Abstract
In response to different environmental stresses, phosphorylation of eukaryotic initiation factor-2 (eIF2) rapidly reduces protein synthesis, which lowers energy expenditure and facilitates reprogramming of gene expression to remediate stress damage. Central to the changes in gene expression, eIF2 phosphorylation also enhances translation of ATF4, a transcriptional activator of genes subject to the integrated stress response (ISR). The ISR increases the expression of genes important for alleviating stress or alternatively triggering apoptosis. One ISR target gene encodes the transcriptional regulator CHOP whose accumulation is critical for stress-induced apoptosis. In this study, we show that eIF2 phosphorylation induces preferential translation of CHOP by a mechanism involving a single upstream ORF (uORF) located in the 5′-leader of the CHOP mRNA. In the absence of stress and low eIF2 phosphorylation, translation of the uORF serves as a barrier that prevents translation of the downstream CHOP coding region. Enhanced eIF2 phosphorylation during stress facilitates ribosome bypass of the uORF due to its poor start site context, and instead it allows scanning ribosomes to translate CHOP. This new mechanism of translational control explains how expression of CHOP and the fate of cells are tightly linked to the levels of phosphorylated eIF2 and stress damage.
Keywords: ER Stress, Protein Synthesis, Translation, Translation Control, Translation Regulation
Introduction
Rapid changes in global and gene-specific translation occur in response to many different environmental stresses. For example, translation is repressed in response to an accumulation of misfolded protein in the endoplasmic reticulum (ER),2 which prevents further overload of the secretory pathway and provides time for the reconfiguration of gene expression with a focus on stress alleviation. A central mechanism for this translational control involves phosphorylation of eIF2 (eIF2∼P) by the protein kinase PERK/PEK. eIF2 is a translation initiation factor that combines with initiator Met-tRNAiMet and GTP and participates in the selection of the start codon (1–3). Phosphorylation of the α subunit of eIF2 at Ser-51 in response to ER stress blocks the exchange of eIF2-GDP to eIF2-GTP, thus reducing global translation initiation and subsequent protein synthesis. In addition to PERK, three other eIF2 kinases respond to other stress conditions, including GCN2 induced by nutritional deprivation, HRI activated by heme deficiency in erythroid cells, and PKR, which functions in an antiviral defense pathway.
Accompanying this repression of global translational initiation, eIF2∼P selectively enhances the translation of ATF4 mRNA, encoding a basic zipper transcriptional activator of stress-related genes involved in metabolism, protection against oxidative damage, and regulation of apoptosis (3–5). The idea that ATF4 is a common downstream target that integrates signaling from PERK and other eIF2 kinases has led to the eIF2∼P/ATF4 pathway being collectively referred to as the integrated stress response (ISR) (4). Preferential translation of ATF4 mRNA during eIF2∼P occurs by a mechanism involving two upstream ORFs (uORFs) (6–8). The 5′-proximal uORF1 is a positive-acting element that enables ribosomes to reinitiate translation at a downstream ORF in the ATF4 transcript (8). When eIF2-GTP is readily available in nonstressed cells, ribosomes completing translation of uORF1 resume scanning downstream and reinitiate at the next coding region, uORF2, which is an inhibitory element that blocks ATF4 expression. During stress conditions, eIF2∼P and the lowered levels of eIF2-GTP increase the time required for the scanning ribosomes to reinitiate translation. Delayed reinitiation allows for ribosomes to scan through the inhibitory uORF2 and instead translate the ATF4 coding region (8). Elevated ATF4 levels induce additional basic zipper transcriptional regulators, such as CHOP/GADD153 and ATF3, which together direct a program of gene expression important for cellular remediation or, alternatively, apoptosis (4, 5, 9, 10). The mechanism of delayed translation reinitiation in response to eIF2∼P is also central for control of GCN4, a transcriptional activator of genes subject to the general amino acid control in the yeast Saccharomyces cerevisiae (11–13).
How are the ATF4-targeted ISR transcripts translated when there is repression of general protein synthesis as a consequence of eIF2∼P? One answer to this question is that ATF4 increases the expression of GADD34, which facilitates feedback control of the eIF2 kinase response by targeting the type 1 Ser/Thr protein phosphatase for dephosphorylation of eIF2∼P (10, 14–17). The resulting lowered eIF2∼P then allows for resumption of protein synthesis following the reprogramming of the stress-related transcriptome. However, many of the ISR gene products are highly expressed coincident with robust eIF2∼P, and their expression is central for implementation of the stress response pathway. For example, there are high levels of CHOP protein during ER or nutritional stress, whereas translation initiation is still repressed (9, 18). Furthermore, CHOP expression has been suggested to be required for the transcriptional activation of GADD34 (17). This suggests that CHOP and other ISR target genes are subject to preferential translation when eIF2∼P levels are high (19, 20). Supporting this idea, CHOP mRNA is associated with polysomes during amino acid starvation (19). The extent and duration of CHOP protein synthesis are thought to be central for altering the ISR from an adaptive pathway that alleviates cellular injury to one that is maladaptive, thus triggering apoptosis (3, 21, 22).
This study demonstrates that CHOP mRNA is subject to preferential translation during stress and describes a new mechanism for preferential translation in response to eIF2∼P. The CHOP mechanism of translational control involves a single uORF that blocks translation in the 5′-leader of the CHOP mRNA. However, with eIF2∼P induced by stress, scanning ribosomes bypass the inhibitory uORF by a process suggested to involve reduced efficiency of translation at initiation codons with a poor Kozak consensus sequence. This study indicates that eIF2∼P can regulate multiple mechanisms of preferential translation involving uORFs, and these mechanisms direct key ISR genes central to cell survival during stress conditions.
EXPERIMENTAL PROCEDURES
Plasmid Constructions
A HindIII-NcoI DNA fragment encoding the 5′-leader of the CHOP mRNA, along with the initiation codon of the CHOP coding region, was inserted between the HindIII and NcoI restriction sites in a derivative of plasmid pGL3. The resulting TK-CHOP-Luc plasmid contains this 5′-leader sequence of CHOP fused to a luciferase reporter downstream of a constitutive TK promoter. Primer sequences used in this construct were as follows: sense 5′-GCTCAAGCTTGTTATCTTGAGCCTAACACGTCGATTAT-3′ and antisense 5′-TCATGAGTGCCATGACTGCACGTGG-3′. The ATG1 and ATG2 codons of the CHOP uORF were mutated individually or in combination to AGG in the PTK-CHOP-Luc plasmid, and this and subsequent mutations were generated using a site-directed mutagenesis kit (Stratagene) following the manufacturer's instructions. To extend the uORF to overlap out-of-frame with the luciferase coding region, the encoded stop codon TGA in the uORF of TK-CHOP-Luc was mutated to GGA. Codon 24 (AGA) in the uORF of PTK-CHOP-Luc was mutated to TGA to generate the luciferase reporter with a shortened version of the uORF, which encoded amino acid residues 1–23. All plasmids were sequenced to ensure that there were only desired changes.
Sequence changes upstream of the CHOP uORF involved the introduction of a PstI restriction site 10 nucleotides upstream of the uORF in PTK-CHOP-Luc plasmid. A previously described stem-loop structure 5′-CTGCAGCCACCACGGCCCCCAAGCTTGGGCCGTGGTGGCTGCAG-3′, with a ΔG value of −41 kcal/mol, was then inserted into the PstI site of the modified PTK-CHOP-Luc plasmid. Alternatively, an extension of the 5′-leader was achieved by inserting a 120-nucleotide sequence into the PstI site. This sequence was devoid of any start and stop codons or predicted strong secondary structures. The initiation codon context from the uORF1 of ATF4 that shares a Kozak consensus was substituted for the first four codons, including the ATG1 and ATG2, of the CHOP uORF in the in PTK-CHOP-Luc plasmid, generating PTK-ATGATF4-CHOP-Luc. This substitution involved replacing the TATATCATGTTGAAGATGA sequence in the CHOP uORF with GCCACCATGG. These substitutions were carried out by the sequence and ligation-independent cloning method (23).
