Abstract
High risk human Papillomavirus (HPV) types are the major causative agents of cervical cancer. Reduced expression of major histocompatibility complex class I (MHC I) on HPV-infected cells might be responsible for insufficient T cell response and contribute to HPV-associated malignancy. The viral gene product required for subversion of MHC I synthesis is the E7 oncoprotein. Although it has been suggested that high and low risk HPVs diverge in their ability to dysregulate MHC I expression, it is not known what sequence determinants of HPV-E7 are responsible for this important functional difference. To investigate this, we analyzed the capability to affect MHC I of a set of chimeric E7 variants containing sequence elements from either high risk HPV16 or low risk HPV11. HPV16-E7, but not HPV11-E7, causes significant diminution of mRNA synthesis and surface presentation of MHC I, which depend on histone deacetylase activity. Our experiments demonstrate that the C-terminal region within the zinc finger domain of HPV-E7 is responsible for the contrasting effects of HPV11- and HPV16-E7 on MHC I. By using different loss- and gain-of-function mutants of HPV11- and HPV16-E7, we identify for the first time a residue variation at position 88 that is highly critical for HPV16-E7-mediated suppression of MHC I. Furthermore, our studies suggest that residues at position 78, 80, and 88 build a minimal functional unit within HPV16-E7 required for binding and histone deacetylase recruitment to the MHC I promoter. Taken together, our data provide new insights into how high risk HPV16-E7 dysregulates MHC I for immune evasion.
Keywords: Antigen, Antigen Presentation, Cellular Immune Response, MHC, Virus
Introduction
Major histocompatibility complex class I (MHC I)3 genes encode transmembrane glycoproteins involved in the processes of immune recognition. They function by binding intracellularly processed peptides and presenting them on the cell surface to cytotoxic T cells (1). These peptides, which arise predominantly from proteolytic cleavage in the cytosol, are delivered into the endoplasmic reticulum by the transporter associated with antigen processing (TAP) (2). During viral infection a spectrum of antigenic peptides is displayed by MHC I molecules, resulting in the elimination of the infected cells. However, many viruses evade the T cell-mediated immune response, primarily by decreasing the levels of surface MHC I, thus, reducing the presentation of pathogen-derived antigens (3, 4) and evading cellular immunosurveillance mechanisms (5).
There are more than 100 recognized genotypes of human papillomaviruses (HPVs), the main etiological agents of human cervical cancer. They can be divided into two major groups; that is, the high risk serotypes, such as HPV16, which are associated with highly invasive anogenital and cervical carcinomas, and the low risk serotypes, such as HPV11, which are the principal causative agents of benign warts. There is clear evidence that surface MHC I presentation is down-regulated in cervical tumors (6), and this phenotype likely results directly from viral gene functions.
E7 proteins of HPV are mostly found in the nucleus of infected cells (7), and regulation of transcription is one of their many functions (8–13). Interestingly, E7 of high risk HPV16 and -18, which is continuously expressed after cellular transformation and in cervical cancer tissues, is able to suppress the activity of MHC I promoters (14). Furthermore, HPV18-E7 also represses activity of a bidirectional promoter that regulates expression of TAP1 and LMP2, whereas HPV16-E7 does not, indicating that E7 proteins from different HPVs employ diverse strategies to modulate antigen presentation. Data from Li et al. (15, 16) showed that HPV16-E7 and specific histone deacetylases (HDACs), which function as transcriptional co-repressors, are physically associated with the MHC I promoter and that the histones bound to the promoter are deacetylated, suggesting that MHC I down-regulation is mediated by repression of chromatin activation. E7 encoded by low risk HPVs, such as HPV11, does not interact with most targets of high risk HPV-E7 or does so much less efficiently (17). On the basis of this and the finding that low risk HPV-E7 does not apparently down-regulate MHC I expression (18), it was speculated that E7 from high and low risk HPVs differ in their ability to modulate MHC I gene activity.
To characterize the molecular properties of high and low risk HPV-E7 that determine the contrasting effects on MHC I expression, we transfected human cells with green fluorescent protein (GFP)-tagged E7 constructs from HPV11 and HPV16. Both HPV-E7 variants were predominately found in the nucleus and showed retinoblastoma protein (pRb) binding properties reflecting the different interaction affinities of high and low risk HPV-E7 (19). In line with previous findings (14, 15, 20) we show that HPV16-E7 causes a significant reduction of MHC I expression and surface presentation, whereas no such dysregulation was observed for HPV11-E7. In addition, we observe that stable transfectants expressing HPV11-E7 or HPV16-E7 have comparable steady-state expression levels of TAP and tapasin, indicating that both HPV-E7 proteins apparently do not modulate the synthesis of components of the peptide-loading complex (PLC). Further experiments show that MHC I down-regulation by HPV16-E7 is largely neutralized by HDAC inhibitors, suggesting that the C-terminal E7 zinc finger domain, which mediates functional complex formation between HPV16-E7 and HDACs (21), is involved in suppression of MHC I. By using different chimeric HPV-E7 constructs in which we mutually exchanged sequences between HPV11- and HPV16-E7, we demonstrate that only the C-terminal region of the protein is responsible for the distinctive effects of HPV11- and HPV16-E7 on MHC I expression. Moreover, substitution of three residues in the C terminus of HPV11-E7 (positions 78, 80 and 88) for the corresponding residues from HPV16-E7 creates a gain-of-function variant that has virtually the same MHC I down-regulation effect as HPV16-E7. Hence, our studies suggest that the identified residues form a functional unit required for binding and HDAC recruitment to the MHC I promoter. Altogether, our experiments provide novel insights into how HPV16-E7 dysregulates MHC I for immune evasion.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
HLA-A3-positive human embryonic kidney (HEK)-293 (ATCC no. CRL 1573) and -293T (ATCC no. CRL 11268) cells (22) were cultivated in Iscove's modified Dulbecco's medium (Invitrogen) containing 10% fetal bovine serum (Biochrom) and penicillin/streptomycin (Invitrogen). Stable transfectants of HEK-293 expressing GFP alone or GFP-tagged HPV-E7-derivatives from HPV11 or HPV16 were cultured in the presence of 0.5 mg/ml G418 (PAA).
Antibodies
Horseradish peroxidase-linked and non-conjugated mouse monoclonal anti-GFP antibody (mAb B-2) as well as rabbit polyclonal anti-pRb antiserum (M-153) were purchased from Santa Cruz. Mouse monoclonal anti-AU1, anti-β-actin, and rabbit polyclonal anti-HDAC1 antibodies are from Covance, Sigma, and Abcam, respectively. W6/32 is a mouse mAb reacting with assembled HLA-A, -B, and -C (23). 3B10.7 is a rat mAb recognizing human MHC I heavy chains (24). The mouse mAbs 148.3 and 435.3 are directed against human TAP1 (25) and TAP2 (26), respectively. R. SinE is a rabbit polyclonal Ab against human tapasin (27). All secondary Abs were purchased from Dianova.
Cell Lysis and Western Blotting
Cells were washed twice in ice-cold NaCl/K/Pi (1.7 mm KH2PO4, 10 mm Na2HPO4, 140 mm NaCl, 2.7 mm KCl) pH 7.5, before solubilization in lysis buffer (NaCl/K/Pi, pH 7.5, containing 1% Triton X-100; Sigma) containing the CompleteTM protease inhibitor mixture (Roche Applied Science). After 30 min of incubation on ice, lysed cells were centrifuged at 1,300 × g for 15 min before postnuclear supernatants were analyzed in Western blots treated with appropriate antibodies. Bands were visualized with enhanced chemiluminescence substrate (Sigma). Fluorographs were quantified using GelEval 1.32 software (FrogDance Software).
Fluorescence Microscopy
Transfected cells were fixed at room temperature for 15 min in 3% paraformaldehyde and quenched with 10 mm glycine for 10 min. GFP signals were enhanced by FITC-conjugated anti-GFP antibody. All images were captured using an Axiovert 200M/Apotome microscope (Zeiss).
