fig. 1B).
Given that PP2Cβ is an IKK2 phosphatase (12), we checked whether the GAS41-PP2Cβ1 complex plays a role in counteracting IKK2-mediated p53 phosphorylation at serine 366. FLAG-p53 was ectopically expressed in 293T HEK cells, and post-translational modifications on the p53 were induced by UV irradiation. FLAG-p53 was purified on M2 agarose under native conditions and used as a substrate in the phosphatase assays in vitro (Fig. 2, D and E). UV irradiation significantly induced phosphorylation at serine 366 of p53, as demonstrated with a phosphospecific p53 antibody (25) (Fig. 2D, lane 2 versus lane 1). As a positive control, calf intestinal phosphatase was used to show that the phosphate group on p53 can be removed in vitro (Fig. 2D, lane 3). The recombinant heterodimeric complexes of either GST-GAS41-His-PP2Cβ1 or FLAG-GAS41-His-PP2Cβ1 efficiently removed the phosphate group on serine 366 of p53, whereas a comparable amount of monomeric His-PP2Cβ1 was phosphatase-inactive (Fig. 2D, compare lanes 5 and 6 with lane 4). In contrast, there was no loss of the phosphate group from serine 15 of p53 with recombinant PP2Cβ1 either in the presence or in the absence of GAS41, demonstrating significant substrate specificity of the GAS41-PP2Cβ complex (Fig. 2E).
GAS41-PP2Cβ Dephosphorylates p53 at Serine 366 in Cells
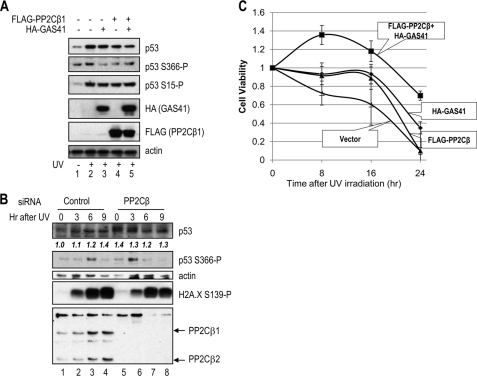
To demonstrate the substrate specificity of the GAS41-PP2Cβ complex in cells, GAS41 and PP2Cβ1 were ectopically expressed in U2OS cells, and the endogenous p53 phosphorylation status at serine 366 was evaluated after UV irradiation. Immunoblot analysis showed that p53 phosphorylation at serine 15 and serine 366 increased in UV-irradiated cells (Fig. 3A, compare lane 2 with lane 1). When either GAS41 or PP2Cβ1 was ectopically expressed, serine 15 phosphorylation of p53 remained unchanged upon UV irradiation, whereas significantly, the level of serine 366 phosphorylation returned to basal levels (Fig. 3A, lanes 3–5). Moreover, coexpression of GAS41 and PP2Cβ1 significantly reduced the amount of stabilized p53, suggesting that the GAS41-PP2Cβ1 complex could be involved in p53 homeostasis after p53 activation (Fig. 3A, top panel, compare lane 5 with lane 2). However, because the GAS41-PP2Cβ1 complex could dephosphorylate other phosphorylation sites of p53, it remains to be determined whether the effect of GAS41-PP2Cβ1 here is solely mediated from inhibiting p53 serine 366 phosphorylation (12).
FIGURE 3.
Coexpression of GAS41 and PP2Cβ affects the phosphorylation status and stability of p53 in cells. A, specific reduction in phosphorylation at serine 366 on p53 in cells. FLAG-PP2Cβ1 and HA-GAS41 were ectopically expressed in U2OS cells for 24 h followed by UV irradiation (lanes 2–5). Whole cell extracts were analyzed by immunoblotting with the indicated antibodies. Representative results from one of the three independent experiments are shown. B, time course of p53 phosphorylation at serine 366. U2OS cells were transfected with control and PP2Cβ siRNAs followed by UV irradiation after 24 h of transfection. Whole cell extracts were analyzed by immunoblotting with the indicated antibodies. The p53 levels are normalized against β-actin bands, and the relative levels are shown in the p53 immunoblot panel. C, effects of GAS41-PP2Cβ1 complex in U2OS cells to UV-induced cytotoxicity. To assess UV sensitivity upon GAS41-PP2Cβ1 overexpression, GAS41 and PP2Cβ1 were ectopically overexpressed individually or together in U2OS cells, and UV-mediated cytotoxicity was evaluated using a Promega CellTiter 96 AQueous One solution and a fluorescence plate reader. The value obtained at 490 nm fluorescence reading after UV irradiation is divided by the value obtained from non-stressed cells, and the average values of triplicate experiments are shown in the graph. ▵, vector control; ◊, GAS41; ▴, PP2Cβ; ■, GAS41+PP2Cβ. Error bars indicate S.D.