The CHOP mRNA sequences from nucleotide 1 to 133 containing the entire coding region of the CHOP uORF were inserted in-frame with the firefly luciferase coding region in a modified version of pGL3. The resulting plasmid PTK-uORF-Luc expressed the uORF-Luc fusion protein from the TK promoter. Deletions in PTK-uORF-Luc were constructed with in-frame deletions of the CHOP uORF codons 14–34 (Δ14–34), 14–23 (Δ14–23), and 23–34 (Δ24–34). A version of the Δ24–34 version of the PTK-uORF-Luc was also constructed with the uORF1 of ATF4 substituted for the first four codons, including the ATG1 and ATG2, of the CHOP uORF portion of the fusion protein.
Cell Culture and Dual-Luciferase Assays
MEF cells that were derived from S/S (wild-type eIF2α) and A/A (mutant eIF2α-S51A) mice were previously described (24, 25). MEF cells were cultured in Dulbecco's modified Eagle's medium (Cellgro) supplemented with 10% FBS, 1 mm nonessential amino acids, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C. ER stress was induced in MEF cells by the addition of either 0.1 or 1 μm thapsigargin to the medium, followed by incubation for up to 12 h, as indicated. Plasmid transfections were performed using the S/S and A/A MEF cells grown to 40% confluency and the FuGENE 6 transfection reagent (Roche Applied Science). Co-transfections were carried out in triplicate using wild-type or mutant versions of the TK-CHOP-Luc or TK-uORF-Luc plasmids and a Renilla luciferase plasmid serving as an internal control (Promega). 24 h after transfection, MEF cells were treated with 0.1 μm thapsigargin for 12 h or with no ER stress. Shorter periods of time, from 4 to 6 h of stress, also showed significant elevation of CHOP-Luc expression in response to ER stress that supported the stated conclusions. Dual-Luciferase assays were carried out as described by the Promega instruction manual. Values are a measure of a ratio of firefly versus Renilla luciferase units (relative light units) and represent the mean values of three independent transfections. Renilla luciferase values did not change significantly in the dual reporter assays. Results are presented as means ± S.D. that were derived from three independent experiments. Parallel to the Dual-Luciferase assays, the amount of firefly luciferase mRNA in each transfected condition was measured by the qRT-PCR method.
Preparation of Protein Lysates and Immunoblot Analyses
MEF cells cultured in the indicated stress conditions or no stress were washed twice with cold phosphate-buffered saline (pH 7.4), followed by lysis using a solution containing 50 mm Tris-HCl (pH 7.9), 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, 100 mm NaF, 17.5 mm β-glycerol phosphate, 10% glycerol supplemented with protease inhibitors (100 μm of phenylmethylsulfonyl fluoride, 0.15 μm aprotinin, 1 μm leupeptin, and 1 μm of pepstatin). Lysates were subjected to sonication for 30 s and precleared by centrifugation. Protein content was determined by using a Bio-Rad protein quantitation kit according to the manufacturer's instructions.
Equal amounts of proteins were separated by SDS-PAGE, and proteins were then transferred to nitrocellulose filters. Molecular weight markers were included for size determination of proteins in the immunoblot analyses. Transferred filters were then incubated in TBS-T solution containing 20 mm Tris-HCl (pH 7.9), 150 mm NaCl, and 0.2% Tween 20 supplemented with 4% nonfat milk, followed by incubation with TBS-T solution containing the primary antibody specific to the indicated protein. ATF4 antibody was prepared against recombinant protein (18). CHOP (sc-7351) antibody was obtained from Santa Cruz Biotechnology, and β-actin monoclonal antibody (A5441) was purchased from Sigma. Polyclonal antibody that specifically recognizes phosphorylated eIF2α at Ser-51 was purchased from BioSource (44-728G). Monoclonal antibody that recognizes either phosphorylated or nonphosphorylated forms of eIF2α was provided by Dr. Scot Kimball (Pennsylvania State University, College of Medicine, Hershey). Cell lysates from MEF cells transfected with the indicated plasmids were blotted for firefly luciferase protein by using antibody obtained from Promega (G7451).
Following incubation of the filters with the indicated antibodies, the filters were then washed three times in TBS-T followed by incubation with horseradish peroxidase-labeled secondary antibody and chemiluminescent substrate. Proteins in the immunoblot were visualized by exposing filters to x-ray film or by imaging using the LI-COR Odyssey system. Images shown in the figures are representative of three independent experiments.
Transcriptional Start Site of CHOP mRNA
The cDNAs corresponding to the 5′-end of the CHOP mRNAs expressed in S/S MEF cells treated with 0.1 μm thapsigargin, or no stress, were amplified by using an RNA ligase-mediated RACE kit (RLM-RACE kit, Ambion) according to the manufacturer's instructions. Antisense primers corresponding endogenous CHOP mRNA that were used in the assays include the outer primer 5′-GGACGCAGGGTCAAGAGTAG-3′ and inner primer 5′-TCATGAGTGCCATGACTGCACGTGG-3′. The outer primer used for amplifying CHOP-Luc mRNA 5′-end was 5′-CGAATTCGAACACGCAGAT-3′, which was combined with the same inner primer listed above. The amplified product was then analyzed using electrophoresis on a 1.2% agarose gel. The prominent DNA bands were excised, gel-purified, and sequenced. The transcriptional start site was determined as the first nucleotide that is 3′ to the adapter sequence ligated to the 5′ of the mRNA transcripts.
RNA Isolation and qRT-PCR
MEF cells were transfected with the indicated plasmids, treated with the designated stress conditions and harvested, and total cellular RNA was prepared using TRIzol reagent (Invitrogen) according to the instruction manual. Contaminating DNA was digested with RNase-free DNase (Promega). Single strand cDNA synthesis was carried out using Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Quantitation of relative mRNA levels was performed using Light Cycler 480 PCR system (Roche Applied Science) and SYBR Green PCR mix from Applied Biosystems. The amount of firefly luciferase mRNA was measured using Renilla luciferase as an internal control. The oligonucleotide primers used were as follows: firefly luciferase 5′-CTCACTGAGACTACATCAGC-3′ and 5′-TCCAGATCCACAACCTTCGC-3′; Renilla luciferase 5′-GGAATTATAATGCTTATCTACGTGC-3′ and 5′-CTTGCGAAAAATGAAGACCTTTTAC-3′. The endogenous CHOP and ATF4 mRNA levels were measured using the following oligonucleotide primers: CHOP 5′-CCTAGCTTGGCTGACAGAGG-3′ and 5′-CTGCTCCTTCTCCTTCATGC-3′; ATF4 5′-GCCGGTTTAAGTTGTGTGCT-3′ and 5′-CTGGATTCGAGGAATGTGCT-3′. Quantitation of target genes was normalized using the reference β-actin. Primers used were 5′-GTATGGAATCCTGTGGCATC-3′ and 5′-AAGCACTTGCGGTGCACGAT-3′. Light Cycler 480 software (version 1.2.9.11) was used to perform quantification and to generate Cp values. Values are a representation of three independent experiments, with standard deviations as indicated.