Cloning and Expression of GFP- and AU1-tagged HPV-E7 Constructs
By using appropriate primers, PCR-generated fragments containing HPV11- and HPV16-E7 were initially cloned into the multiple cloning site (HindIII/XhoI for HPV11-E7 and EcoRI/XhoI for HPV16-E7) of pcDNA3 (Stratagene). To generate GFP-tagged constructs, E7-containing EcoRI/BamHI restriction fragments of the resulting pcDNA3 plasmids were inserted into the multiple cloning site of pEGFP-C1 (Clontech). All constructs were fully sequenced. To generate double-tagged constructs (HPV11-E7/dtag and HPV16-E7/dtag) site-directed mutagenesis with primer pairs 5′-CCCATCTGCGCACCAAAACCAGACACCTATCGATATATATAAGGATCCACCGGATCTAGATAAC-3′/5′-GTTATCTAGATCCGGTGGATCCTTATATATATCGATAGGTGTCTGGTTTTGGTGCGCAGATGGG-3′ and 5′-CCCATCTGTTCTCAGAAACCAGACACCTATCGATATATATAAGGATCCACCGGATCTAGATAAC-3′/5′-GTTATCTAGATCCGGTGGATCCTTATATATATCGATAGGTGTCTGGTTTCTGAGAACAGATGGG-3′ was employed to insert an AU1 tag (DTYRYI, amino acid single-letter code) (underlined) at the C terminus of HPV11-E7 and HPV16-E7, respectively. To generate chimeric E7 variants HPV11/16-E7 and HPV16/11-E7, we introduced a silent MfeI site at position 843 and 840 (position 1 is the A of the first AUG of the GFP constructs) of HPV11- and HPV16-E7 via site-directed mutagenesis using primer pair 5′-GCTATGAGCAATTGGAAGACAGCTCAGAAGATGAGG-3′/5′-CCTCATCTTCTGAGCTGTCTTCCAATTGCTCATAGC-3′ and 5′-CTACTGTTATGAGCAATTGAATG ACAGCTCAGAGGAGG-3′/5′-CCTCCTCTGAGCTGTCATTCAATTGCTCATAACAGTAG-3′, respectively. The chimeric constructs HPV11/16-E7 and HPV16/11-E7 were then generated by swapping MfeI fragments between these vectors. For generation of chimeric constructs HPV11/16/11-E7 and HPV16/11/16-E7, we mutually exchanged amino acid sequences 29–75 and 28–75 between HPV11- and HPV16-E7. Therefore, a silent EcoRV site was introduced into HPV11/16-E7 (nucleotide position 987) and HPV16/11-E7 (nucleotide position 981) by site-directed mutagenesis using primer pair 5′-CGTACAAAGCACACACGTAGATATCCGTACTTTGGAAGACC-3′/5′-GGTCTTCCAAAGTACGGATATCTACGTGTGTGCTTTGTACG-3′ and 5′-GGAGTGCACAGACGGAGATATCAGACAACTACAAGACC-3′/5′GGTCTTGTAGTTGTCTGATATCTCCGTCTGTGCACTCC-3′, respectively. The chimeric constructs HPV11/16/11-E7 and HPV16/11/16-E7 were then generated by swapping EcoRV/StuI fragments between these vectors. For inserting single point mutations in HPV11- and HPV16-E7, the mutagenesis primers 5′-GGAGACATCAGACAACTAGAAGATCTTTTGCTGGGCACAC-3′/5′-GTGTGCCCAGCAAAAGATCTTCTAGTTGTCTGATGTCTCC-3′ (HPV11-E7/E; Q80E) and 5′-CGTAGACATTCGTACTTTGCAAGATCTGTTAATGGGCACAC-3′/5′-GTGTGCCCATTAACAGATCTTGCAAAGTACGAATGTCTACG-3′ (HPV16-E7/Q; E80Q) were used. Further amino acid exchanges in HPV11-E7 and HPV11-E7/E were introduced using primer pairs 5′-GGAGTGCACAGACGGAGATATCAGAACACTACAAGACCTTTTGCTGG-3′/5′-CCAGCAAAAGGTCTTGTAGTGTTCTGATATCTCCGTCTGTGCACTCC-3′ (HPV11-E7/T; Q78T), 5′-GGAGTGCACAGACGGAGATATCAGAACACTAGAAGATCTTTTGCTGG-3′/5′-CCAGCAAAAGATCTTCTAGTGTTCTGATATCTCCGTCTGTGCACTCC-3′ (HPV11-E7/TE; Q78T, Q80E), 5′-CAAGACCTTTTGCTGGGCACCCTAGGTATTGTGTGTCCCATCTGC-3′/5′-GCAGATGGGACACACAATACCTAGGGTGCCCAGCAAAAGGTCTTG-3′ (HPV 11-E7/G; N88G), and 5′-GAAGATCTTTTGCTGGGCACCCTAGGTATTGTGTGTCCCATCTGC-3′/5′-GCAGATGGGACACACAATACCTAGGGTGCCCAGCAAAAGATCTTC-3′ (HPV11-E7/EG, Q80E, N88G; HPV11-E7/TEG, Q78T, Q80E, N88G). All mutant vectors were sequenced. GFP-tagged HPV-E7 constructs were transfected into HEK-293 and/or -293T cells by using the lipofection reagent DOTAP from Roche Applied Science. After selection with G418 (0.5 mg/ml) for 4–6 weeks, stable transfectants of HEK-293 were subcloned and screened for HPV-E7 expression by Western blotting.
Immunoprecipitation
Cells were solubilized in lysis buffer (NaCl/K/Pi, pH 7.5) containing 1% Triton X-100 (Sigma) and CompleteTM protease inhibitor mixture from Roche Applied Science. Immunoprecipitations of pRb/E7 complexes were performed for 2 h in the cold with anti-pRb Abs covalently coupled to protein A-Sepharose by dimethylpimelimidate. After extensive washing, isolated immune complexes were eluted by 0.5% sodium dodecyl sulfate (SDS) in the presence of 2 mm urea. Eluates were separated by SDS-PAGE under reducing conditions and analyzed by Western blot using anti-pRb and anti-GFP antibodies.
Quantitative Real-time PCR
For SYBR Green-based quantitative real-time PCR, mRNA was isolated from transfected and non-transfected HEK-293 cells using the TRIzol method. The analysis was performed on a Cepheid SmartCycler (Cepheid) in two triplicates using the primer pairs 5′-GGACCAGGAGACACGGAATGTGAAGGCC-3′/5′-GCCGTCGTAGGCGTCCTGCCGGTAC-3′ (HLA-A3) and 5′-ATGACAACTTTGGTATCGTGGAAGG-3′/5′-GAAATGAGCTTGACAAAGTGGTCGT-3′ (GAPDH).
Chromatin Immunoprecipitation (ChIP) Assay
HEK-293T cells were transiently transfected with HPV16-E7 and different HPV11-E7 constructs. After 24 h transfectants were fixed with 1% formaldehyde for 10 min at room temperature and quenched with glycine at a final concentration of 125 mm for 5 min. Fixed cells were sequentially washed at room temperature for 15 min with 10 mm Tris/HCl, pH 8.0, 10 mm EDTA, 0.5 mm EGTA, 0.25% Triton X-100. The cells were then suspended in 1 ml of lysis buffer containing 50 mm Tris/HCl, pH 8.0, 1% SDS, 10 mm EDTA, and 5 μg/ml sonicated salmon sperm DNA. After 30 min of incubation on ice, samples were sonicated for 20 s with at least 5-min intervals on ice. For ChIP assays with appropriate Abs, cell lysates were sonicated twice for 5 s each. Samples were then spun for 10 min at 15,000 × g at 4 °C, and the supernatants were collected. 50 μl of the supernatant was used as an input control during the PCR. Cell lysates were diluted to 5 ml with 20 mm Tris/HCl, pH 8.0, 1% Triton X-100, 2 mm EDTA, 150 mm NaCl, 0.01% SDS, and 5 μg/ml sonicated salmon sperm DNA. Protein-chromatin complexes were immunoprecipitated overnight with Abs and rotated at 4 °C. The chromatin was collected on protein G-Sepharose beads (Roche Applied Science). The beads were washed sequentially with 1 ml of the following buffers: (a) 10 mm Tris/HCl, pH 8.0, 0.1% SDS, 1% Triton X-100, 1 mm EDTA, 0.5 mm EGTA, 0.1% sodium deoxycholate, 150 mm NaCl, (b) 10 mm Tris/HCl, pH 8.0, 0.1% SDS, 1% Triton X-100, 1 mm EDTA, 0.1% sodium deoxycholate, 500 mm NaCl, and (c) 10 mm Tris/HCl, pH 8.0, 250 mm LiCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 1 mm EDTA followed by washing 3 times with cold Tris/HCl, 1 m EDTA, pH 8.0. Precipitates were then extracted by incubating with elution buffer (1% SDS, 100 mm NaHCO3) at 65 °C overnight and with 1/25 volume of 5 m NaCl for 6 h at 65 °C. DNA was purified, and PCR amplification was carried out using primers for human MHC I promoter (forward primer, 5′-CCAGTTCAGGGACAGAGATTACGGG-3′; reverse primer, 5′-AGAGGGAGAAAAGAAACTGCGGAG-3′ (15, 28). Amplification products were separated on a 2% agarose gel.
Flow Cytometry
Cells were carefully detached by mild trypsin treatment, stained with mAb W6/32 for 30 min on ice, and analyzed on a FACScan flow cytometer (BD Biosciences).
RESULTS
Construction and Cellular Expression of GFP-tagged E7 Constructs from HPV11 and HPV16
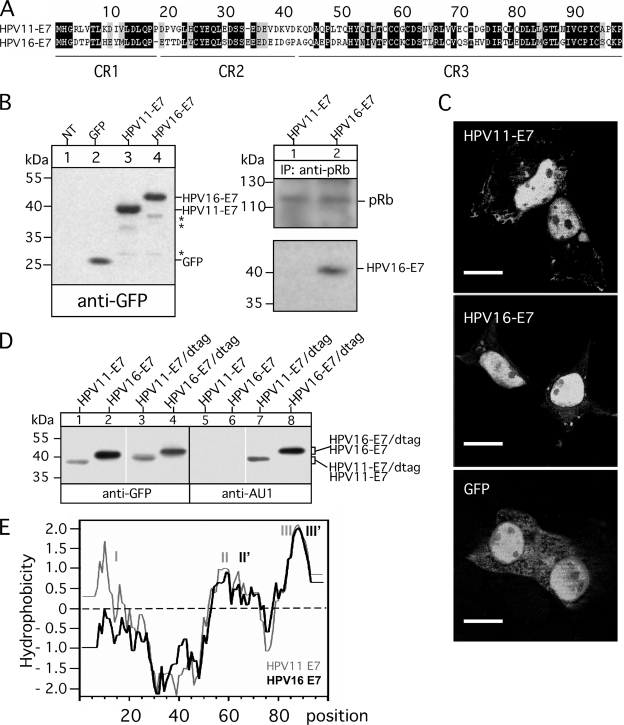
The HPV16-E7 protein (29) has been shown to be the major viral protein in cervical cancers and cancer-derived cell lines containing HPV16 DNA (30). Previous studies suggest that HPV16-E7 is involved in the transcriptional regulation of MHC I in infected cells to avoid T lymphocyte attacks (14), whereas in contrast HPV11-E7 apparently has no effect on MHC I expression (18). The E7 proteins of low risk HPV11 and high risk HPV16 comprise 98 amino acids and have an overall sequence similarity with each other of 64%. Regions of low identity that might be involved in functional diversity of HPV-E7 are prominently present in the N-terminal part of the two proteins, at the flanking regions of CR1, CR2, whereas the C-terminal regions containing CR3 display a rather moderate sequence diversity (Fig. 1A).
FIGURE 1.