We next examined whether phosphorylation of p53 at serine 366 is regulated by endogenous PP2Cβ. To exclude the possibility of other p53-modifying complexes such as GAS41-containing TIP60 complexes, we first used PP2Cβ siRNA transfection for selective knockdown of the phosphatase (Fig. 3B). Transfection of U2OS cells with PP2Cβ siRNA reduced the two human isoforms of PP2Cβ to undetectable levels as compared with a non-targeting control siRNA (Fig. 3B, bottom panel, compare lanes 5–8 with lanes 1–4). Consistent with a previous report (25), phosphorylation of p53 at serine 366 in U2OS cells treated with control siRNA was relatively slow with maximal phosphorylation being observed 6 h after UV treatment (Fig. 3B, lane 3). However, in the PP2Cβ knockdown, phosphorylation of serine 366 was much more rapid with a strong signal detected at 3 h after UV treatment (Fig. 3B, lane 6). In comparison, H2A.X phosphorylation, which is mediated by ATM/ATR (26), was not changed by knockdown of PP2Cβ during the time course (Fig. 3B, H2A.X S139-P immunoblot). Given that p53 phosphorylation at serine 366 in the PP2Cβ knockdown shows the normal pattern of down-regulation between 6 and 9 h (Fig. 3B, lanes 7 and 8), phosphatases other than PP2Cβ must also be involved in regulating serine 366 dephosphorylation. Because the removal of PP2Cβ from IKK2 can possibly enhance the IKK2-mediated p53 phosphorylation, the change of the p53 phosphorylation kinetics by the PP2Cβ knockdown could be mediated by both up-regulation of the upstream kinase IKK2 and/or down-regulation of the GAS41-PP2Cβ complex. Interestingly, like a GAS41 knockdown effect that has been shown to increase p53 level (27), the PP2Cβ knockdown also increased the p53 level significantly in the absence of UV irradiation (Fig. 3B, lane 5). The result suggests that GAS41-PP2Cβ complex plays a role in destabilizing p53 through multiple mechanisms, not solely relying on the dephosphorylation of serine 366.
GAS41-PP2Cβ Enhances Cell Survival after Genotoxic UV Irradiation
To assess cellular proliferation and UV sensitivity, we ectopically coexpressed GAS41 and PP2Cβ1 in U2OS cells and evaluated UV-mediated cytotoxicity (Fig. 3C). Ectopic coexpression of GAS41 and PP2Cβ1 significantly protected cells from UV-mediated cell death, with 70% of cells still viable 24 h after a lethal dose of UV irradiation. In contrast, expression of either GAS41 or PP2Cβ1 alone showed a modest protection up to 16 h after UV irradiation. These results are consistent with the p53 levels observed from ectopic expression of GAS41 and PP2Cβ1 (Fig. 3A), suggesting that a reduced level of p53 stabilization by the GAS41-PP2Cβ complex results in a decreased UV-induced cell death.
DISCUSSION
We have found that GAS41 is stably associated with PP2Cβ and plays a role in determining the substrate specificity of PP2Cβ for p53. GAS41-PP2Cβ is an independent complex that is distinct from other GAS41-containing chromatin-remodeling complexes such as the TIP60 or SRCAP complex. This conclusion is supported by the fact that DMAP1, a common subunit of the TIP60 and SRCAP complexes, is not associated with PP2Cβ and that a GAS41 mutant with an inability to interact with chromatin-remodeling complexes still retains an interaction with PP2Cβ. The GAS41-PP2Cβ heterodimer interacts with and dephosphorylates p53 specifically at serine 366 both in vitro and in cells. Serine 366 of p53 is one of several amino acid residues that are inducibly phosphorylated by Chk1 and Chk2 upon DNA damage (25). The same phosphorylation of p53 is also mediated by IKK2, which serves as a p53 degradation mark for the SCFβ-TrCP E3 ubiquitin ligase (12). IKK2-dependent phosphorylation occurs specifically at serine 362 and 366 of p53 because IKK2−/− mouse embryonic fibroblast cells or siRNA-mediated IKK2 knockdown result in the loss of serine 366 phosphorylation and further stabilization of p53 (12). However, we have found that GAS41 and PP2Cβ coexpression decreases the p53 stabilization in UV-stressed cells. Given the conflicting role of serine 366 phosphorylation in p53 stability, we examined cell viability using two different single point mutations at serine 366. Under ectopic transient expression conditions, p53 serine 366 mutants did not show any significant difference in expression levels and cell viabilities as compared with wild type after lethal UV irradiation (supplemental Fig. 2). Human p53 is known to have more than 20 different phosphorylation and dephosphorylation sites. Because there are further possibilities for the post-translational modification of p53 through interaction with the GAS41-PP2Cβ complex, this issue is currently being investigated using in vitro phosphatase assays coupled with phosphopeptide mapping.