Polysome Analysis and Translational Control of CHOP mRNA
MEF cells were cultured as described above and treated with 1 μm thapsigargin for 6 h or no stress. 10 min prior to harvesting, cells were treated with 50 μg/ml cycloheximide. Cells were washed with cold phosphate-buffered saline (pH 7.4) solution containing 50 μg/ml cycloheximide, and cell lysates were prepared in a solution of 20 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 100 mm NaCl, and 0.4% Nonidet P-40 supplemented with 50 μg/ml cycloheximide. Cell lysates were passed though a 23-gauge needle and incubated on ice for 10 min. The cell lysate was precleared with brief centrifugation (10,000 rpm for 10 min at 4 °C) and then layered onto a 10–50% sucrose gradient solution containing 20 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 100 mm NaCl, and 50 μg/ml cycloheximide. The sucrose gradients were then subjected to centrifugation in a Beckman SW-41Ti rotor for 2 h at 40,000 rpm at 4 °C. A portion of unfractionated cell lysate was used for determining total mRNA levels of CHOP, ATF4, and β-actin. Gradients were fractionated using a Biocomp Gradient Station, and absorbance of RNA at 254 nm was recorded using an in-line UV monitor. Equivalent amounts of synthetic poly(A)+ luciferase RNA (10 ng/ml) purchased from Promega were added to each collected fraction. RNA was isolated from each fraction using the TRIzol LS reagent (Invitrogen), and synthesis of single-stranded cDNA was performed using Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Prepared cDNA was used to measure the relative mRNA levels for CHOP, ATF4, and encoded β-actin. qRT-PCR data were normalized with the luciferase mRNA that was added prior to RNA isolation. Data represented are the result of three independent experiments with standard deviations as indicated. Alternatively, MEF cells cultured to 40% confluency were transfected with wild-type TK-CHOP-Luc plasmid or mutant variants with ΔATG1 and ΔATG2, individually or in combination, using the FuGENE 6 (Roche Applied Science) transfection reagent. After 24 h of transfection, cells were treated with 0.1 μm thapsigargin up to 6 h or no stress. Cell lysates prepared were then subjected to sucrose gradient analyses and fractionated as described above. Equivalent amounts of bacterial control RNA (Affymetrix) were added to each sucrose fraction to serve as internal control for the RNA isolation and in qRT-PCR analysis. RNA isolated from each fraction was used in preparation of single strand cDNA synthesis as described. The amount of firefly luciferase in each fraction was quantitated and normalized to the THR mRNA of bacterial control added in the RNA mixture. Oligonucleotide primers used for the THR mRNA measurement were as follows: forward 5′-AGGATGACGAGACCCAAATG-3′ and reverse 5′-TGATCGCAGCAATGAGGATA-3′.
RESULTS
eIF2∼P Is Required for CHOP Transcription and Translation
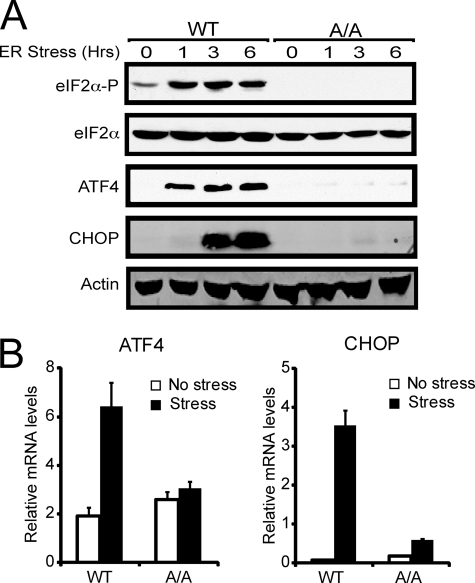
In response to ER stress, eIF2∼P triggers preferential translation of ATF4 mRNA concurrent with repressed global translation initiation. This is illustrated by treatment of MEF cells with thapsigargin, a potent ER stress agent (5). Within 1 h of thapsigargin exposure, wild-type MEF cells displayed an enhanced eIF2∼P accompanied by increased expression of ATF4 protein (Fig. 1A). By contrast, MEF cells containing Ala for the eIF2α phosphorylation site Ser-51 (A/A) showed no eIF2∼P and minimal levels of ATF4 protein. In addition to translational control, ATF4 was reported to be subject to transcriptional regulation, with a 3-fold increase in ATF4 mRNA following 6 h of the ER stress (Fig. 1B) (26). This increase in ATF4 mRNA levels in response to ER stress is substantially blocked in the A/A cells.
FIGURE 1.
Phosphorylation of eIF2α increases CHOP expression in response to ER stress. A, wild-type MEF cells (WT) and mutant cells expressing the nonphosphorylated eIF2α-S51A (A/A) were treated with the ER stress agent, thapsigargin, for up to 6 h, as indicated, or to no stress treatment (0 h). Lysates were prepared, and the levels of phosphorylated eIF2α, total eIF2α, ATF4, CHOP, and β-actin were measured by immunoblot analysis using antibody that specifically recognizes each protein. B, total RNA was isolated from the wild-type and A/A MEF cells treated with thapsigargin for 6 h (stress), or to no stress, and the relative levels of ATF4 and CHOP mRNA were measured by qRT-PCR.
ATF4 is a transcriptional activator of ISR genes, such as CHOP mRNA. Levels of CHOP protein and mRNA are sharply increased in response to ER stress by a mechanism requiring eIF2∼P (Fig. 1, A and B). Given that increased expression of CHOP protein occurs despite high levels of eIF2∼P, we reasoned that translation of CHOP mRNA may be favored even when global translation initiation is severely restricted. To test this idea, we carried out a polysome analysis using sucrose gradient centrifugation. Thapsigargin treatment of MEF cells significantly reduced polysome levels, concomitant with elevated levels of free ribosomes and monosomes, which is consistent with repressed translation initiation (Fig. 2). This reduction in translation initiation is dependent on eIF2∼P, as the polysome profile was largely unchanged when A/A cells were treated with thapsigargin (supplemental Fig. 1).
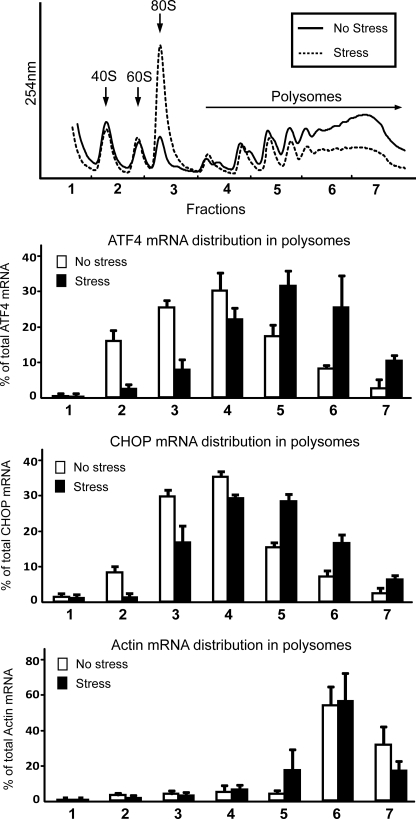
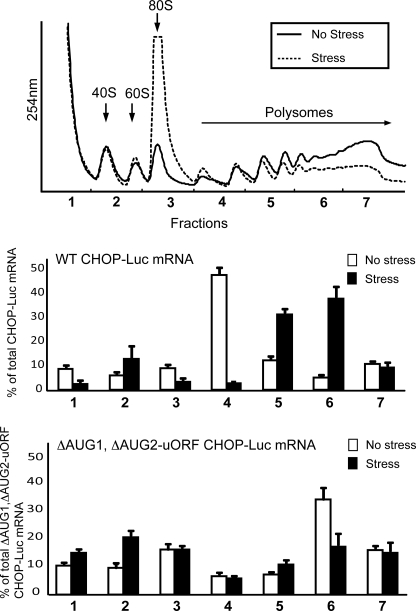
FIGURE 2.
