Abstract
The widespread, long sought-after bacterial aerobic phenylalanine/phenylacetate catabolic pathway has recently been elucidated. It proceeds via coenzyme A (CoA) thioesters and involves the epoxidation of the aromatic ring of phenylacetyl-CoA, subsequent isomerization to an uncommon seven-membered C-O-heterocycle (oxepin-CoA), and non-oxygenolytic ring cleavage. Here we characterize the hydrolytic oxepin-CoA ring cleavage catalyzed by the bifunctional fusion protein PaaZ. The enzyme consists of a C-terminal (R)-specific enoyl-CoA hydratase domain (formerly MaoC) that cleaves the ring and produces a highly reactive aldehyde and an N-terminal NADP+-dependent aldehyde dehydrogenase domain that oxidizes the aldehyde to 3-oxo-5,6-dehydrosuberyl-CoA. In many phenylacetate-utilizing bacteria, the genes for the pathway exist in a cluster that contains an NAD+-dependent aldehyde dehydrogenase in place of PaaZ, whereas the aldehyde-producing hydratase is encoded outside of the cluster. If not oxidized immediately, the reactive aldehyde condenses intramolecularly to a stable cyclic derivative that is largely prevented by PaaZ fusion in vivo. Interestingly, the derivative likely serves as the starting material for the synthesis of antibiotics (e.g. tropodithietic acid) and other tropone/tropolone related compounds as well as for ω-cycloheptyl fatty acids. Apparently, bacteria made a virtue out of the necessity of disposing the dead-end product with ring hydrolysis as a metabolic branching point.
Keywords: Antibiotics, Bacterial Metabolism, Coenzyme A, Enzyme Catalysis, NMR, MaoC, PaaZ, Tropone, Phenylacetic Acid, Tropodithietic Acid
Introduction
Aromatics like phenylacetic acid constitute the second most abundant class of natural organic compounds that serve as substrates mainly for microorganisms. Oxygen availability is key to how bacteria utilize such inert substrates (1). Under aerobic conditions oxygen is used to hydroxylate and cleave the ring (2, 3). In contrast, under anaerobic conditions the inert substrates become activated to CoA-thioesters followed by shortening of the side chain and energy-driven ring reduction; furthermore, ring cleavage occurs hydrolytically rather than by oxygenation. This strategy also applies to anaerobic phenylacetate catabolism (Ref. 4 and literature cited therein) (Fig. 1A).
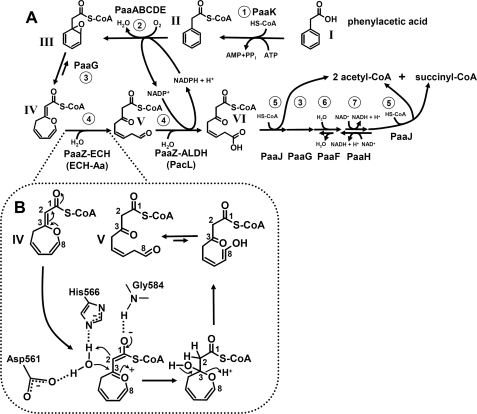
FIGURE 1.