GAS41 was first identified as one of several genes that are amplified in human glioblastoma multiforme cell lines and subsequently has been shown to be frequently amplified in all grades of gliomas (16). More importantly, as the majority of gene amplifications in gliomas tend to occur exclusively in late stage tumors, the early amplification of GAS41 suggests that it has significant oncogenic activity during early tumor progression. We previously reported that GAS41 plays a role as a negative regulator of p53 that is independent of chromatin-remodeling/modifying complexes (19). Subsequently, a large scale loss-of-function screen has identified GAS41 as a negative regulator of p53 stability (27). Coexpression of GAS41 with p53 significantly reduces p53 levels comparable with those observed with MDM2 and p53 coexpression (27). Our novel finding that GAS41 forms a complex with a member of the PP2C family of phosphatases establishes a new mechanism to down-regulate p53 activation.
PP2Cβ plays diverse roles in cell proliferation, differentiation, and stress signaling. On the one hand, PP2Cβ suppresses a cell survival signal derived from the NFκB signaling pathway and activates a proapoptotic signal by dephosphorylating the pro-apoptotic protein, BAD. Therefore, PP2Cβ seems to facilitate growth arrest and cell death through multiple targets and mechanisms (28, 29). On the other hand, PP2Cβ is a negative regulator of stress signaling pathways that include the p38 and c-Jun pathways (30). Stress-activated p38 and c-Jun kinases phosphorylate multiple p53 residues (including serine 15, serine 33, serine 46, threonine 81, and serine 392) and promote p53 stability and functional activities including transcription and apoptosis (6). Therefore, PP2Cβ may represent one of several negative feedback regulators for p53 activation. Consistent with a role as a negative feedback regulator, PP2Cβ is known to be significantly up-regulated by p53 in leukemic cells (31). Thus, PP2Cβ is a p53-regulated gene, although it remains to be determined whether p53 can directly regulate transcription of the PP2Cβ gene. Taken together, our results reveal a negative regulatory mechanism for p53 through an interaction of PP2Cβ with GAS41. Based on the results of this work, it will also be important to determine whether there is a relationship between the levels of GAS41-PP2Cβ and p53 function in a range of human tumor samples.
Supplementary Material
Acknowledgments
We thank Drs. Tracy Hale, Barry Scott, and Natisha Magan for critical reading of this manuscript. We are grateful to Dr. Sara Lavi for human PP2Cβ antibody and Donna Montesclaros for the technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant CA129325 (to R. G. R.) and by a Massey University Research Fund grant (to J. H. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- PP2C
- protein phosphatase 2C
- Ni-NTA
- nickel-nitrilotriacetic acid
- SRCAP
- snf2-related CREB-binding protein activator protein.