Expression and characterization of GFP-tagged E7 proteins from HPV11 and HPV16. A, aligned amino acid sequences of E7 polypeptides from HPV11 and HPV16 are shown. Sequences were retrieved from the GenBankTM data base (GenBankTM AAA21704.1 and NP_041326.1) and aligned using the software Vector NTI (Invitrogen). Identical residues are marked by black boxes, whereas gray boxes indicate similar or weakly similar residues. The conserved sequence regions CR1, CR2, and CR3 are underlined. B, expression of the N-terminal GFP-tagged E7 proteins from HPV11 and HPV16 (left panel) is shown. Cell lysates of stably transfected HEK-293 cells were separated on a 15% SDS-Gel and blotted onto nitrocellulose as described under “Experimental Procedures.” Western blots were probed for the GFP tag by using mAb B-2. Complex formation with pRb and intracellular localization of the GFP-tagged HPV-E7 proteins was analyzed by immunoprecipitations (IP; B, right panel) and fluorescence microscopy (C). Anti-pRb immunoisolated precipitates from Triton X-100-lysed transfectants were separated on a 15% SDS gel and analyzed in Western blots probed with Abs to pRb and GFP. Asterisks indicate minor bands detected in the immunoblot stained with anti-GFP antibody. For fluorescence microscopy-transfected cells (GFP and GFP-tagged HPV11- and HPV16-E7) were grown on coverslips, fixed at room temperature with paraformaldehyde, and analyzed by using an Axiovert 200M/Apotome microscope. Scale bars indicate 10 μm. D, SDS-PAGE analysis of GFP/AU1-double-tagged E7 proteins from HPV11 and HPV16 are shown. Cell lysates of HEK-293T cells transiently transfected with single and double-tagged (dtag) E7 constructs from HPV11 and HPV16 were separated on a 15% SDS-Gel and blotted onto nitrocellulose as described under “Experimental Procedures.” Western blots were probed for the GFP and the AU1 tag. E, Kyte Doolittle hydrophobicity plots for HPV11- and HPV16-E7 were constructed using a window size of nine residues (32). Positive values indicate regions of hydrophobicity. Identified hydrophobic regions are marked by I, II, II′, III, and III′.
To investigate the amino acid sequence properties of HPV-E7 required for the dysregulation of MHC I, we first constructed N-terminal GFP-tagged E7 variants of HPV11- and HPV16-E7 and stably transfected them into HEK-293 cells. The major advantage of this system is that a single Ab (anti-GFP) can be used to detect E7 proteins regardless of sequence variations between HPV11- and HPV16-E7, allowing a direct comparison of HPV-E7 expression in transfected cells. As can be seen from the Western blot experiment shown in Fig. 1B (left panel, lanes 3 and 4), both HPV-E7-constructs are expressed by the HEK-293 transfectants at comparable levels with apparent molecular masses of 39 (HPV11-E7) and 41 kDa (HPV16-E7), respectively. Analysis of the intracellular distribution of the GFP-tagged HPV-E7 proteins by fluorescence microscopy (Fig. 1C) revealed that both are predominantly localized in the nucleus, whereas GFP alone additionally showed a more diffuse location pattern throughout the cell. Consistent with results obtained for non-tagged HPV-E7 proteins E7 (19), we observed in immunoprecipitation experiments with anti-pRb Ab (Fig. 1B, right panel) that the GFP-tagged variant of high risk HPV16-E7 has much higher binding affinity for pRb than the corresponding low risk HPV11-E7 construct. In contrast to the well detectable pRb/HPV16-E7 complexes, no signal of pRb-co-isolated HPV11-E7 could be observed in Western blots with exposure times of 1 min (Fig. 1B, right panel). Only at much longer exposures (>10 min) of the Western blots a very weak band corresponding to HPV11-E7 could be detected (data not shown). Thus, in line with the results of other groups (31), it seems that the N-terminal addition of the GFP moiety to HPV-E7 results in fusion proteins that can be readily expressed/visualized and apparently does not alter cellular localization or biochemical interaction properties of the original proteins. In the Western blot depicted in Fig. 1B only a few additional faint bands with molecular masses between 26 and 38 kDa were recognized, indicating that only a very small amount (1–2%) of the two HPV-E7 proteins is apparently sensitive to cellular and/or postlytic degradation. However, to distinguish whether the distinct electrophoretic migration behavior observed for the two GFP-tagged HPV-E7 proteins (Fig. 1B, left panel, lanes 3 and 4) is caused by C-terminal degradation of HPV11-E7 or due to differences in SDS loading within the E7 part of the fusion proteins, we inserted an additional small AU1 tag (DTYRYI, single-letter code) at the C terminus of the GFP constructs of HPV11- and HPV16-E7. Western blot analysis of cell lysates from transiently transfected HEK-293 cells showed that the double-tagged HPV-E7 constructs (HPV11-E7/dtag and HPV16-E7/dtag) retained their different electrophoretic mobility and were both detectable by GFP-specific as well as by AU1-specific Abs (Fig. 1D, compare lanes 3 and 4 with 7 and 8), indicating that the GFP-stained 39- and 41-kDa bands of HPV11-E7 and HPV16-E7 (Fig. 1, B and D) represent the full-length forms of the two GFP-tagged proteins. The Kyte Doolittle hydrophobicity plot (32) shown in Fig. 1E revealed that HPV11-E7 contains three hydrophobic regions (indicated by I, II, and III), whereas in contrast, HPV16-E7 comprises only two of these (indicated by II′ and III′), reflecting the pronounced sequence divergence at the N termini of the two HPV-E7 proteins. In view of the fact that SDS generally loads at hydrophobic sites (33) and variations in sequence hydrophobicity influence SDS binding and electrophoretic protein mobility (34), it is tempting to assume that differences in detergent binding and SDS/weight ratios are indeed responsible for the distinctive SDS-PAGE migration behavior of the two HPV-E7 proteins. In summary, both GFP-tagged HPV-E7 variants are expressed as intact stable polypeptides possessing the anticipated cell biological and biochemical properties required to analyze their functional influence on MHC I presentation.
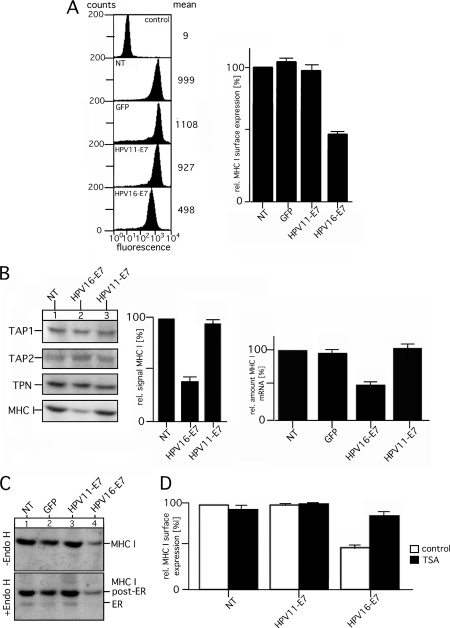
Distinctive Effects of HPV11- and HPV16-E7 on MHC I Expression
During the last years, different aspects of the immune evasion mechanisms of HPV-E7 have been investigated (35). However, it is still unclear whether and to what extent the E7 proteins from high and low risk HPV types differ in their influence on MHC I processing. To assess the effects of GFP-tagged HPV11- and HPV16-E7 on MHC I-mediated antigen presentation, we determined the steady-state surface levels of MHC I in stably transfected HEK-293 cells by flow cytometry (Fig. 2A). Compared with the phenotypes of non-transfected cells or transfectants expressing GFP, HPV11-E7 did not alter MHC I surface presentation (Fig. 2A, left and right panel). In contrast, however, HPV16-E7 expression caused a substantial decrease (50–60%) in MHC I surface levels, supporting the notion that HPV11- and HPV16-E7 differ in their ability to interfere with MHC I processing (18). To ensure that differences observed in MHC I presentation between the HPV11 and HPV16-E7 transfectants were not the result of clonal variation, we measured MHC I surface expression of additional transfectants with comparable HPV-E7 synthesis (data not shown) and found the MHC I levels to be in agreement with those of the cell lines depicted in Fig. 2A. Our experiments on the steady-state expression of components of the PLC showed that in HPV16-E7 transfectants the amount of MHC I was clearly reduced by 60% compared with the amount detected for control cells or transfectants expressing HPV11-E7 (Fig. 2B, left and middle panel). Neither HPV11- nor HPV16-E7 had any measurable influence on the steady-state expression of TAP1, TAP2, or tapasin (TPN) (Fig. 2B, left panel). Moreover, none of the HPV-E7 variants showed any detectable physical interaction with TAP and/or the PLC (data not shown) or any influence on the intracellular transport and maturation of MHC I (Fig. 2C, compare lanes to 1–4). In all cases, the majority of MHC I molecules is efficiently exported from the endoplasmic reticulum. Further experiments in which we analyzed MHC I-mRNA expression by quantitative RT-PCR (Fig. 2B, right panel) revealed that HPV16-E7 mediates a transcriptional down-regulation of MHC I, whereas no such effect was observed for HPV11-E7. Finally, we tested whether the observed effect of HPV16-E7 on MHC I is HDAC-dependent (15) and, thus, sensitive to trichostatin A (TSA), a specific HDAC inhibitor (36). Therefore, control cells and HPV-E7 transfectants were incubated overnight in the absence or presence of TSA and then immunostained with anti-MHC I Ab to assess surface expression of MHC I by flow cytometry. The results shown in Fig. 2D demonstrate that in the presence of the inhibitor MHC I, surface levels are largely restored (75–80%) in HPV16-E7-transfectants, indicating that histone deacetylation and the C-terminal HPV-E7 zinc finger domain, which mediates functional complex formation between HPV16-E7 and HDACs (21), seems to be involved in MHC I repression. In contrast, control cells and transfectants expressing HPV11-E7 showed no measurable TSA effect on MHC I surface presentation (Fig. 2D). Taken together, the analyzed constructs of HPV11- and HPV16-E7 differed in their ability to dysregulate MHC I expression and surface presentation. HPV16-E7 caused significant diminution of mRNA synthesis and surface presentation of MHC I, which depends on HDAC activity, whereas no such effects were observed for HPV11-E7.