Aerobic phenylacetate catabolic pathway. A, reactions and intermediates of the pathway and proposed enzyme names are shown. 1, phenylacetate-CoA ligase (AMP forming); 2, ring-1,2-phenylacetyl-CoA epoxidase (NADPH); 3, ring-1,2-epoxyphenylacetyl-CoA isomerase (oxepin-CoA forming), postulated 3,4-dehydroadipyl-CoA isomerase; 4, oxepin-CoA hydrolase/3-oxo-5,6-dehydrosuberyl-CoA semialdehyde dehydrogenase (NADP+); 5, 3-oxoadipyl-CoA/3-oxo-5,6-dehydrosuberyl-CoA thiolase; 6, 2,3-dehydroadipyl-CoA hydratase; 7, 3-hydroxyadipyl-CoA dehydrogenase (NAD+) (probably (S)-3-specific). Later steps resemble β-oxidation and lead to acetyl-CoA and succinyl-CoA. Compounds: I, phenylacetate; II, phenylacetyl-CoA; III, ring-1,2-epoxyphenylacetyl-CoA; IV, 2-oxepin-2(3H)-ylideneacetyl-CoA (oxepin-CoA); V, 3-oxo-5,6-dehydrosuberyl-CoA semialdehyde, VI, 3-oxo-5,6-dehydrosuberyl-CoA. B, shown is the mechanism of hydrolytic ring fission by PaaZ-ECH or ECH-Aa. Proposed hydrolytic ring-cleavage of oxepin-CoA (IV) by PaaZ-ECH is via an acid-base mechanism. A conserved histidine and an aspartic acid (His-566 and Asp-561 in E. coli) may function as catalytic residues, whereas a conserved glycine (Gly-584) may form an oxyanion hole. This step is also catalyzed by ECH-Aa from A. aromaticum.
Phenylacetate (I) is a key intermediate in the degradation of various substrates like phenylalanine, lignin-related aromatic compounds, or environmental contaminants (5, 6). The first studies from more than 20 years ago reported the induction of a phenylacetate-CoA ligase under aerobic conditions in Pseudomonas putida, suggesting an unconventional strategy for aerobic phenylacetic acid (Paa)2 degradation (7, 8). A total of 14 genes in 3 transcriptional units were identified in the corresponding paa gene cluster (9). However, the underlying biochemical conversions remained obscure until they were recently elucidated in Escherichia coli K12 and Pseudomonas sp. strain Y2 (10). This novel metabolic strategy involves the usage of oxygen as well as CoA-thioester intermediates throughout the pathway, hence, showing typical features of aerobic as well as anaerobic strategies. Phenylacetyl-CoA (II) becomes epoxidized by PaaABC(D)E, a new type of bacterial multicomponent monooxygenase. The reactive and unstable epoxide intermediate (III) is further isomerized by enoyl-CoA isomerase PaaG to an unorthodox C-O heterocyclic enolether (IV), 2-oxepin-2(3H)-ylideneacetyl-CoA (oxepin-CoA). This is followed by PaaZ-mediated ring cleavage, and β-oxidation-like steps finally yield central intermediates (acetyl-CoA and succinyl-CoA) (Fig. 1, A and B) (10). Interestingly, a similar route applies to aerobic benzoate metabolism in numerous bacteria either as the only pathway or as an additional strategy under certain conditions (11). In contrast to the case of benzoate metabolism, there are no hints for alternative aerobic phenylacetate degradation strategies in bacteria that may also be reflected by the large percentage (>16%) of bacteria harboring key genes of this pathway (10).
Here we investigated the intriguing ring-cleavage reaction catalyzed by the fusion protein PaaZ that functions as an oxepin-CoA hydrolase/3-oxo-5,6-dehydrosuberyl-CoA semialdehyde dehydrogenase and is important for the removal of toxic early pathway intermediates (10). It consists of a C-terminal (R)-specific Hotdog-fold enoyl-CoA hydratase domain (PaaZ-ECH) and an N-terminal aldehyde dehydrogenase domain (PaaZ-ALDH) (10, 12). We addressed the question of how 50% of bacteria that utilize the pathway lack a gene for the multifunctional PaaZ in their paa gene cluster. Instead, a gene is present in the cluster whose product demonstrates aldehyde dehydrogenase activity but lacks hydratase activity, e.g. PacL from Aromatoleum aromaticum. To elucidate the unprecedented ring-cleavage reaction, we isolated and characterized recombinant PaaZ, performed 18O-labeling studies, and analyzed the products by mass spectrometry (MS). Furthermore, we identified by 13C NMR the spontaneous breakdown product of the reactive (PaaZ-ECH produced) aldehyde intermediate. A literature search indicated that this pathway is used by other microorganisms for the synthesis of various primary and secondary metabolites and signaling compounds.