Both ATF4 and CHOP mRNAs are preferentially associated with large polysomes during ER stress. Wild-type MEF cells were exposed to the ER stress agent thapsigargin for 6 h (stress) or no stress treatment. Cell lysates were then analyzed by centrifugation in a 10–50% sucrose gradient, and the profiles were measured by absorbance at 254 nm. The top panel highlights the 40 S and 60 S ribosomal subunits, 80 S monosomes, and polysomes. Total RNA was prepared from the fractions collected from the sucrose gradients, and the percentage of ATF4, CHOP, and actin-encoding transcripts present in each of the seven fractions derived from the ER stress (Stress) or the nontreated cells (No stress) were determined by qRT-PCR. Values are represented as histograms for each fraction. Three independent experiments were carried out for each measurement, with the S.D. indicated for each. The top panel is representative of three independent experiments.
During nonstressed conditions, the levels of ATF4 transcript, measured as the percentage of total ATF4 mRNA, were most abundant in the monosome and small polysome fractions of the sucrose gradient. In this condition, only 28% of the ATF4 mRNAs were associated with large polysomes consisting of transcripts associated with four or more ribosomes. By comparison, upon ER stress, there was a substantial shift of ATF4 transcripts to the large polysome fractions (67% associated with large polysomes), consistent with earlier reports that ATF4 mRNA is preferentially translated upon eIF2∼P (6–8). CHOP mRNA displayed a similar distribution pattern in the polysome profiles as the ATF4 transcripts (Fig. 2). In the absence of stress, CHOP mRNA was most abundant in the monosomes and small polysomes, whereas ER stress triggered increased association of this transcript with the large polysomes (25% associated with large polysomes in nonstressed conditions compared with 52% during ER stress). As a control, we also measured actin mRNA among the fractions in the sucrose gradient and found that this transcript was largely insensitive to ER stress.
CHOP Translational Control Is Facilitated by an uORF in the 5′-Leader of the CHOP mRNA
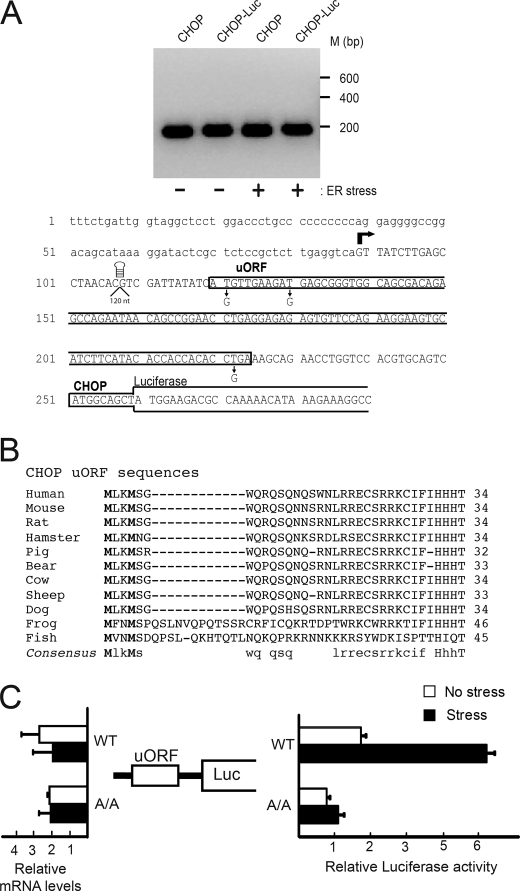
We next addressed whether the 5′-leader of the CHOP mRNA directs translational control in response to ER stress. The transcriptional start site of the CHOP transcript was determined in MEF cells in the presence or absence of stress by 5′-RACE and DNA sequencing (Fig. 3A). Transcription of CHOP occurs at the same site independent of stress conditions, leading to a 5′-leader sequence 162 nucleotides in length. The CHOP leader sequence encodes a single uORF representing a 34-residue polypeptide that is highly conserved among vertebrates (Fig. 3B) (27). Notable among the conserved residues are Met residues at positions 1 and 4 (encoded by codons designated ATG1 and ATG2), providing for two possible initiation codons and basic amino acid residues in the carboxyl terminus of the uORF.
FIGURE 3.
5′-Leader of the CHOP mRNA contains an uORF and is sufficient for translational control in response to eIF2∼P. A, top panel, 5′-RACE was carried out for endogenous CHOP and CHOP-Luc using RNA prepared from wild-type MEF cells expressing the PTK-CHOP-Luc reporter, which were treated with the ER stress agent thapsigargin (+) or no stress agent (−). DNA products were separated and visualized by electrophoresis using a 1.2% agarose gel, with markers of the indicated size in base pairs represented on the right. A, bottom panel, sequence of the 5′-leader of the mouse CHOP mRNA is presented with the boxes indicating the uORF and the coding region of the CHOP-luciferase fusion. Residues mutated in the analysis of CHOP translational control are indicated below the box. The bold arrow indicates the transcription start site of the CHOP gene as determined by sequencing of the 5′-RACE products. A stem-loop structure or 120-bp segment was inserted at the indicated position upstream of the uORF. B, polypeptide sequences encoded by the uORF in the CHOP mRNAs from different vertebrates. The uORF polypeptide sequences were from representative cDNAs derived from the indicated CHOP orthologs, including human (GenBankTM accession number BC003637), mouse (BC013718), rat (BC100664), hamster (M29238), pig (AK346731), bear (GW278660), cow (BC122721), sheep (DY499855), dog (DN431044), frog Xenopus tropicalis (BC153679), and fish Danio rerio (BC134052). The number of polypeptide residues encoded by each uORF is listed following the sequences. Residues conserved among the uORF sequences are listed in the consensus, with invariant residues in capital letters and those conserved in lowercase letters. C, CHOP translational control in response to ER stress was measured by a Dual-Luciferase assay. The PTK-CHOP-Luc reporter and a Renilla luciferase plasmid, which served as an internal control, were transfected into wild-type MEF cells (WT) or A/A cells expressing eIF2α-S51A, and treated with thapsigargin (Stress), or no stress. The PTK-CHOP-Luc reporter contains cDNA sequences corresponding to the entire 5′-leader of the CHOP mRNA, which is illustrated along with the luciferase reporter gene. Three independent experiments were carried out for each measurement, and relative values are represented as histograms for each, with the S.D. indicated. In parallel, the levels of the CHOP-Luc mRNA were measured by qRT-PCR, and relative values of the reporter transcripts were presented as histograms, with error bars representing the S.D.
The role of the 5′-leader sequence of the CHOP transcript in translation control was investigated by using a PTK-CHOP-Luc reporter, which contained a cDNA segment encoding the mouse CHOP 5′-leader segment fused to firefly luciferase downstream of the minimal TK promoter. CHOP-Luc expression was increased by 3-fold in the wild-type MEF cells in response to ER stress (Fig. 3C). In contrast, there was low luciferase activity in the A/A cells devoid of eIF2∼P, which did not appreciably change in response to ER stress. The transcriptional start site of the a PTK-CHOP-Luc reporter was the same as that determined for the endogenous CHOP (Fig. 3A), and there were no significant changes in CHOP-Luc mRNA levels in the tested conditions, consistent with the 5′-leader of the CHOP mRNA directing translational expression in response to eIF2∼P (Fig. 3C).
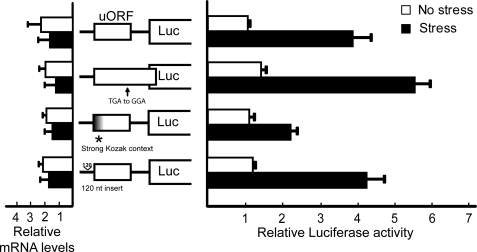
Earlier studies suggested that translation of the uORF can repress translation of the downstream CHOP coding region (27). To identify the underlying mechanisms by which eIF2∼P and stress leads to preferential translation of CHOP, we constructed a series of mutations in the 5′-leader portion of the PTK-CHOP-Luc reporter and analyzed their effects on expression in response to ER stress. First, we addressed whether initiation of CHOP mRNA translation occurs by cap-dependent ribosome scanning or is rather facilitated by an internal translation initiation process such as internal ribosome entry site (IRES)-mediated initiation (28, 29). A stem-loop structure (ΔG value of −41 kcal/mol) was inserted 5′ to the uORF (Fig. 3A). Minimal CHOP-Luc expression was observed in MEF cells irrespective of stress conditions (Fig. 4). This result supports the idea that CHOP translation involves the processive scanning of ribosomes from the 5′-end of the CHOP transcript.