FIGURE 2.
Comparison of MHC I expression and surface presentation in stable HEK-293 transfectants expressing GFP-tagged E7 from HPV11 and HPV16. A, surface expression of MHC I was assessed by flow cytometry using the mAb W6/32 (second to fifth analysis, from the top). Background staining was analyzed by incubating with secondary Ab alone (control, upper analysis). Data are representative of three independent experiments (see the histogram, right panel). NT, non-transfected. B, analysis of the expression of components of the PLC in HEK-293 transfectants expressing GFP-tagged E7 from HPV11 and HPV16 is shown. Cell lysates of stably transfected and non-transfected HEK-293 cells were analyzed by Western blot using the indicated Abs (left panel). One representative experiment of two independent experiments is shown. Signals obtained for MHC I were quantitated by densitometric scanning (middle panel). Expression MHC I HLA-A3-mRNA was analyzed by quantitative real-time PCR (right panel). Expression levels of GAPDH-mRNA were used for normalizing the obtained results. C, intracellular transport and maturation of MHC I are shown. Cell lysates of HEK-293 transfectants were digested or not with endoglycosidase H (Endo H), separated by SDS-PAGE, and analyzed by Western blots probed for MHC I. D, TSA sensitivity of MHC I surface expression in HEK-293 cells expressing GFP-tagged E7 from HPV11 and HPV16 is shown. Cells were incubated overnight in the presence of 0.24 μm TSA. Subsequently, cells were immunostained with the mAb W6/32 to assess surface expression of MHC I by flow cytometry. Surface MHC I staining in the presence or absence of the inhibitor is plotted as a percentage of MFI compared with the mean fluorescent intensity obtained for the untreated NT HEK-293 (set to 100%). One representative result of three separate experiments is shown.
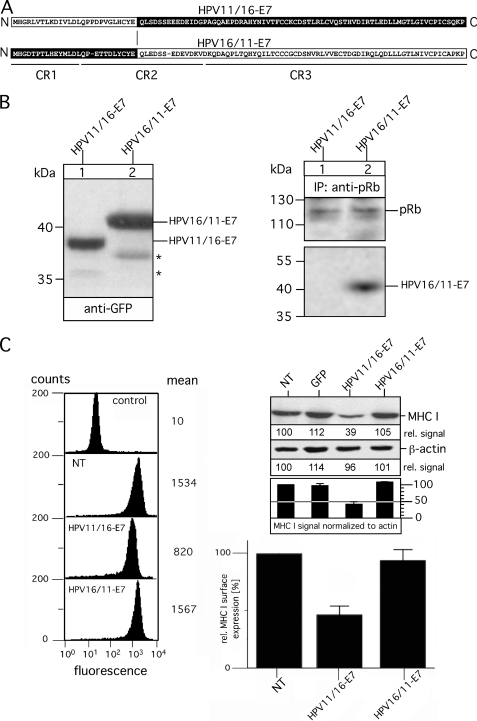
The C Terminus of HPV-E7 Controls Dysregulation of MHC I Expression and Surface Presentation
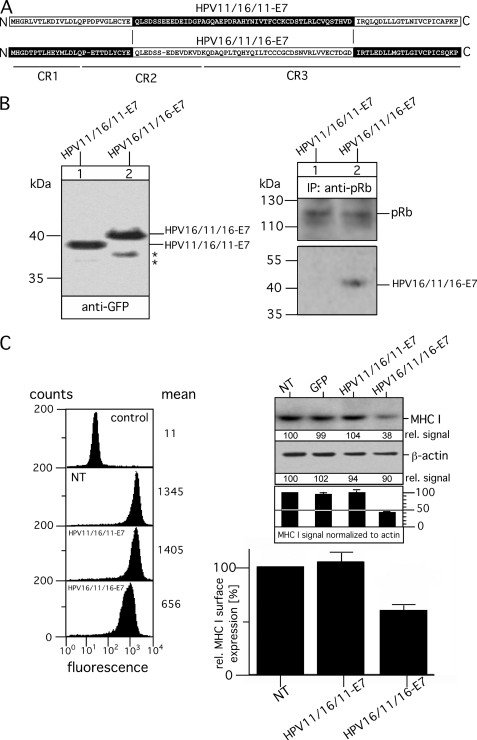
To identify the sequence of HPV16-E7 that is functionally involved in the modulation of MHC I expression, we created a pair of GFP-tagged chimeric HPV-E7 variants with the N-terminal part of HPV11-E7 (comprising the CR1 sequence up to the LXCXE motif of CR2) and the C-terminal part of HPV16-E7 (containing sequence regions of CR2 and CR3 downstream the pRb binding motif, LXCXE) (referred to as HPV11/16-E7) and vice versa (referred to as HPV16/11-E7). A pictorial overview of the two chimeric HPV-E7 constructs is shown in Fig. 3A. HPV11/16-E7 and HPV16/11-E7 were stably transfected into HEK-293 cells, and their expression was analyzed by immunoblots probed for the GFP-tag. Fig. 3B shows a well balanced expression of the two HPV-E7 chimeras. Moreover, the migration properties of HPV11/16-E7 and HPV16/11-E7 strongly suggest that the distinct electrophoretic mobilities observed for HPV11- and HPV16-E7 (Fig. 1D) are indeed determined by different hydrophobicity within their N termini. In agreement with previous findings (19), our immunoprecipitation experiments using anti-pRb verified that the N-terminal half of HPV-E7 also determines the affinity for pRb binding, indicating that the sequence switch between HPV11- and HPV16-E7 preserves the functional folding of the two HPV-E7 chimeras. Indeed, analysis of MHC I surface expression revealed, in agreement with our TSA experiments (Fig. 2D), that HPV-E7-mediated effects on MHC I are strictly confined to the sequence region containing the zinc finger domain of HPV11- and HPV16-E7. Thus, only cells expressing HPV11/16-E7 showed a reduction of MHC I expression (Fig. 3C, upper right panel) and surface presentation (Fig. 3C, left and lower right panel) between 50–60%, which is in the range observed for the original HPV16-E7-transfectants (Fig. 2A), whereas no such MHC I dysregulation was seen for cells expressing HPV16/11-E7. To narrow down the sequence region within the C-terminal halves of HPV11- and HPV16-E7 that imposes the different effects on MHC I, we constructed on the basis of HPV16/11-E7 and HPV11/16-E7 two additional HPV-E7 chimeras (referred to as HPV16/11/16-E7 and HPV11/16/11-E7, see Fig. 4A) by reciprocal exchange of the sequence stretch between the LXCXE motif and amino acid position 76. As can be seen from the Western blot in Fig. 4B, left panel, both HPV-E7 chimeras were stably expressed in HEK-293 at comparable steady-state levels. Again, anti-pRb immunoisolation (Fig. 4B, right panel) demonstrated that pRb binding affinities are solely determined by the sequence characteristics of the N terminus of HPV11- and HPV16-E7, respectively. Flow cytometry (Fig. 4C, left and lower right panel) and Western blot analysis (Fig. 4C, upper right panel) showed that HPV16/11/16-E7, which contains the very C terminus of HPV16-E7, is able to down-regulate MHC I by 50–60%, whereas in contrast HPV11/16/11-E7 comprising the C-terminal amino acids of HPV11-E7 had no detectable effects on MHC I expression and surface presentation. It should be noted that individual cell lines of the same transfection type showed no significant clonal variation for MHC I presentation (data not shown). Hence, although the C-terminal regions of HPV11-and HPV16-E7 exhibit a much higher sequence identity to each other than the N-terminal parts of the two proteins (Fig. 1A), our findings suggest that the sequence properties of the last C-terminal 22 amino acids of HPV16-E7 are functionally critical for the observed dysregulation of MHC I.
FIGURE 3.
Analysis of chimeric variants HPV11/16-E7 and HPV16/11-E7. A, shown is a schematic overview of chimeric variants HPV11/16-E7 and HPV16/11-E7. Polypeptide regions of HPV11-E7 are indicated in white, whereas the regions of HPV16-E7 are shown in black. Sequence regions CR1, CR2, and CR3 are underlined. B, expression of HPV11/16-E7 and HPV16/11-E7 in stably transfected HEK-293 cells (left panel) is shown. Cell lysates of the transfectants were separated on a 15% SDS-Gel and blotted onto nitrocellulose as described under “Experimental Procedures.” Western blots were probed for the GFP tag by using mAb B-2. Complex formation with pRb was analyzed by immunoprecipitations (IP; right panel). Anti-pRb immunoisolated precipitates from Triton X-100-lysed transfectants were separated on a 15% SDS gel and analyzed in Western blots probed with Abs to pRb and GFP. Asterisks indicate minor bands detected with anti-GFP antibody. C, surface expression of MHC I was assessed by flow cytometry using the mAb W6/32 (left panel, second to fourth analysis, from the top). Background staining was analyzed by incubating with secondary Ab alone (control, upper analysis). Data are representative of three independent experiments (see the histogram, lower right panel). Steady-state expression of MHC I was analyzed by Western blot probed with mAb 3B10.7 (upper right panel). Anti-β-actin staining served as the internal control for equal protein loading. MHC I and β-actin signals were quantitated by densitometric scanning. Four different exposures of the immunoblot were taken to ensure linearity, and the obtained MHC I signals were normalized to the corresponding β-actin signals (see the histogram below the immunoblot). One representative exposure of the Western blot is shown in C, upper right panel. NT, non-transfected.
FIGURE 4.