EXPERIMENTAL PROCEDURES
Materials
Oxygen-18O (normalized, > 97 atom%) and water-18O (normalized with respect to hydrogen, > 97 atom%) were obtained from Campro Scientific (Berlin, Germany). [1-14C]Phenylacetic acid and l-[U-14C]phenylalanine were purchased from American Radiolabeled Chemicals (Cologne, Germany) and GE Healthcare, respectively. l-[U-13C,15N]phenylalanine was obtained from Spectra (Andover, MA). Other chemicals were obtained from Amersham Biosciences, Fluka (Neu-Ulm, Germany), Sigma, Merck, Serva (Heidelberg, Germany), or Roth (Karlsruhe, Germany). Biochemicals were from Roche Diagnostics, Applichem (Darmstadt, Germany), or Gerbu (Craiberg, Germany). Materials for cloning and expression were purchased from MBI Fermentas (St. Leon-Rot, Germany), New England Biolabs (Frankfurt, Germany), Novagen (Schwalbach, Germany), Finnzymes (Espoo, Finnland), MWG Biotech AG (Ebersberg, Germany), Biomers (Ulm, Germany), or Qiagen (Hilden, Germany). Materials and equipment for protein purification were obtained from New England Biolabs or Millipore (Eschborn, Germany).
Syntheses
[U-13C]Phenylacetic acid was synthesized from 40 mg of l-[U-13C,15N]phenylalanine with 740 kBq of l-[U-14C]phenylalanine as a tracer. Phenylalanine was oxidatively deaminated by l-amino acid oxidase to phenylacetic acid as described before (13).
Phenylacetyl-CoA was synthesized from phenylacetyl succinimide modified after Schachter and Taggart (14). Phenylacetyl succinimide was synthesized as described before (10). β-Methylcrotonyl-CoA was purchased from Sigma. (R)/(S)-3-hydroxybutyryl-CoA, acryloyl-CoA, butyryl-CoA, and methacryloyl-CoA were synthesized from the free acid (15). Oxepin-CoA was enzymatically synthesized and purified as described below. CoA thioesters were quantified by determining the absorption at 260 nm, assuming the same molar extinction coefficient as for CoA with ϵ = 16,400 m−1cm−1 or for enoyl-CoA thioesters with ϵ = 22,000 m−1 cm−1 (16). The purity was analyzed by RP-HPLC (see below).
Purification of Products by Reverse Phase HPLC
Aliquots were applied to a column of LiChrospher 100 RP 18E, 5.0 μm, 125 × 4 mm (Wicom, Heppenheim, Germany) equilibrated with 40 mm ammonium acetate (NH4Ac), pH 6.8, containing 2% (v/v) acetonitrile at a flow rate of 1 ml min−1. The column was developed at a flow rate of 1 ml min−1 by a linear gradient from 2% acetonitrile in 40 mm ammonium acetate (pH 6.8) to 30% acetonitrile in the same buffer within 15 min. Elution was monitored with an UV diode array detector routinely at 260 nm.
HPLC Separation of Substrates and Ring-cleavage Products of Aerobic Phenylacetate Catabolism
To observe the formation of ring-cleavage products, an enzymatic assay (0.3 ml) containing 50 mm Tris-HCl (pH 8.0), 0.5 mm phenylacetyl-CoA, 2 mm NADPH, 1 mm NADP+, and 30 μg PaaG was started by the addition of 160 μg of PaaABC(D)E and stirred at 30 °C. A sample was taken after 2 min, before the assay was split and 1.5 μg of PaaZ or PaaZ-E256Q were added. Further samples were taken after additional incubation for 1.5 min. A similar assay (0.3 ml), which contained 1 mm NAD+ instead of NADP+, was incubated for 2 min, before the assay was split and incubated for additional 1.5 min with 35 μg of ECH-Aa or 35 μg of ECH-Aa and 41 μg of PacL. The assays were stopped by adding 1% formic acid to obtain pH 4.5. The samples were analyzed by RP-HPLC in an acetonitrile gradient as described above and detected at 260 nm.