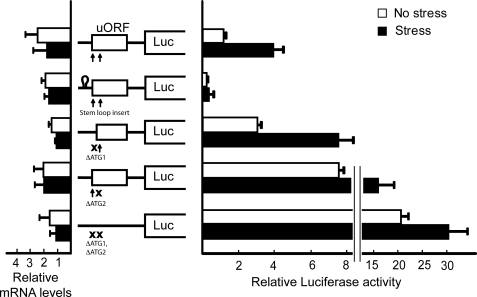
FIGURE 4.
uORF is inhibitory to CHOP translation. The illustrated wild-type and mutant versions of the PTK-CHOP-Luc reporter were transfected into wild-type MEF cells, and Dual-Luciferase assays were carried out in response to thapsigargin (stress) or no stress. Mutant versions of the CHOP-Luc reporter include an insertion of a stem-loop structure upstream of the uORF, and X indicates ATG1 and ATG2 of the uORF substituted to AGG, individually or combined. Additionally, the relative amounts of CHOP-Luc mRNA were measured by qRT-PCR. Three independent experiments were carried out for each of the measurements, and relative values are represented as histograms, in conjunction with the indicated S.D.
We next mutated the ATG1 and ATG2 of the uORF to AGG, individually or in combination, in the PTK-CHOP-Luc reporter. Mutation of ATG1 led to elevated CHOP-Luc expression during both stressed and nonstressed conditions compared with the wild-type reporter (Fig. 4). The increase was greatest in the nonstressed conditions, with over a 3-fold increase in CHOP-Luc expression in the ΔATG1 mutant compared with the wild-type reporter, whereas during ER stress there was about a 2-fold elevation (Fig. 4). Together, these results indicate a diminished induction during ER stress, with about a 2-fold enhancement in the MEF cells expressing the ΔATG1 mutant and a 4-fold increase in the wild-type version. Loss of ATG2 led to an even higher increase in CHOP-Luc expression compared with the wild-type counterpart, with increases of 7- and 4-fold during the nonstressed and stressed conditions, respectively. Furthermore, the highest luciferase activity was observed when both ATG1 and ATG2 were mutated (Fig. 4). Levels of CHOP-Luc mRNA were comparable between the different ATG mutant arrangements and stress arrangements (Fig. 4). These results support the idea that both ATG1 and ATG2 are able to serve as initiation codons for the uORF, with ATG2 being predominant. The uORF is suggested to be inhibitory to CHOP translation, and this repressing function can be overcome with stress and eIF2∼P. Further supporting this model, the wild-type CHOP-Luc mRNA was found to be preferentially associated with large polysomes in MEF cells treated with thapsigargin, and this transcript was the most abundant in the disome fraction in the absence of stress (Fig. 5). By comparison, mRNA expressed from the PTK-CHOP-Luc reporter devoid of both ATG1 and ATG2 was most prevalent in polysome fraction 6 of the sucrose gradient in the absence of stress. During ER stress, the mutant CHOP-Luc transcript was less abundant in the large polysome fractions, with a broad distribution among several fractions of the sucrose gradient (Fig. 5).
FIGURE 5.
CHOP-Luc mRNA is preferentially associated with large polysomes in response to ER stress. Wild-type MEF cells were transfected with a wild-type version of the PTK-CHOP-Luc reporter or a version with mutations in both ATG1 and ATG2 (ΔATG1 and ΔATG2). Transfected cells were exposed to thapsigargin (Stress) for 6 h or to no stress treatment. Cell lysates were then analyzed by sucrose gradient centrifugation, and fractions were monitored by absorbance at 254 nm (top panel) with the indicated 40 S and 60 S ribosomal subunits, 80 S monosomes, and polysomes. Total RNA was prepared from the fractions, and the percentage of CHOP-Luc mRNA present in each of the seven fractions obtained from the ER stress (Stress) or the nontreated cells (No stress) was determined by qRT-PCR. Three independent experiments were carried out, with measurements for each fraction illustrated, along with the S.D. values.
CHOP Translational Control Is Mediated by Leaky Scanning of Ribosomes through the Inhibitory uORF
We considered two models by which stress and eIF2∼P can overcome the inhibitory functions of the uORF in CHOP translational control. The first is a “Reinitiation” model, which suggests that following translation of the inhibitory uORF during low eIF2∼P, ribosomes dissociate from the CHOP mRNA, and therefore CHOP expression is repressed. Upon stress and eIF2∼P, ribosomes translating the uORF would resume scanning and reinitiate translation at the CHOP coding region. Alternatively, in a “Bypass” model, ribosomes initiate translation at the uORF in nonstressed conditions (44). In this latter model, translation of the uORF would preclude expression at the downstream CHOP coding region. In response to stress and increased eIF2∼P, the scanning ribosome would bypass or scan through the inhibitory uORF and instead initiate translation at the downstream CHOP coding region. To delineate between these two models, we mutated the stop codon of the CHOP uORF to a sense codon (TGA to GGA), resulting in an extended uORF that overlaps by 19 nucleotides out-of-frame with the coding region of the downstream reporter (Figs. 3A and 6). Luciferase activity from the PTK-CHOP-Luc with the extended uORF was induced in response to ER stress similar to that measured for the wild-type reporter. This result strongly supports the Bypass model, as the extended uORF would not be expected to interfere with the induced CHOP translation. By contrast, in the Reinitiation model ribosomes terminating translation of the extended uORF would be 3′ of the initiation codon of the CHOP coding region and require protracted 3′ to 5′ scanning to express CHOP, an unlikely event (29–33).
FIGURE 6.
Strong start codon context for initiation of uORF translation thwarts bypass of the inhibitory element in response to ER stress. Wild-type and the indicated mutant versions of the PTK-CHOP-Luc reporter were transfected into wild-type MEF cells, and Dual-Luciferase assays were carried out in response to thapsigargin (Stress) or no stress. Mutant versions include substitution of the encoded stop codon of the uORF (TGA to GGA), leading to an extension of the uORF so that it overlaps out-of-frame with the Luc coding region. Additionally, a strong Kozak context was substituted for ATG2 of the uORF, in the absence of ATG1. Alternatively, a 120-nucleotide segment devoid of initiation codons and strong predicted secondary structure was inserted upstream of the uORF. The relative amounts of CHOP-Luc mRNA were also measured by qRT-PCR. Three independent experiments were carried out for each assay, and the relative values are represented with the S.D.
We next addressed the basis for the ribosome bypass of the uORF in response to eIF2∼P. We considered two ideas for the ribosome bypass as follows: the poor initiation context of ATG1 and ATG2 encoded in the uORF, and the short length (31 nucleotides) from the 5′-end of the CHOP mRNA to the ATG1 of the CHOP uORF, both of which may reduce translation of the uORF during eIF2∼P. Start codon context is an important contributor to the efficiency of translation initiation (34–36). The uORF of CHOP has a less than optimal start codon context at ATG1 (TATATCATGT) and the primary ATG2 (TTGAAGATGA) compared with the Kozak consensus sequence, gcc(A/G)ccATGG, where the capital letters at −3 and +4 in the consensus are most critical for translation initiation. The translation initiation context of ATF4 uORF1 matches this consensus (GCCACCATGG), and this sequence was substituted into the CHOP uORF, replacing the predominant ATG2 in the absence of ATG1 in the PTK-CHOP-Luc reporter (Fig. 6). Substitution of the strong Kozak consensus led to significantly lowered CHOP-Luc expression in response to ER stress, with about a 2-fold reduction compared with the wild-type reporter. Levels of CHOP-Luc mRNA were comparable between the Kozak consensus and its wild-type counterpart (Fig. 4A). These results suggest that less than optimal initiation site contexts in the uORF contribute to a leaky scanning mechanism by which the inhibitory uORF is bypassed during stress conditions and high levels of eIF2∼P.