Analysis of chimeric variants HPV11/16/11-E7 and HPV16/11/16-E7. A, shown is a schematic overview of the chimeric variants HPV11/16/11-E7 and HPV16/11/16-E7. Polypeptide regions of HPV11-E7 are indicated in white, and the corresponding regions of HPV16-E7 are shown in black. Regions CR1, CR2, and CR3 are underlined. B, expression of HPV11/16/11-E7 and HPV16/11/16-E7 in stably transfected HEK-293 cells (left panel) is shown. Cell lysates of the transfectants were analyzed by Western blot as described above. Complex formation with pRb was analyzed by immunoprecipitations (IP, right panel). Anti-pRb immunoisolated precipitates from Triton X-100-lysed transfectants were separated on a 15% SDS gel and analyzed in Western blots probed with Abs to pRb and GFP. Asterisks indicate minor bands detected with anti-GFP antibody. C, surface expression of MHC I was assessed by flow cytometry using the mAb W6/32 (left panel, second to fourth analysis from the top). Background staining was analyzed by incubating with secondary Ab alone (control, upper analysis). Data are representative of three independent experiments (see the histogram, lower right panel). Steady-state expression of MHC I was analyzed by Western blot probed with mAb 3B10.7 (upper right panel). Anti-β-actin staining served as the internal control for equal protein loading. MHC I and β-actin signals were quantitated by densitometric scanning. Four different exposures of the immunoblot were taken to ensure linearity, and the obtained MHC I signals were normalized to the corresponding β-actin signals (see the histogram below the immunoblot). One representative exposure of the Western blot is shown in Fig. 4C, upper right panel. NT, non-transfected.
Distinct Residues within the C-terminal Region of HPV-E7 Are Critical for MHC I Down-regulation
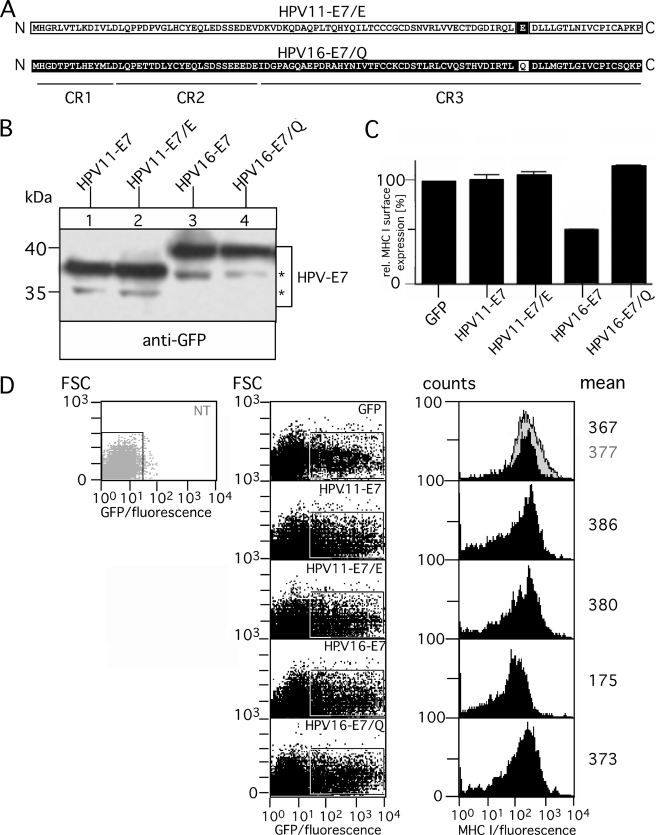
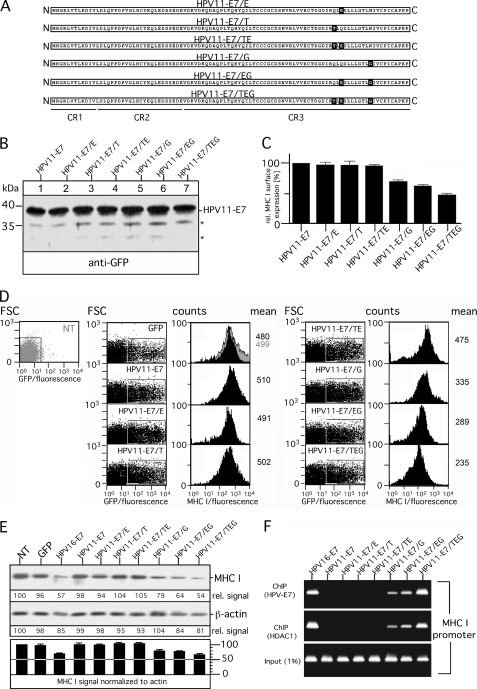
The polypeptide alignment in Fig. 1A shows that the C-terminal 22-amino acid-long region is only slightly different in sequence between HPV11- and HPV16-E7. In particular, the very C terminus of HPV16-E7 contains six amino acids that are different from that encoded by HPV11-E7, two similar and four dissimilar residues (Fig. 1A). Most interestingly, the group of residues (leucine residues 82 and 83, cysteine residue 91) that have been characterized previously for their importance in HDAC binding by HPV16-E7 (21) do not differ between the two HPV-E7 proteins. Hence, to identify residue variations responsible for the functional non-equivalence of the C termini of the two HPV-E7 proteins, we constructed single point mutants of HPV11- and HPV16-E7 where we exchanged in HPV11-E7 glutamine at position 80 by glutamate that is located at the equivalent position of HPV16-E7 (and vice versa, see Fig. 5A) next to residues that are thought to be critical for HDAC interaction (21). Expression of the wild type and mutant HPV-E7 (referred to as HPV11-E7/E and HPV16-E7/Q) was analyzed in transiently transfected HEK-293T cells. As shown in Fig. 5B, both mutants are expressed at levels comparable with that of the original HPV-E7 wild type constructs (Fig. 5B, compare lanes 1–4). The same transfectants were used to analyze surface expression of MHC I. Therefore, GFP- and HPV-E7-positive cells were determined by flow cytometry based on their GFP fluorescent intensity (Fig. 5D, left and middle panel). The histogram plots of MHC I surface expression were gated on the population non-transfected/GFP-negative and GFP-positive HEK-293T cells (gray and black histograms, Fig. 5D, right panel). Our findings (summarized in Fig. 5C) show that HPV16-E7/Q represents a loss-of-function mutant that in comparison to the original construct has lost its ability to modulate MHC I expression. In contrast, however, mutant HPV11-E7/E did not gain the ability to interfere with MHC I presentation. Hence, we conclude that glutamate at position 80 is clearly necessary for HPV16-E7-mediated dysregulation of MHC I but not sufficient to cause the observed differences between HPV11- and HPV16-E7. To assess which additional residues within the C terminus of HPV16-E7 might be required to transform HPV11-E7 into a modulator of MHC I expression, we constructed on the basis of HPV11-E7 five additional variants (Fig. 6A). HPV11-E7/T, HPV11-E7/TE, HPV11-E7/G, and HPV11-E7/EG contain the amino acid exchanges Q78T, Q78T/Q80E, N88G, and Q80E/N88G, respectively, whereas in the case of HPV11-E7/TEG, all three substitutions (Q78T/Q80E/N88G) were present in the mutant. After transient transfection into HEK-293T cells, we verified expression of the different HPV11-E7 constructs by Western blot (Fig. 6B) and analyzed in parallel the amount of surface MHC I (Fig. 6D) by flow cytometry as described above (see Fig. 5D). Our experiments revealed that cells expressing HPV11-E7/G, HPV11-E7/EG, and HPV11-E7/TEG are characterized by increasing down-regulation of MHC I (30, 40, and 50%, respectively) (Fig. 6C), whereas the other three HPV-E7 variants (HPV11-E7/E, HPV11-E7/T, and HPV11-E7/TE) had no detectable effect on MHC I surface expression (Fig. 6C). Moreover, we also analyzed the steady-state levels of MHC I in the different transient transfectants (Fig. 6E). All transfectants with reduced MHC I surface presentation showed a diminution of MHC I expression. However, although the HPV-E7 variants are very highly expressed (Figs. 5B and 6B), the reduced steady-state levels of MHC I observed in the detergent extracts of the transiently transfected cells (HPV16-E7, HPV11-E7/G, HPV11-E7/EG, and HPV11-E7/TEG; see Fig. 6E) are not as pronounced (20–30% when normalized to β-actin) as the MHC I reduction in the cell lysates of stable transfectants (Figs. 2B, 3C, and 4C) as the HPV-E7 variants act only on the transfected cell population in the transient system, which covers 50–60%. Taken together, residue characteristics at position 88 seem to have the most critical influence on the ability of HPV-E7 to down-regulate MHC I. Nevertheless, it should be noted that the different properties of HPV11- and HPV16-E7 are not exclusively confined to a single residue variation but, rather, depend on the joint influence of different C-terminal amino acids.
FIGURE 5.
Analysis of chimeric variants HPV11-E7/E and HPV16-E7/Q. A, shown is a schematic overview of chimeric variants HPV11-E7/E and HPV16-E7/Q. Polypeptide regions of HPV11-E7 are indicated in white, whereas the corresponding regions of HPV16-E7 are shown in black. Regions CR1, CR2, and CR3 are underlined. B, expression of HPV11-E7/E and HPV16-E7/Q in transiently transfected HEK-293T cells (left panel) is shown. Cell lysates of the transfectants were analyzed by Western blot as described above. Asterisks indicate minor bands detected with antibody to GFP. C, surface expression of MHC I was assessed by flow cytometry using the mAb W6/32. Data are representative of three independent experiments of which one study is shown in D. D, 24 h after transient transfection GFP- and GFP-tagged HPV-E7-positive HEK-293T cells were determined by flow cytometry based on their GFP-fluorescent intensity (forward scatter (FSC) versus GFP-fluorescence dot plot; left panel). The histogram plots of MHC I surface expression (right panel) were gated on the population non-transfected (NT)/GFP-negative (filled gray histogram) and GFP-positive HEK-293T cells (filled black histograms).
FIGURE 6.