HPLC Time Course of Product Formation
To show substrate consumption and product formation by PaaZ, PaaZ-E256Q, ECH-Aa, and PacL, reaction assays (0.2 ml) containing 50 mm Tris-HCl, 0.25 mm oxepin-CoA, and 1.5 mm NADP+ (for PaaZ/PaaZ-E256Q) or 1.5 mm NAD+ (for ECH-Aa/PacL) were started by the addition of 0.5 μg of PaaZ, 0.8 μg of PaaZ-E256Q, or 46 μg of ECH-Aa. For ECH-Aa, product formation was followed either in the absence or in the presence of (28 μg) PacL. Before the reactions were started, samples were incubated for 2 min without enzymes (except for PacL). Then, first samples were taken, and enzymes were added. Further samples were taken after 30, 60, 90, and 180 s. All samples were stopped and analyzed as described above.
Analysis by MS
Samples of RP-HPLC fractions were transferred at 5 μl/min by a syringe pump into the nano-electrospray ionization source of a Finnigan LTQ-FT mass spectrometer (Thermo Electron Corp., Waltham, MA) for online mass detection assembled from a linear ion trap and an ion cyclotron (7 tesla magnet) with Fourier transform ion cyclotron resonance mass spectrometry. Measurements covered the range of m/z from 600 to 1,300 at a resolution of 100,000.
NMR Analysis
NMR spectra were measured at 10 °C using Bruker AVANCE 500 spectrometers equipped with an inverse HC probehead or a QNP cryoprobe. The lyophilized sample was dissolved in methanol-D4 directly before measurement. 13C,1H INADEQUATE, HSQC and HSQC-TOCSY experiments were measured using the pulse programs and experimental settings implemented in the TOPSPIN 3.0 software package. The mixing time in the HSQC-TOCSY was 60 ms. Due to the 13C enrichment of the [U-13C]phenylacetyl-CoA precursor, the one-dimensional 13C NMR spectrum of the product as well as the two-dimensional INADEQUATE, HSQC, and HSQC-TOCSY spectra were dominated by the signals due to the 13C-enriched positions. Signals of the unlabeled CoA-ester residue were clearly detected in the one-dimensional 1H NMR spectrum. Moreover, the presence of the thioester moiety was gleaned from the downfield shifts of the carbonyl-CoA atom (195 ppm). The multiple 13C-labeling was also conducive to scalar couplings between adjacent 13C atoms. The chemical shifts of the 13C-labeled moiety as well as the multiplicities, the coupling constants, and the correlation observed in the two-dimensional experiments are listed in Table 1. In combination with the mass data, the NMR chemical shifts, couplings, and correlations provided solid evidence for the proposed structure.
TABLE 1.
NMR data of [U-13C8]2-hydroxycyclohepta-1,4,6-triene-1-formyl-CoA (VII)
Coupling constants were observed in the one-dimensional 13C NMR spectrum (coupling partners are in parentheses). ND, not determined due to signal overlap; d, doublet; dd, double-doublet; t, pseudo-triplet; qua, pseudo-quartet; m, multiplet. The compound was enzymatically synthesized from [U-13C8]phenylacetyl-CoA. Note that in the table the C-atom numbering is according to Fig. 4 and not to nomenclature, meaning that e.g. the hydroxy group at position C3 represents C2 (2-hydroxy-) according to nomenclature.