To determine whether the abbreviated leader length preceding the uORF is important for stress-induced CHOP translation, a heterologous 120-nucleotide sequence (8), devoid of any start and stop codons and without predicted strong secondary structures, was inserted upstream of the uORF in the PTK-CHOP-Luc reporter (Fig. 3A). Insertion of this sequence did not change the regulation of CHOP-Luc expression, with a 4-fold increase in luciferase activity in response to ER stress, which was similar to the wild-type reporter (Fig. 6). There were also no significant changes in CHOP-Luc mRNA levels with the 120-nucleotide insertion. These results indicate that bypass of the inhibitory uORF in response to stress is not dependent upon the relatively short length of CHOP mRNA situated between the 5′-end and the initiation codons of the uORF.
Carboxyl-terminal Portion of the uORF Is Inhibitory to the Downstream CHOP ORF Translation
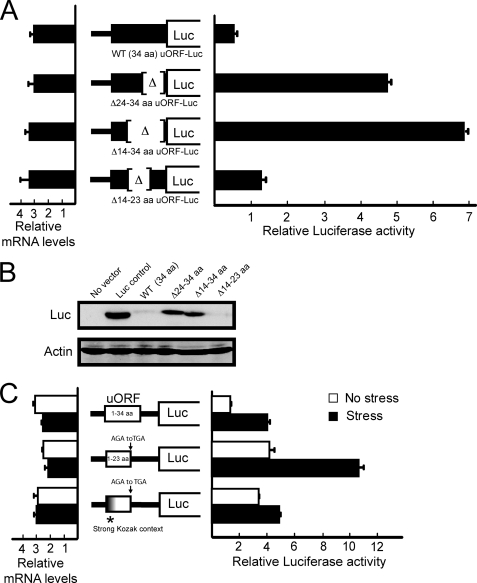
We next investigated the mechanism by which the uORF represses translation of the downstream CHOP coding region. The basis for the inhibitory function of the uORF in the Bypass model may be that following termination of the uORF translation ribosomes would dissociate from the CHOP mRNA; alternatively, translation of the uORF may lead to a block in translation elongation or termination that prevents translation at the downstream CHOP ORF. Arrested ribosomes may not only stall synthesis of the nascent polypeptide but also impede subsequent scanning from the 5′-end of the CHOP mRNA. To address the inhibitory properties of the uORF in translation, we constructed an in-frame fusion between the uORF and firefly luciferase expressed from the TK promoter. We transfected the resulting PTK-uORF-Luc plasmid into wild-type MEF cells and found minimal luciferase activity and low levels of fusion protein as judged by immunoblot analyses (WT in Fig. 7, A and B). Deletion analysis of the uORF portion of the fusion gene, including an in-frame deletion of uORF codons 14–34 (Δ14–34) and 24–34 (Δ24–34), led to significant increases in the luciferase activity and measurable fusion proteins by immunoblot. By comparison, removal of the uORF codons 14–23 (Δ14–23) showed low uORF-Luc expression, as judged by luciferase activity and an immunoblot measurement of the fusion protein (Fig. 7, A and B). The levels of uORF-Luc mRNA were similar among the MEF cells expressing the fusion protein with the full-length uORF and the deletion mutants (Fig. 7A). These results support the idea that the 3′-portion of the uORF, including codons 24 to 34, can block translation.
FIGURE 7.
Carboxyl-terminal portion of the uORF is inhibitory to CHOP translation. A, PTK-uORF-Luc plasmid, encoding an in-frame fusion between the uORF and firefly luciferase, was transfected into wild-type MEF cells, and dual reporter assays were carried out for each. The PTK-uORF-Luc plasmid contained the full-length uORF or versions containing deletions of codons 24–34, 14–34, or 14–23, as illustrated. In parallel, the relative amounts of CHOP-Luc mRNA were also measured by qRT-PCR. Three independent experiments were carried out for each of the reporter construct, and the relative values are represented for each, with the S.D. indicated. B, levels of the uORF-Luc fusion protein containing the full-length uORF, or the indicated deletion mutants, were determined by immunoblot analysis using antibody that recognizes firefly luciferase. Actin levels were also measured for normalization between the lysates. As control, the immunoblot analysis was also carried out with lysates prepared from MEF cells containing no vector or with vector expressing only firefly luciferase. C, illustrated wild-type and mutant versions of the PTK-CHOP-Luc reporter were transfected into wild-type MEF cells, and Dual-Luciferase assays were carried out in response to ER stress or during no stress. The mutant versions include a mutation of codon 24 (AGA) of the uORF to TGA to generate the luciferase reporter with shortened version of the uORF. Additionally, the shortened version of the uORF was combined with the strong Kozak context, as illustrated. Relative levels of CHOP-Luc mRNA were also determined by qRT-PCR. Three independent experiments were conducted for each, and the relative values are represented, along with the S.D.
The uORF was next truncated in the PTK-CHOP-Luc reporter by introducing a stop codon at residue 24 (AGA to TGA) (Figs. 3A and 7C). Expression of the CHOP-Luc was significantly increased during both the stressed and nonstressed conditions compared with the wild-type reporter (Fig. 7C). These results suggest that translation of the carboxyl-terminal portion of the uORF can lead to a block in translation elongation or termination, which can effectively prevent subsequent initiation at the downstream CHOP coding region. Removal of the inhibitory portion of the uORF is suggested to allow for a significant amount of the ribosomes translating the uORF to resume scanning along the CHOP mRNA and reinitiate at the downstream ORF coding region.
We previously suggested that substituting the Kozak consensus for ATG1 and ATG2 in the uORF prevents bypass of the inhibitory uORF in response to ER stress (Fig. 6). Taking this into consideration, we further reasoned that if the inhibitory portion of the uORF was removed that this would at least in part overcome the inclusion of the optimized initiation codon context in the PTK-CHOP-Luc reporter. This was indeed the outcome with elevated levels of CHOP-Luc expression in the MEF cells during nonstressed conditions that were similar to that measured in the ER-stressed MEF cells expressing the wild-type reporter (Fig. 7C). Note that during ER stress luciferase activity expressed from the PTK-CHOP-Luc reporter containing the combined Kozak consensus and Δ24–34 deletion did not match that with the carboxyl-terminal deletion alone. This suggests that in this stress context, reinitiation at the downstream CHOP coding region is limited and that bypass of the inhibitory uORF in response to eIF2∼P is central for preferential translation of CHOP mRNA.
DISCUSSION
In this study, we addressed the underlying mechanisms by which eIF2∼P can recalibrate the protein synthetic machinery, such that mRNAs are individually evaluated, leading to prescribed changes in their translation efficiencies. The 5′-leader of the CHOP mRNA has a single uORF, which is a significant barrier to CHOP translation in nonstressed conditions (Fig. 8). However, in response to ER stress, induced eIF2∼P facilitates bypass of the repressing uORF, allowing scanning ribosomes to instead initiate translation at the CHOP coding region (Fig. 8). Underlying this translation bypass mechanism is the initiating context of the CHOP uORF. The uORF has two encoded AUGs, which are conserved at positions 1 and 4 throughout vertebrates (Fig. 3). Either AUG1 or AUG2 can serve as an initiation codon, although AUG2 is predominant, as viewed by the finding that loss of this second AUG in the uORF leads to the high expression of a CHOP-Luc reporter in the absence of stress, i.e. the greatest suppression of the inhibitory function of the uORF (Fig. 4). Central to the ability of eIF2∼P to direct the bypass of AUG1 and AUG2 is their less than optimal sequence context for translation initiation, a feature that was conserved among each of the mammalian orthologs of CHOP illustrated in Fig. 3B. Substitution of the Kozak consensus sequence for AUG2, in the absence of AUG1, significantly reduced the ability of ER stress and eIF2∼P to overcome the inhibitory properties of the uORF (Fig. 6).