REFERENCES
- 1. Vousden K. H., Prives C. (2009) Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 2. Scheffner M., Huibregtse J. M., Vierstra R. D., Howley P. M. (1993) Cell 75, 495–505 [DOI] [PubMed] [Google Scholar]
- 3. Poyurovsky M. V., Prives C. (2006) Genes Dev. 20, 125–131 [DOI] [PubMed] [Google Scholar]
- 4. Bode A. M., Dong Z. (2004) Nat. Rev. Cancer 4, 793–805 [DOI] [PubMed] [Google Scholar]
- 5. Olsson A., Manzl C., Strasser A., Villunger A. (2007) Cell Death Differ. 14, 1561–1575 [DOI] [PubMed] [Google Scholar]
- 6. Toledo F., Wahl G. M. (2006) Nat. Rev. Cancer 6, 909–923 [DOI] [PubMed] [Google Scholar]
- 7. Shieh S. Y., Ikeda M., Taya Y., Prives C. (1997) Cell 91, 325–334 [DOI] [PubMed] [Google Scholar]
- 8. Chehab N. H., Malikzay A., Stavridi E. S., Halazonetis T. D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13777–13782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bech-Otschir D., Kraft R., Huang X., Henklein P., Kapelari B., Pollmann C., Dubiel W. (2001) EMBO J. 20, 1630–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Katayama H., Sasai K., Kawai H., Yuan Z. M., Bondaruk J., Suzuki F., Fujii S., Arlinghaus R. B., Czerniak B. A., Sen S. (2004) Nat. Genet. 36, 55–62 [DOI] [PubMed] [Google Scholar]
- 11. Li H. H., Li A. G., Sheppard H. M., Liu X. (2004) Mol. Cell 13, 867–878 [DOI] [PubMed] [Google Scholar]
- 12. Xia Y., Padre R. C., De Mendoza T. H., Bottero V., Tergaonkar V. B., Verma I. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moorhead G. B., Trinkle-Mulcahy L., Ulke-Lemée A. (2007) Nat. Rev. Mol. Cell Biol. 8, 234–244 [DOI] [PubMed] [Google Scholar]
- 14. Lammers T., Lavi S. (2007) Crit. Rev. Biochem. Mol. Biol. 42, 437–461 [DOI] [PubMed] [Google Scholar]
- 15. Klumpp S., Thissen M. C., Krieglstein J. (2006) Biochem. Soc. Trans. 34, 1370–1375 [DOI] [PubMed] [Google Scholar]
- 16. Fischer U., Heckel D., Michel A., Janka M., Hulsebos T., Meese E. (1997) Hum. Mol. Genet. 6, 1817–1822 [DOI] [PubMed] [Google Scholar]
- 17. Schulze J. M., Wang A. Y., Kobor M. S. (2009) Biochem. Cell Biol. 87, 65–75 [DOI] [PubMed] [Google Scholar]
- 18. Zeisig D. T., Bittner C. B., Zeisig B. B., García-Cuéllar M. P., Hess J. L., Slany R. K. (2005) Oncogene 24, 5525–5532 [DOI] [PubMed] [Google Scholar]
- 19. Park J. H., Roeder R. G. (2006) Mol. Cell. Biol. 26, 4006–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park J., Wood M. A., Cole M. D. (2002) Mol. Cell. Biol. 22, 1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Doyon Y., Selleck W., Lane W. S., Tan S., Côté J. (2004) Mol. Cell. Biol. 24, 1884–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jin J., Cai Y., Yao T., Gottschalk A. J., Florens L., Swanson S. K., Gutiérrez J. L., Coleman M. K., Workman J. L., Mushegian A., Washburn M. P., Conaway R. C., Conaway J. W. (2005) J. Biol. Chem. 280, 41207–41212 [DOI] [PubMed] [Google Scholar]
- 23. Tamura S., Toriumi S., Saito J., Awano K., Kudo T. A., Kobayashi T. (2006) Cancer Sci. 97, 563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Le Guezennec X., Bulavin D. V. (2010) Trends Biochem. Sci. 35, 109–114 [DOI] [PubMed] [Google Scholar]
- 25. Ou Y. H., Chung P. H., Sun T. P., Shieh S. Y. (2005) Mol. Biol. Cell 16, 1684–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Attikum H., Gasser S. M. (2009) Trends Cell Biol. 19, 207–217 [DOI] [PubMed] [Google Scholar]
- 27. Llanos S., Efeyan A., Monsech J., Dominguez O., Serrano M. (2006) Cell Cycle 5, 1880–1885 [DOI] [PubMed] [Google Scholar]
- 28. Sun W., Yu Y., Dotti G., Shen T., Tan X., Savoldo B., Pass A. K., Chu M., Zhang D., Lu X., Fu S., Lin X., Yang J. (2009) Cell. Signal. 21, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klumpp S., Selke D., Krieglstein J. (2003) Neurochem. Int. 42, 555–560 [DOI] [PubMed] [Google Scholar]
- 30. Hanada M., Ninomiya-Tsuji J., Komaki K., Ohnishi M., Katsura K., Kanamaru R., Matsumoto K., Tamura S. (2001) J. Biol. Chem. 276, 5753–5759 [DOI] [PubMed] [Google Scholar]
- 31. Lotem J., Benjamin H., Netanely D., Domany E., Sachs L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16022–16027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Prajapati S., Verma U., Yamamoto Y., Kwak Y. T., Gaynor R. B. (2004) J. Biol. Chem. 279, 1739–1746 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.