| Position | Chemical shifts |
Coupling constant | Correlations observed in |
|||
|---|---|---|---|---|---|---|
| 1H (HSQC) | 13C | HSQC | HSQC-TOCSY | INADEQUATE | ||
| ppm | Hz | |||||
| 1 | 194.9 (d) | 61 (2) | ||||
| 2 | 110.1 (qua) | 62 (1, 3, 8) | 8 | |||
| 3 | 166.4 (dd) | 44 (4), 69 (2) | ||||
| 4 | 2.67 | 33.9 (t) | 42 (3, 5) | 4 | 4, 5, 6 | |
| 5 | 5.5 | 119.7 (dd) | 38 (4), 67 (6) | 5 | 4, 5, 6, 7, 8 | 6 |
| 6 | 6.21 | 128.3 (m) | ND | 6 | 4, 5, 6, 7, 8 | 5 |
| 7 | 6.35 | 124.9 (m) | ND | 7 | 6, 7, 8 | 8 |
| 8 | 6.87 | 129.3 (m) | ND | 8 | 5, 6, 7, 8 | 7, 2 |
Molecular Biological Techniques
The cloning procedures for paaZ from E. coli K12, paaABCDE/paaG from Pseudomonas sp. strain Y2, and boxD from Azoarcus evansii were described before (10, 17). Chromosomal DNA from A. aromaticum was extracted using standard techniques. The genes for ech-Aa were amplified by PCR from chromosomal DNA using the forward primer 5′-GCTATGAATTCGCCAAAACGGGATGCAAG-3′ and reverse primer 5′-CTATCGAAGCTTCAACTATCTTGACCCAGAGG-3′. Genes for pacL were amplified using forward primer 5′-GAGTGAATTCACCCATCCGCTGTTCG-3′ and reverse primer 5′-CCGCAAGCTTGCACGGCGAGAGGCGTTAC-3′. Introduced restriction sites are underlined; the forward primers contained EcoR1 restriction sites replacing the start codon, and the reverse primers contained HindIII restriction sites. PCR was performed with Red Taq polymerase (Genaxxon) for 30 cycles, including denaturation for 45 s at 94 °C, annealing for 80 s at 65 °C (ech-Aa) or for 30 s at 68 °C (pacL), polymerization for 5 min at 72 °C, and a final extension at 72 °C for 5 min. The PCR products were isolated and cloned into the expression vector pMAL-c2X (New England Biolabs), which adds a maltose-binding protein (MBP) to the N terminus of the expressed proteins.
For mutation of paaZ from E. coli, a PCR was carried out with the pMAL-c2X-derived expression plasmid for PaaZ (10) as a template, according to published procedures (18). For this purpose, the primer 5′-CTTCACTATGCAAGCTGATTCCCTGAACTG-3′ was used, which contains a cytosine (shown in bold) instead of guanine, compared with native paaZ E. coli. PCR was performed with Phusion DNA polymerase (Finnzymes) for 25 cycles including denaturation for 10 s at 98 °C, annealing for 20 s at 65 °C, and polymerization for 4.5 min at 72 °C with a final extension at 72 °C for 7 min. Expression with the mutated construct yielded PaaZ-E256Q.
Bacterial Strains and Growth Conditions
PaaABC(D)E, PaaG, and BoxD were produced as described before (10, 17). For overproduction of the heterologous proteins PaaZ, PaaZ-E256Q, PacL, and ECH-Aa, transformed E. coli DH5α cells were grown at 37 °C on lysogeny broth medium (19) with 100 μg of ampicillin ml−1 up to an A578 of 0.6 before induction with 0.5 mm isopropylthiogalactopyranoside. The temperature was lowered to 22 °C, and the cells were harvested after additional growth for 3 h and stored at −20 °C until use.