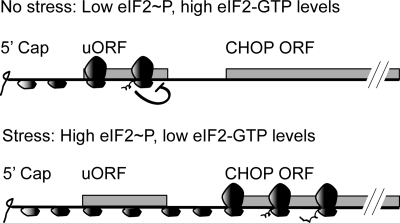
FIGURE 8.
Phosphorylation of eIF2α facilitates ribosome bypass of the inhibitory uORF, enhancing translation of the CHOP coding region. In the absence of stress, there is low eIF2∼P and elevated levels of eIF2-GTP. Ribosomes are suggested to bind the 5′-end of the CHOP mRNA, and the scanning ribosomes translate the uORF, leading to a block in translation elongation or termination and low translation of the CHOP coding region. During stress, there is robust eIF2∼P that reduces exchange of eIF2-GTP to eIF2-GDP that is proposed to reduce translation initiation at the uORF due to the poor initiation codon context. As a consequence, the scanning ribosome bypasses the inhibitory uORF and instead translates the CHOP coding region, which has an initiation codon with a strong Kozak consensus.
A second feature central to this model is the idea that translation of the uORF is suggested to block translation elongation or termination (Fig. 8). This idea was supported by the observation that the uORF-Luc fusion gene was poorly translated, with minimal expression of the fusion protein or luciferase activity (Fig. 7, A and B). Critical to this translation block is uORF residues 24–34, as deletion of this segment of the uORF allowed for translation of the fusion protein. When this region of the uORF was removed in the 5′-leader of the PTK-CHOP-Luc reporter, luciferase activity was significantly elevated during both stressed and nonstressed conditions (Fig. 7C). This was observed even when Δ24–34 was combined with a start codon containing the Kozak initiation context that was substituted into the uORF. In this case, eIF2α∼P would not facilitate bypass of the uORF, but the loss of this inhibitory segment is compromised, which could allow for some translation reinitiation to occur at the downstream CHOP coding region.
The importance of the polypeptide sequence for the inhibitory function of the uORF is also supported by Jousse et al. (27), who first reported that the uORF can repress CHOP expression. Although the mechanism of alleviation of this inhibition was not addressed in this earlier study, it was found that the repressing properties of the uORF were significantly overcome by shortening of the uORF to three residues in length or by introducing a frameshift that alters the sequence but not the length of the encoded polypeptide. Together these results support the model that translation of the carboxyl-terminal portion of the uORF polypeptide is critical for the repressing function of the uORF. Although the RNA sequence or structure per se does not appear to serve as a barrier to translation of the downstream CHOP coding region, it is possible that these RNA elements could serve as a contributing factor. The uORF is suggested to prevent ribosomal reinitiation at the downstream coding region in the mRNA and also potentially interfere with scanning of subsequent ribosomes proceeding from the 5′-end of the CHOP mRNA.
CHOP and ATF4 Translational Controls Differ in Fundamental Ways
The mechanism of CHOP translational control is different from that described for ATF4 in several fundamental ways. Although both CHOP and ATF4 regulation involve uORFs programmed into the 5′-leaders of their mRNAs, the configuration of the uORFs and their function differ. ATF4 and its yeast counterpart GCN4 require two or more uORFs (1, 8, 11). The 5′-proximal uORF serves as a positive regulatory element that facilitates subsequent ribosome scanning and translation reinitiation at a downstream ORF. The levels of eIF2-GTP directed by induced eIF2∼P determine how rapid this reinitiation event will be. During periods of no stress, when eIF2∼P is reduced and eIF2-GTP is plentiful, ribosome reinitiation occurs rapidly at an adjacent inhibitory uORF, thus blocking expression of the transcription factor. In the case of ATF4, a single inhibitory uORF is sufficient, whereas GCN4 mRNA contains three short inhibitory uORFs, and translation of any one is thought to be sufficient to release ribosomes from the transcript. When eIF2∼P is enhanced during stress, the resulting lowered eIF2-GTP levels delay translation reinitiation. As a result, reinitiating ribosomes can scan through the more immediate downstream inhibitory uORFs and instead initiate translation at the ATF4 coding sequence.
Although the CHOP regulatory model shares with ATF4 translational control the idea that eIF2∼P can bypass an inhibitory uORF, it accomplishes this without the aid of a positive-acting uORF that facilitates translation reinitiation. Instead CHOP has devised a single uORF with a poor translation initiation context that can be bypassed in response to eIF2∼P (Fig. 8). In this way translational control induced by eIF2∼P is no longer viewed as requiring two or more uORFs, but rather a single uORF in the specified context will suffice.
eIF2∼P induced by stress does not appear to significantly reduce the binding of the 43 S preinitiation complex (consisting of the 40 S ribosomal subunit and translation initiation factors including eIF2-GTP-Met-tRNAiMet) to the 5′-end of the CHOP mRNA. This is a feature shared with the ATF4 and GCN4 translational control models. Supporting this idea is the finding that the expression of CHOP-Luc deleted for both ATG1 and ATG2 was significantly elevated irrespective of stress conditions (Fig. 4). Furthermore, CHOP-Luc mRNA devoid of both initiation codons was broadly distributed among the fractions of the sucrose gradient during ER stress (Fig. 5). Rather eIF2∼P is suggested to enhance the bypass of scanning ribosomes through an uORF with a poor initiation codon context. We do not yet understand the biochemical basis for the eIF2∼P bypass of the uORF in our model of CHOP translational control. Lowered eIF2-GTP levels may contribute to the reduced recognition of the CHOP uORF. Additional contributors to this bypass may be eIF2∼P itself, or eIF2∼P regulation of the expression of other critical translation factors, or regulators that would then facilitate the bypass of the CHOP uORF during stress conditions. It is noted that the translation factor eIF1 along with eIF2α and specific segments of 18 S rRNA are important for recognition of the initiation codon context (29, 37). It was reported that high levels of eIF1 can enhance the stringency for selection of gene start codons (38). However, our preliminary studies suggest that eIF2∼P during ER stress does not significantly change the levels of eIF1 in MEF cells, with only a modest reduction during the early phase of thapsigargin treatment (data not shown).
eIF2∼P inhibits general translation concurrent with preferential translation of select mRNAs, such as ATF4 and CHOP. Inhibition of global translation by eIF2∼P can differentially lower translation of mRNAs genome-wide, with some gene transcripts being severely repressed, although others are more resistant to eIF2∼P. This gradient model for translational repression suggests that the translational machinery can delineate between transcripts genome-wide to determine the degree of repression. The degree of repression may involve a myriad of features in the 5′-leaders for each gene transcript, as well as possibly the 3′-untranslated regions. Given the proposal that eIF2∼P can enhance bypass of gene coding regions with initiation codons with less than optimal initiation codon context, it is inviting to speculate that this feature may be a distinguishing feature between those mRNAs whose translations are severely repressed. By contrast, optimal Kozak context for initiation may contribute to enhanced translation resistance of gene transcripts during eIF2∼P.