Analysis of chimeric variants HPV11-E7/E, HPV11-E7/T, HPV11-E7/TE, HPV11-E7/G, HPV11-E7/EG, and HPV11-E7/TEG. A, shown is a schematic overview of chimeric variants HPV11-E7/E, HPV11-E7/T, HPV11-E7/TE, HPV11-E7/G, HPV11-E7/EG, and HPV11-E7/TEG. Polypeptide regions of HPV11-E7 are indicated in white, and the corresponding regions of HPV16-E7 are shown in black. Regions CR1, CR2, and CR3 are underlined. B, shown is expression of HPV11-E7 variants in transiently transfected HEK-293T cells (left panel). Cell lysates of the transfectants were analyzed by Western blot as described above. Asterisks indicate minor bands detected with antibody to GFP. C, surface expression of MHC I was assessed by flow cytometry using the mAb W6/32. Data are representative of three independent experiments, of which one study is shown in D. D, 24 h after transient transfection, GFP- and GFP-tagged HPV-E7-positive HEK-293T cells were determined by flow cytometry based on their GFP fluorescent intensity (forward scatter (FSC) versus GFP fluorescence dot plot; left panels). The histogram plots of MHC I surface expression were gated on the population non-transfected (NT)/GFP-negative (filled gray histogram) and GFP-positive HEK-293T cells (filled black histograms) (right panels). E, steady-state expression of MHC I was analyzed by Western blot probed with mAb 3B10.7 (upper right panel). Anti-β-actin staining served as the internal control for equal protein loading. MHC I and β-actin signals were quantitated by densitometric scanning. Four different exposures of the immunoblot were taken to ensure linearity, and the obtained MHC I signals were normalized to the corresponding β-actin signals (see the histogram below the immunoblot). One representative exposure of the Western blot is shown in E. F, HPV16-E7 and C-terminal mutants of HPV11-E7 are associated with the MHC I promoter. Sonicated chromatin from transfected HEK-293T cells were immunoprecipitated with anti-GFP and anti-HDAC1 Abs. Samples were then amplified by PCR using human MHC I promoter-specific primers, and the PCR products were resolved on a 2% agarose gel. The depicted image is a representative of multiple independent experiments.
Our own experiments and that of others suggest that in HPV16-E7-expressing cells MHC I is affected on the level of mRNA synthesis (Fig. 2B and Ref. 14). Furthermore, studies of Li et al. (15) provided evidence that HPV16-E7 recruits HDACs (e.g. HDAC1) to the MHC I promoter and, thereby, suppresses MHC I transcription. To determine whether and to what extent HPV16-E7 and the different HPV11-E7 variants diverge in terms of MHC I promoter interaction and HDAC recruitment, we performed ChIP assays by incubating Abs directed to GFP-tagged HPV-E7 and HDAC1 with supernatants of sonicated cross-linked chromatin from the different transfectants. Immunoisolated chromatin fragments were analyzed by PCR using specific primers for the human MHC I promoter (Fig. 6F). In line with our results depicted in Figs. 5C and 6, C and E, we found that HPV16-E7, HPV11-E7/G, HPV11-E7/EG, and HPV11-E7/TEG reside in the MHC I promoter complex and recruit also HDAC1, as indicated by the PCR signals obtained for co-isolated chromatin fragments. In contrast, no such DNA amplification was observed for the transfectants expressing HPV11-E7, HPV11-E7/E, HPV11-E7/T, or HPV11-E7/TE, suggesting that these HPV-E7 variants do apparently not interact with the MHC I promoter.
In conclusion, our findings from the ChIP analysis are consistent with the results obtained by flow cytometry and Western blot (Fig. 6, C and E), suggesting that the observed effect on MHC I expression in the context of mutant HPV11-E7/G (N88G) is increased in the presence of mutations Q78T and/or Q80E, which alone have apparently no detectable influence. Thus, it is tempting to speculate that both act in a supportive manner on N88G and that the three residues build a minimal functional unit within the C terminus of HPV16-E7 critically involved in the HDAC-mediated down-regulation of MHC I.
DISCUSSION
Because dysregulation of MHC I surface expression has been observed in up to 90% of cervical cancers patients (37, 38), the ability of HPV16-E7 to down-regulate MHC I expression and surface presentation has been discussed as a critical feature within the multistep pathway to cervical carcinogenesis (18). E7 proteins from high and low risk HPV types have been shown to differ in their influence on MHC I expression (14). Thus, E7 from high risk HPV types has been shown to repress the MHC I gene promoter (e.g. HPV16-E7) (14), whereas no such MHC I down-regulation has been found for E7 from low risk HPV types (e.g. HPV11-E7) (18). As virus infections are predominantly controlled by MHC I-restricted T cells, this might contribute to differential effects on the anti-viral immune response. However, it is not clear what molecular properties of high and low risk HPV-E7 are responsible for differences in MHC I immune evasion. To address this, we constructed GFP-tagged E7 variants of HPV16 and HPV11 (Fig. 1). Both HPV-E7 constructs could be stably expressed in HEK-293 cells (Fig. 1B) and, in agreement with previous findings, showed different properties concerning pRb binding (Fig. 1B and Ref. 19) as well as a different influence on MHC I expression (Fig. 2 and Ref. 14). Thus, HPV16-E7 caused a significant reduction of MHC I synthesis and surface expression, whereas in contrast no such dysregulation was observed for HPV11-E7 (Fig. 2A and 2B). Our experiments with chimeric HPV-E7 constructs containing the N-terminal half of HPV16-E7 and the C-terminal region of HPV11-E7 and vice versa (Figs. 3 and 4) demonstrate that C-terminal sequence properties of HPV-E7 control down-regulation of MHC I, whereas the different pRb binding affinities are determined by the N-terminal regions. This indicates that the different effects of HPV11- and HPV16-E7 on MHC I are not linked to pRb-dependent functions of HPV-E7, which might play a role in the suppression of TNF-α/NF-κB-mediated (39) MHC I induction (16). Although it has been demonstrated that pRb is implicated in the control of the NF-κB pathway (39), it was also suggested that HPV-E7 might directly interfere with NF-κB functions by targeting the IκB kinase complex (40). As for the structurally and functionally related adenovirus 12 E1A protein (41, 42), a dual strategy of HPV16-E7 might provide a kind of fail-safe mechanism to ensure that surface MHC I remains down-regulated under different physiological conditions. However, the molecular necessities for the interaction between HPV-E7 and the IκB kinase complex are not known and will require further experimental clarification.
Viruses, which are not recognized by the immune system, may have a substantial selection advantage in view of replication und spreading. This is especially true for viral pathogens that cause a weak stimulus of the immune system, for example, by subversion of MHC I or by their propagation in immune privileged tissues. On the basis of our results one could ask, Why do HPV11- and HPV16-E7 differ in their ability to down-regulate MHC I antigen presentation? Unlike HPV16, which is a major causative agent for cervical carcinomas, HPV11 is most commonly associated with benign lesions such as genital warts and with laryngeal papillomatosis with low oncogenic potential (43). Most importantly, it has recently been shown that the laryngeal epithelium of humans is characterized by significant low MHC I expression (44) and, thus, is much less susceptible to the immune control, as it appears to have a reduced ability to present MHC I-restricted antigens. An interesting idea is that HPV11 does not essentially require E7-mediated down-regulation of MHC I to escape the immune surveillance during replication. Thus, in contrast to HPV16, HPV11 might evade (rather than subvert) the cellular immune system by its tropism for immune privileged tissues.
Our experiments in Fig. 2 showed that HPV16-E7 causes a ≥50% reduction of MHC I presentation, which is in the range of HPV-E7-mediated MHC I down-regulation sufficient to increase susceptibility to natural killer (NK) cells (20) normally prevented from action by inhibitory signals provided through self-MHC I ligands (45). This strategy is advantageous in that it permits detection of virus-infected cells that have down-regulated MHC I and impaired cytotoxic T cell recognition. Nevertheless, previous studies provide evidence that HPV16 might also be able to escape immune detection by NK cells (46, 47) via subversion mechanisms controlled by HPV16-E5. HPV16-E5 interacts with the 16-kDa pore-forming subunit of the vacuolar H(+)-ATPase (V-ATPase). It inhibits endosome acidification (48) and interferes with the pH homeostasis of the Golgi complex (49). It has been proposed that HPV16-E5-mediated alkalinization of the Golgi apparatus and endosomes leads to disruption of the exo- and endocytic trafficking, including transport of the MHC I (50). Thus, HPV16-E5 sequesters MHC I in the Golgi and causes a 50% reduction in MHC I presentation (46). Most interestingly, it has been suggested that HPV16-E5 affects surface expression of MHC I in a highly selective manner to down-modulate antigen-presenting MHC I (for evasion of a cytotoxic T cell-mediated response) while simultaneously maintaining critical inhibitory NK cell ligands (for evasion of an NK cell-mediated response) (50). Our own studies4 suggest that HPV16-E5 also blocks endosomal recycling of MHC I and thereby increases the half-lives of pre-existing self-peptide MHC I complexes at the plasma membrane. Furthermore, Suprynowicz et al. (47) speculated that HPV16-E5 might also impair the ability to form immune synapses between target and NK cells by preventing the rearrangement of lipid raft-associated antigens. Hence, it is tempting to assume that HPV16-E7 and -E5 build a cooperative alliance in the dysregulation of MHC I to undermine immune recognition of virus-infected cells by cytotoxic T cells as well as NK cells. Thus, HPV16-E5 and -E7 help the establishment of a successful infection not only through cell transformation but also by controlling antigen presentation. It has been discussed that there is a correlation between their transforming strength and the extent to which they interfere with MHC I (18). In contrast to HPV16-E5, HPV11-E5 seems to be primarily localized in the nucleus and/or the nuclear membrane (51). Although HPV11-E5 might also interact with the vacuolar H(+)-ATPase (52), its influence on antigen presentation has been not analyzed so far. In view of the idea that HPV16-E7 and -E5 may act as allies in MHC I evasion, it would be interesting to see whether and to what extent HPV11- and HPV16-E5 differ in their ability to affect intracellular transport of MHC I and/or NK inhibitory ligands and thereby reflect the observed functional differences between HPV11- and HPV16-E7.