Purification of MBP-tagged Proteins PaaZ, PaaZ-E256Q, PacL, ECH-Aa, PaaABC(D)E, PaaG, and BoxD
PaaABC(D)E, PaaG, and BoxD were purified and stored as described before (10, 17). The purifications of the other proteins were performed aerobically at 10 °C. For the preparation of cell extracts, one weight part of E. coli cells was suspended in one volume part of 20 mm Tris-HCl (pH 8.0) containing 0.1 mg of DNase I ml−1 and 200 mm KCl. The suspension was passed twice through a cooled French pressure cell at 137 megapascals and centrifuged (100,000 × g) at 4 °C for 1 h. The supernatant (cell extract) was directly applied to an amylose resin column (New England Biolabs, volume 15 ml). The cell extract was applied at a flow rate of 1 ml min−1 to the amylose resin column, which had been equilibrated with buffer (20 mm Tris-HCl (pH 8.0), 200 mm KCl). After binding of the enzymes, the column was washed with 2 column volumes of buffer at a rate of 2 ml min−1. The enzymes were eluted with buffer containing 10 mm maltose and stored with 30% glycerol (v/v) at −20 °C.
Protein Analyzing Methods
Enzyme fractions were analyzed by SDS-12.5% polyacrylamide gel electrophoresis (20). Proteins were visualized by Coomassie Brilliant Blue R-250 staining. Protein concentrations were determined by the Bradford method (21) with bovine serum albumin as a standard.
Preparation of Oxepin-CoA and Unlabeled and [U-13C]-labeled 2-Hydroxycyclohepta-1,4,6-triene-1-formyl-CoA
For photometric tests and RP-HPLC assays, oxepin-CoA was enzymatically synthesized and purified in a large scale assay. A reaction mixture (10 ml) contained 4 mg of PaaABC(D)E, 0.1 mg of PaaG, 1 mm NADPH, 50 mm Tris-HCl (pH 8.0), and 0.4 mm phenylacetyl-CoA. For MS analysis, the derivative 2-hydroxycyclohepta-1,4,6-triene-1-formyl-CoA was synthesized in an assay (0.3 ml) containing 0.03 mg PaaZ-E256Q, 50 mm Tris-HCl (pH 8.0), and 0.1 mm purified oxepin-CoA. For 13C NMR analysis of the derivative, a large scale assay (5 ml) was used to produce [U-13C]2-hydroxycyclohepta-1,4,6-triene-1-formyl-CoA. The reaction mixture contained 1.5 mm NADPH, 1 mg of PaaZ-E256Q, 3.5 mg of PaaABC(D)E, 0.2 mg of PaaG, 50 mm Tris-HCl (pH 8.0), and 1 mm [U-13C]phenylacetyl-CoA. All enzymatic assays were stirred for 5 min at 30 °C and stopped by adding 1% formic acid to obtain pH 4.5. The samples were centrifuged (14,000 × g) for 5 min at 4 °C. Unlabeled derivative was purified by HPLC and frozen in liquid nitrogen until MS measurement. The supernatants of the large scale assays were applied to C18-E columns (end-capped, bed size 200 mg, reservoir volume 3 ml, Phenomenex) that had been equilibrated with two reservoir volumes of 2% methanol in 20 mm ammonium acetate (pH 4.5). The columns were washed with 1 reservoir volume of equilibration buffer, and the reaction products were eluted with 1 reservoir volume 100% methanol. The methanol solutions were concentrated with a rotary evaporator at 50 millibar low pressure and 25 °C. The concentrates were loaded stepwise onto a C18-E column using RP-HPLC for separation. The respective peaks were collected, immediately frozen in liquid nitrogen, lyophilized, and kept frozen (−20 °C). The samples were dissolved in cold D2O or methanol-D4 directly before NMR measurement (for 13C-labeled 2-hydroxycyclohepta-1,4,6-triene-1-formyl-CoA) or in 10 mm MES-KOH (pH 4.5) (for oxepin-CoA) before use.