Role of CHOP Translational Control in Stress Responses
Expression of CHOP is central to the ISR and regulation of cell survival in response to environmental stress. Although the ISR can serve essential adaptive functions, chronic stress conditions and unabated expression of key target genes, such as CHOP, can contribute to morbidity (10, 22, 39). In this way, the ISR, which directs critical adaptive functions that can ameliorate cellular injury occurring during environmental stresses, becomes maladaptive. High levels of CHOP can direct the transcription of many genes that can promote caspase activation and apoptosis (3, 17, 22, 40–42). In addition to transcriptional and translational regulation, CHOP mRNA and protein are subject to rapid turnover, with half-lives between 2 and 4 h (21). Therefore, the levels of CHOP protein are tightly linked to the current levels of stress insult and eIF2∼P. The combination of transcriptional and translational regulation allows for a rapid increase in CHOP during ER stress, and with alleviation of the stress and diminished eIF2∼P levels, CHOP translation levels are sharply reduced and CHOP mRNA and protein subject to decay. This model emphasizes that CHOP levels are tightly linked to the amount of eIF2∼P, which is adjusted rapidly to the extent of cellular stress. When a stress transcends from acute to chronic, these regulatory mechanisms would direct sustained elevated levels of CHOP protein and the products of its target genes, triggering apoptotic pathways.
Multiple stress pathways are thought to contribute to the levels of CHOP transcriptional activity achieved during a given environmental stress. Chen et al. (20) suggested that, along with eIF2∼P, CHOP translation can be regulated by phosphorylated eIF4E and its association with eIF4G. In this earlier report, treatment with low concentrations of anisomycin increased CHOP mRNA levels and enhanced CHOP translation by a mechanism suggested to involve activation of the p38 MAPK/Mnk1 and mammalian target of rapamycin pathways. This alternative signaling scheme may also facilitate CHOP translation by a process involving the uORF in the CHOP mRNA, suggesting that multiple signaling pathways regulated by stress may converge on CHOP translation. Phosphorylation of CHOP protein by p38 protein kinase has also been proposed to modulate its transcriptional activity, so this MAPK may contribute to regulation of CHOP function by multiple mechanisms (43). In the future, it will be important to determine whether these additional stress pathways function in conjunction with eIF2∼P through the proposed bypass mechanism or entail alternative translational control processes involving the uORF or other features of the 5′-leader of the CHOP mRNA.
Acknowledgments
We thank Souvik Dey and Brian Teske for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM49164 and GM64350 (to R. C. W.).
This article was selected as a Paper of the week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- ER
- endoplasmic reticulum
- ISR
- integrated stress response
- uORF
- upstream ORF
- qRT
- quantitative RT
- RACE
- rapid amplification of cDNA ends
- MEF
- mouse embryonic fibroblast
- TK
- thymidine kinase.
REFERENCES
- 1. Sonenberg N., Hinnebusch A. G. (2009) Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wek R. C., Jiang H. Y., Anthony T. G. (2006) Biochem. Soc. Trans. 34, 7–11 [DOI] [PubMed] [Google Scholar]
- 3. Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 4. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 5. Schröder M., Kaufman R. J. (2005) Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 6. Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 7. Lu P. D., Harding H. P., Ron D. (2004) J. Cell Biol. 167, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vattem K. M., Wek R. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang H. Y., Wek S. A., McGrath B. C., Lu D., Hai T., Harding H. P., Wang X., Ron D., Cavener D. R., Wek R. C. (2004) Mol. Cell. Biol. 24, 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marciniak S. J., Ron D. (2006) Physiol. Rev. 86, 1133–1149 [DOI] [PubMed] [Google Scholar]
- 11. Hinnebusch A. G. (2005) Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 12. Abastado J. P., Miller P. F., Jackson B. M., Hinnebusch A. G. (1991) Mol. Cell. Biol. 11, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., Hinnebusch A. G. (1992) Cell 68, 585–596 [DOI] [PubMed] [Google Scholar]
- 14. Novoa I., Zeng. H., Harding H. P., Ron D. (2001) J. Cell Biol. 153, 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Connor J. H., Weiser D. C., Li S., Hallenbeck J. M., Shenolikar S. (2001) Mol. Cell. Biol. 21, 6841–6850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ma Y., Hendershot L. M. (2003) J. Biol. Chem. 278, 34864–34873 [DOI] [PubMed] [Google Scholar]
- 17. Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P., Ron D. (2004) Genes Dev. 18, 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou D., Palam L. R., Jiang L., Narasimhan J., Staschke K. A., Wek R. C. (2008) J. Biol. Chem. 283, 7064–7073 [DOI] [PubMed] [Google Scholar]
- 19. Dang Do A. N., Kimball S. R., Cavener D. R., Jefferson L. S. (2009) Physiol. Genomics 38, 328–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Y. J., Tan B. C., Cheng Y. Y., Chen J. S., Lee S. C. (2010) Nucleic Acids Res. 38, 764–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rutkowski D. T., Arnold S. M., Miller C. N., Wu J., Li J., Gunnison K. M., Mori K., Sadighi Akha A. A., Raden D., Kaufman R. J. (2006) PLoS Biol. 4, e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rutkowski D. T., Kaufman R. J. (2007) Trends Biochem. Sci. 32, 469–476 [DOI] [PubMed] [Google Scholar]
- 23. Li M. Z., Elledge S. J. (2007) Nat. Methods 4, 251–256 [DOI] [PubMed] [Google Scholar]
- 24. Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R. J. (2001) Mol. Cell 7, 1165–1176 [DOI] [PubMed] [Google Scholar]
- 25. Jiang H. Y., Wek S. A., McGrath B. C., Scheuner D., Kaufman R. J., Cavener D. R., Wek R. C. (2003) Mol. Cell. Biol. 23, 5651–5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dey S., Baird T. D., Zhou D., Palam L. R., Spandau D. F., Wek R. C. (2010) J. Biol. Chem. 285, 33165–33174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jousse C., Bruhat A., Carraro V., Urano F., Ferrara M., Ron D., Fafournoux P. (2001) Nucleic Acids Res. 29, 4341–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gilbert W. V. (2010) J. Biol. Chem. 285, 29033–29038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jackson R. J., Hellen C. U., Pestova T. V. (2010) Nat. Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dever T. E. (2002) Cell 108, 545–556 [DOI] [PubMed] [Google Scholar]
- 31. Kozak M. (2001) Nucleic Acids Res. 29, 5226–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spirin A. S. (2009) Biochemistry 48, 10688–1069219835415 [Google Scholar]
- 33. Costa-Mattioli M., Sossin W. S., Klann E., Sonenberg N. (2009) Neuron 61, 10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kozak M. (1984) Nucleic Acids Res. 12, 857–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kozak M. (1986) Cell 44, 283–292 [DOI] [PubMed] [Google Scholar]
- 36. Kozak M. (1987) Mol. Cell. Biol. 7, 3438–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pisarev A. V., Kolupaeva V. G., Pisareva V. P., Merrick W. C., Hellen C. U., Pestova T. V. (2006) Genes Dev. 20, 624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ivanov I. P., Loughran G., Sachs M. S., Atkins J. F. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 18056–18060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wek R. C., Anthony T. G. (2009) Cell Metab. 10, 1–2 [DOI] [PubMed] [Google Scholar]
- 40. McCullough K. D., Martindale J. L., Klotz L. O., Aw T. Y., Holbrook N. J. (2001) Mol. Cell. Biol. 21, 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li G., Mongillo M., Chin K. T., Harding H., Ron D., Marks A. R., Tabas I. (2009) J. Cell Biol. 186, 783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Song B., Scheuner D., Ron D., Pennathur S., Kaufman R. J. (2008) J. Clin. Invest. 118, 3378–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang X. Z., Ron D. (1996) Science 272, 1347–1349 [DOI] [PubMed] [Google Scholar]
- 44. Hood H. M., Neafsey D. E., Galagan J., Sachs M. S. (2009) Annu. Rev. Microbiol. 63, 385–409 [DOI] [PubMed] [Google Scholar]