To identify the C-terminal sequence region of HPV16-E7 affecting MHC I expression and to better understand the different abilities of HPV11- and HPV16-E7 to subvert antigen presentation, we created a series of different chimeric HPV-E7 variants by systematically exchanging sequences and amino acids between HPV11- and HPV16-E7. Analysis of HEK-293 transfectants expressing the chimeric constructs (Fig. 4) demonstrated that the crucial determinant that controls the contrasting behavior of HPV11- and HPV16-E7 is located within the C-terminal 22-amino acid-long region of the zinc finger domain. This sequence region has been shown to bind to HDACs through a “linker protein” called Mi2β (21). Mi2β is a member of the nucleosome remodeling and histone deacetylation (NURD) complex that has the ability to modify chromatin structure through deacetylation of histones (53). In the last years it has become increasingly clear that cellular transcription is regulated at the chromatin level. Histone modifications, including acetylation, determine transcription factor accessibility (54–57) and regulate gene expression (54). Histone acetylation produces a weakened interaction between histones and DNA, leading to a more open and dynamic chromatin structure (56) that is conducive to transcription. Conversely, histone deacetylation leads to a tightened compact chromatin structure that suppresses transcription (56). It has been suggested that binding of HPV-E7 to the Mi2β subunit of the NURD complex results in the deregulation of different host genes, including those that play important roles in the anti-viral immune response (15, 58). Studies of Li et al. (15, 16) provide evidence that HPV16-E7 mediates MHC I down-regulation via HDAC-dependent chromatin repression.
By the use of loss- and gain-of-function mutants (Figs. 5 and 6) we were able to identify three distinct residue variations between HPV11- and HPV16-E7 at sequence positions 78, 80, and 88 that are critical for the HPV-E7-mediated suppression of MHC I (Figs. 5 and 6) and seem to control HPV-E7/HDAC-interaction with the MHC I promoter (Fig. 6). These residues are in proximity to conserved amino acids (leucine residues 82 and 83, cysteine residue 91) that have been previously shown to be essential for HDAC binding of HPV16-E7 (21). In view of the fact that these “HDAC binding residues” do not differ between HPV11- and HPV16-E7 (see Fig. 1A), it is tempting to speculate that they might represent an important molecular prerequisite for HDAC-interaction but that adjacent residues clearly affect the competence of HPV-E7 to suppress MHC I expression. Our studies suggest that the residue properties at sequence position 88 seem to have the most critical influence on the ability of HPV-E7 to modulate MHC I expression (Fig. 6). Most interestingly, this sequence position is occupied by asparagine (or glutamine) in low risk HPV-E7 proteins (e.g. HPV11-, HPV6b-, HPV1a-, HPV5-, and HPV8-E7), whereas glycine (or serine) is present at the corresponding sites of HPV16- and HPV18-E7 as well as other high risk HPV-E7 proteins (e.g. HPV31-E7 and HPV45-E7). Moreover, in the case of position 78, glutamine or glutamate is present in low risk HPV11-, HPV6-, and HPV1a-E7, whereas threonine or alanine can be found in high risk HPV16-, HPV18-, and HPV45-E7. Results from structure analysis of different HPV-E7 proteins provide evidence that the respective residues are both surface-exposed (59, 60). It is, thus, conceivable that the presence of bulky polar amino acids at positions 78 and 88 in HPV11-E7 could sterically impair its ability to perform protein interactions required to bind HDACs and/or to gain access to the MHC I promoter. In contrast, residues corresponding to position 80 point inside HPV-E7 (59, 60) and, thus, may contribute to the observed mutational effects on HPV11 and HPV16 (Figs. 5 and 6) by a rather conformational constraint on the C terminus. Hence, our current interest is focused on the functional role of the identified amino acids as well as on the exact molecular mechanisms involved in the recruitment of HPV-E7 and HDACs to the MHC I promoter.
Our own studies (Figs. 2D and 6F) and that of others (15, 16) provide evidence that HPV16-E7-mediated down-regulation of MHC I is caused by HDAC-dependent chromatin repression. However, in view of the experimental observation that TSA treatment of the HPV16-E7-transfectants did not lead to a full reconstitution of MHC I surface expression (Fig. 2D), it might be that additional mechanisms are co-involved in the observed suppression of MHC I. In this context it is interesting to note that HPV16-E7 is able to interact with different AP-1 (activator protein-1) transcription factors (e.g. c-Jun, JunB, JunD, and c-Fos) (61) and trans-activates c-Jun-induced transcription from responsive promoters. Most interestingly, the interaction with c-Jun has been shown to involve the zinc finger domain of HPV-E7, whereas pRb does not seem to be required for the complex formation between HPV-E7 and c-Jun. It was discussed that binding of c-Jun by HPV-E7 alters the tertiary structure of the c-Jun DNA binding region or that HPV-E7 may modulate the ability of c-Jun to form complexes with cellular proteins, which are known to heterodimerize with c-Jun. By modulating the affinity of c-Jun for any of these factors, HPV-E7 might activate the transcription of genes containing specific promoter sequences. Such alterations could affect c-Jun DNA binding activity or may target c-Jun to different DNA recognition sequences. Because, it has been shown that c-Jun acts as a negative regulator of MHC I transcription (62), it is possible that trans-activation of c-Jun by HPV16-E7 contributes to dysregulation of MHC I expression and surface presentation.
Based on in vitro experiments, it has been postulated by one study that HPV11- and HPV16-E7 inhibits ATP-dependent peptide transport by complex formation with TAP1 (63). Nevertheless, it is yet unclear whether HPV-E7-mediated TAP-inhibition also happens in intact cells and to what extent the proposed HPV-E7 effect is restricted by the host genetic background (64) and the polymorphism of TAP1 (65). In contrast to Vambutas and et al. (63), we did not observe any physical complex formation between HPV11-E7 (or HPV16-E7) and TAP chains (data not shown), suggesting that in the cell transfectants used in our experiments none of the two HPV-E7 proteins forms an inhibitory complex with TAP or the PLC. Because nothing is known about the sequence region and the molecular mechanism by which HPV11-E7 might impart inhibition of peptide translocation, further detailed studies are necessary to clarify the ability of HPV-E7 to impair peptide transporter function.
Our results in Fig. 2, B and C, demonstrated that expression of HPV11-E7 or HPV16-E7 has no detectable influence on the cellular synthesis of TAP1, TAP2, and tapasin as well as the intracellular transport and maturation of MHC I, which is known to depend on the ATP-dependent peptide supply by TAP. Thus, in contrast to HPV18-E7, which has been reported to repress promoter activity of MHC I as well as TAP1 (14), neither HPV11-E7 nor HPV16-E7 seem to dysregulate synthesis of accessory components associated with MHC I antigen presentation. The ability of HPV18-E7 to impede MHC I and TAP might confer an increased oncogenic potential on HPV18 compared with HPV16, which only represses MHC I transcription. Indeed, although HPV16 is the most commonly detected virus type in cervical carcinoma, HPV18 is associated with more-advanced tumors (66), increased risk for progression to malignancy, poorer prognosis in early-stage cervical cancer (67), and rapidly progressing tumors (68).
In conclusion, our results on the different gain-of-function mutants of low risk “non-active“ HPV11-E7 enabled us to discern the functional importance of individual residues in high risk “active” HPV16-E7 for the dysregulation of MHC I. For the first time we identified three key residues within the C terminus of HPV-E7 at positions 78, 80, and 88 that seem to build a minimal functional unit that is essentially required for the HPV16-E7/HDAC-mediated down-regulation of MHC I expression. In view of the current hypothesis that HPVs associated with cancer have an enhanced ability to avoid the immune response compared with their benign-disease-causing counterparts, our present findings provide important new insights into the molecular requirements of high risk HPV-E7 for the subversion of the MHC I pathway in virus-infected cells.
Acknowledgments
We are indebted to Dr. Frank Stubenrauch for the kind donation of the DNA of HPV11-E7 and HPV16-E7. We thank Madeleine Queiser, Philipp Steimle, and Svenja Fengler for technical assistance. We further thank Drs. Gregor Meyers and Achim Rziha for helpful discussions and for critically reading the manuscript.
This work was supported by the Friedrich-Loeffler-Institut.
A. Mueller-Schickert, C. Heller, T. Weisser, E. Rufer, A. Hoh, R. M. Leonhardt, and M. R. Knittler, unpublished data.
- MHC I
- major histocompatibility complex class I
- HPV
- human Papillomavirus
- TAP
- transporter associated with antigen processing
- PLC
- peptide-loading complex
- TSA
- trichostatin A
- HDAC
- histone deacetylase
- mAb
- monoclonal antibody
- pRb
- retinoblastoma protein
- NK
- natural killer.
REFERENCES
- 1. Williams D. B., Vassilakos A., Suh W. K. (1996) Trends Cell Biol. 6, 267–273 [DOI] [PubMed] [Google Scholar]
- 2. Elliott T. (1997) Immunol. Today 18, 375–379 [DOI] [PubMed] [Google Scholar]
- 3. Garcia-Lora A., Algarra I., Garrido F. (2003) J. Cell. Physiol. 195, 346–355 [DOI] [PubMed] [Google Scholar]
- 4. Hewitt E. W. (2003) Immunology 110, 163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heemels M. T., Ploegh H. (1995) Annu. Rev. Biochem. 64, 463–491 [DOI] [PubMed] [Google Scholar]
- 6. Stern P. L. (1996) Adv. Cancer Res. 69, 175–211 [PubMed] [Google Scholar]
- 7. Greenfield I., Nickerson J., Penman S., Stanley M. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 11217–11221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Phelps W. C., Yee C. L., Münger K., Howley P. M. (1988) Cell 53, 539–547 [DOI] [PubMed] [Google Scholar]
- 9. Nishida M., Miyamoto S., Kato H., Miwa T., Imamura T., Miwa K., Yasumoto S., Barrett J. C., Wake N. (1995) Mol. Carcinog. 13, 157–165 [DOI] [PubMed] [Google Scholar]
- 10. Hiraiwa A., Kiyono T., Suzuki S., Ohashi M., Ishibashi M. (1996) Virus Genes 12, 27–35 [DOI] [PubMed] [Google Scholar]
- 11. Armstrong D. J., Roman A. (1997) Virology 239, 238–246 [DOI] [PubMed] [Google Scholar]
- 12. Massimi P., Banks L. (1997) Virology 227, 255–259 [DOI] [PubMed] [Google Scholar]
- 13. zur Hausen H. (2000) J. Natl. Cancer Inst. 92, 690–698 [DOI] [PubMed] [Google Scholar]
- 14. Georgopoulos N. T., Proffitt J. L., Blair G. E. (2000) Oncogene 19, 4930–4935 [DOI] [PubMed] [Google Scholar]
- 15. Li H., Ou X., Xiong J., Wang T. (2006) Biochem. Biophys. Res. Commun. 349, 1315–1321 [DOI] [PubMed] [Google Scholar]
- 16. Li H., Zhan T., Li C., Liu M., Wang Q. K. (2009) Biochem. Biophys. Res. Commun. 388, 383–388 [DOI] [PubMed] [Google Scholar]
- 17. Duensing A., Chin A., Wang L., Kuan S. F., Duensing S. (2008) Virology 372, 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Brien P. M., Campo M. S. (2003) Trends Microbiol. 11, 300–305 [DOI] [PubMed] [Google Scholar]
- 19. Münger K., Yee C. L., Phelps W. C., Pietenpol J. A., Moses H. L., Howley P. M. (1991) J. Virol. 65, 3943–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bottley G., Watherston O. G., Hiew Y. L., Norrild B., Cook G. P., Blair G. E. (2008) Oncogene 27, 1794–1799 [DOI] [PubMed] [Google Scholar]
- 21. Brehm A., Nielsen S. J., Miska E. A., McCance D. J., Reid J. L., Bannister A. J., Kouzarides T. (1999) EMBO J. 18, 2449–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dulat H. J., von Grumbkow C., Baars W., Schröder N., Wonigeit K., Schwinzer R. (2001) Eur. J. Immunol. 31, 2217–2226 [DOI] [PubMed] [Google Scholar]
- 23. Brodsky F. M., Lem L., Solache A., Bennett E. M. (1999) Immunol. Rev. 168, 199–215 [DOI] [PubMed] [Google Scholar]
- 24. Lutz P. M., Cresswell P. (1987) Immunogenetics 25, 228–233 [DOI] [PubMed] [Google Scholar]
- 25. Meyer T. H., van Endert P. M., Uebel S., Ehring B., Tampé R. (1994) FEBS Lett. 351, 443–447 [DOI] [PubMed] [Google Scholar]
- 26. van Endert P. M., Tampé R., Meyer T. H., Tisch R., Bach J. F., McDevitt H. O. (1994) Immunity 1, 491–500 [DOI] [PubMed] [Google Scholar]
- 27. Peaper D. R., Cresswell P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10477–10482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mora-García, Mde L., Duenas-González A., Hernández-Montes J., De la Cruz-Hernández E., Pérez-Cárdenas E., Weiss-Steider B., Santiago-Osorio E., Ortíz-Navarrete V. F., Rosales V. H., Cantú D., Lizano-Soberón M., Rojo-Aguilar M. P., Monroy-García A. (2006) J. Transl. Med. 4, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Androphy E. J., Hubbert N. L., Schiller J. T., Lowy D. R. (1987) EMBO J. 6, 989–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilczynski S. P., Bergen S., Walker J., Liao S. Y., Pearlman L. F. (1988) Hum. Pathol. 19, 697–704 [DOI] [PubMed] [Google Scholar]
- 31. Hwang S. G., Lee D., Kim J., Seo T., Choe J. (2002) J. Biol. Chem. 277, 2923–2930 [DOI] [PubMed] [Google Scholar]
- 32. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) The Proteomics Protocols Handbook, Humana Press (Walker J. M. ed) pp. 571–607, Humana Press Inc., Totowa, NJ [Google Scholar]
- 33. Imamura T., Konishi K., Konishi K. (2006) J. Pept. Sci. 12, 403–411 [DOI] [PubMed] [Google Scholar]
- 34. de Jong W. W., Zweers A., Cohen L. H. (1978) Biochem. Biophys. Res. Commun. 82, 532–539 [DOI] [PubMed] [Google Scholar]
- 35. Kanodia S., Fahey L. M., Kast W. M. (2007) Curr. Cancer Drug Targets 7, 79–89 [DOI] [PubMed] [Google Scholar]
- 36. Vanhaecke T., Henkens T., Kass G. E., Rogiers V. (2004) Biochem. Pharmacol. 68, 753–760 [DOI] [PubMed] [Google Scholar]
- 37. Keating P. J., Cromme F. V., Duggan-Keen M., Snijders P. J., Walboomers J. M., Hunter R. D., Dyer P. A., Stern P. L. (1995) Br. J. Cancer 72, 405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koopman L. A., Corver W. E., van der Slik A. R., Giphart M. J., Fleuren G. J. (2000) J. Exp. Med. 191, 961–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garcia M. A., Gallego P., Campagna M., González-Santamaría J., Martínez G., Marcos-Villar L., Vidal A., Esteban M., Rivas C. (2009) PLoS One 4, e6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spitkovsky D., Hehner S. P., Hofmann T. G., Möller A., Schmitz M. L. (2002) J. Biol. Chem. 277, 25576–25582 [DOI] [PubMed] [Google Scholar]
- 41. Zhao B., Ricciardi R. P. (2006) Virology 352, 338–344 [DOI] [PubMed] [Google Scholar]
- 42. Guan H., Jiao J., Ricciardi R. P. (2008) J. Virol. 82, 40–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gissmann L., Diehl V., Schultz-Coulon H. J., zur Hausen H. (1982) J. Virol. 44, 393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hobbs C. G., Rees L. E., Heyderman R. S., Birchall M. A., Bailey M. (2006) Clin. Exp. Immunol. 145, 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kärre K. (1997) Immunol. Rev. 155, 5–9 [DOI] [PubMed] [Google Scholar]
- 46. Ashrafi G. H., Brown D. R., Fife K. H., Campo M. S. (2006) Virus. Res. 120, 208–211 [DOI] [PubMed] [Google Scholar]
- 47. Suprynowicz F. A., Disbrow G. L., Krawczyk E., Simic V., Lantzky K., Schlegel R. (2008) Oncogene 27, 1071–1078 [DOI] [PubMed] [Google Scholar]
- 48. Straight S. W., Herman B., McCance D. J. (1995) J. Virol. 69, 3185–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schapiro F., Sparkowski J., Adduci A., Suprynowicz F., Schlegel R., Grinstein S. (2000) J. Cell. Biol. 148, 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ashrafi G. H., Haghshenas M. R., Marchetti B., O'Brien P. M., Campo M. S. (2005) Int. J. Cancer 113, 276–283 [DOI] [PubMed] [Google Scholar]
- 51. Chen S. L., Mounts P. (1990) J. Virol. 64, 3226–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen S. L., Tsai T. Z., Han C. P., Tsao Y. P. (1996) J. Virol. 70, 3502–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Y., LeRoy G., Seelig H. P., Lane W. S., Reinberg D. (1998) Cell 95, 279–289 [DOI] [PubMed] [Google Scholar]
- 54. Zhang W. H., Srihari R., Day R. N., Schaufele F. (2001) J. Biol. Chem. 276, 40373–40376 [DOI] [PubMed] [Google Scholar]
- 55. Li J., Lin Q., Wang W., Wade P., Wong J. (2002) Genes Dev. 16, 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Orlando V., Jones K. A. (2002) Genes Dev. 16, 2039–2044 [DOI] [PubMed] [Google Scholar]
- 57. Yamamoto Y., Verma U. N., Prajapati S., Kwak Y. T., Gaynor R. B. (2003) Nature 423, 655–659 [DOI] [PubMed] [Google Scholar]
- 58. Park J. S., Kim E. J., Kwon H. J., Hwang E. S., Namkoong S. E., Um S. J. (2000) J. Biol. Chem. 275, 6764–6769 [DOI] [PubMed] [Google Scholar]
- 59. Liu X., Clements A., Zhao K., Marmorstein R. (2006) J. Biol. Chem. 281, 578–586 [DOI] [PubMed] [Google Scholar]
- 60. Ohlenschläger O., Seiboth T., Zengerling H., Briese L., Marchanka A., Ramachandran R., Baum M., Korbas M., Meyer-Klaucke W., Dürst M., Görlach M. (2006) Oncogene 25, 5953–5959 [DOI] [PubMed] [Google Scholar]
- 61. Antinore M. J., Birrer M. J., Patel D., Nader L., McCance D. J. (1996) EMBO J. 15, 1950–1960 [PMC free article] [PubMed] [Google Scholar]
- 62. Howcroft T. K., Richardson J. C., Singer D. S. (1993) EMBO J. 12, 3163–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vambutas A., DeVoti J., Pinn W., Steinberg B. M., Bonagura V. R. (2001) Clin. Immunol. 101, 94–99 [DOI] [PubMed] [Google Scholar]
- 64. Bonagura V. R., Vambutas A., DeVoti J. A., Rosenthal D. W., Steinberg B. M., Abramson A. L., Shikowitz M. J., Gjertson D. W., Reed E. F. (2004) Hum. Immunol. 65, 773–782 [DOI] [PubMed] [Google Scholar]
- 65. Vambutas A., Bonagura V. R., Reed E. F., Abramson A. L., Mullooly V., DeVoti J., Gjertson D. W., Steinberg B. M. (2004) J. Infect. Dis. 189, 871–879 [DOI] [PubMed] [Google Scholar]
- 66. Arends M. J., Donaldson Y. K., Duvall E., Wyllie A. H., Bird C. C. (1993) Hum. Pathol. 24, 432–437 [DOI] [PubMed] [Google Scholar]
- 67. Burger R. A., Monk B. J., Kurosaki T., Anton-Culver H., Vasilev S. A., Berman M. L., Wilczynski S. P. (1996) J. Natl. Cancer Inst. 88, 1361–1368 [DOI] [PubMed] [Google Scholar]
- 68. Cox J. T. (1995) Baillieres. Clin. Obstet. Gynaecol. 9, 1–37 [DOI] [PubMed] [Google Scholar]