Enzyme Measurements
All described enzymatic reactions were carried out at 22 °C, and at least two independent measurements were performed. A spectrophotometric assay was used for the measurement of PaaZ, in which the formation of NADPH was followed spectrophotometrically at 365 nm (ϵ365 nm for NADPH of 3500 m−1 cm−1). The reaction mixture (0.3 ml) contained 0.5 μg of PaaZ, 50 mm Tris-HCl (pH 8.0), and 1.0–1.5 mm NADP+ (or NAD+ to determine cofactor-specificity) and was started by the addition of 0.1 mm purified oxepin-CoA. The Km value for oxepin-CoA was determined by adding variable amounts of oxepin-CoA (10–100 μm) to the assay while keeping the NADP+ concentration constant (1.5 mm). The Km value for NADP+ was determined by adding variable amounts of NADP+ (0.3–3 mm) to the assay while keeping the oxepin-CoA concentration constant (0.1 mm).
To determine the hydrolysis rate of oxepin-CoA through the ECH domain of PaaZ, a spectrophotometric assay with PaaZ-E256Q was used in which NADH formation through the auxiliary enzyme PacL was followed at 365 nm (ϵ365 nm for NADH of 3400 m−1 cm−1). The reaction mixture (0.3 ml) contained 0.5 μg of PaaZ-E256Q, an excess of 0.14 mg of PacL, 1.0 mm NAD+, and 50 mm Tris-HCl (pH 8.0) and was started by the addition of 0.1 mm oxepin-CoA. A similar assay was used to measure ECH-Aa activity. The reaction mixture (0.3 ml) contained 0.03 mg of ECH-Aa, an excess of 0.4 mg of PacL, 1.0 mm NAD+, 50 mm Tris-HCl (pH 8.0), and was started by the addition of 0.1 mm oxepin-CoA. This photometric assay was also used to determine the Km value of ECH-Aa for oxepin-CoA by recording the time course of complete NADH formation after addition of 0.1 mm oxepin-CoA followed by a graphic determination of the curve point at which half-maximal velocity was attained.
To determine the pH optima of PaaZ and ECH-Aa, (N-(2-acetamido)-2-aminoethanesulfonic acid-tris-ethanolamine (ATE) buffer was used, which shows a constant ionic strength between pH 6 and 10 (22). The specific activities were compared in 0.5 steps between pH 6 to 10 by using the above described photometric assays.
To determine the stereospecificity of the PaaZ-ECH and ECH-Aa catalyzed hydration, the backward dehydration reaction of 3-hydroxybutyryl-CoA to crotonyl-CoA was measured in a coupled assay with crotonyl-CoA carboxylase/reductase as an auxiliary enzyme (23). The reaction mixtures contained 1 mm NADPH, 100 mm Tris-HCl (pH 8.0), 0.03 mg of crotonyl-CoA carboxylase/reductase, 40 mm NaHCO3 (as a source of CO2), 0.5 mm concentrations of either the (R)- or the (S)-enantiomer of 3-hydroxybutyryl-CoA and 0.05 mg of PaaZ-E256Q or 0.24 mg of ECH-Aa, respectively.
A spectrophotometric assay was used to measure the crotonyl-CoA hydration rate to (R)-3-hydroxybutyryl-CoA by PaaZ-ECH or ECH-Aa, respectively, in which the absorption decrease at 263 nm was quantified due to the removal of the enoyl-thioester bond (ϵ263 nm for the enoyl-thioester bond of 6700 m−1 cm−1 (24)). The assays (220 μl) contained 50 mm Tris-HCl (pH 8.0), 0.1 mg of PaaZ or 0.03 μg of ECH-Aa and were started by the addition of 0.1–0.5 mm crotonyl-CoA. For PaaZ, also butyryl-CoA, β-methylcrotonyl-CoA, acryloyl-CoA, and methacryloyl-CoA were tested as substrates.
A photometric test to measure PacL activity would require an excess of ECH-Aa or PaaZ-E256Q. This was not possible due to overlapping spectra of the derivative (which is inevitably formed once the labile semialdehyde accumulates) and NADH; therefore, a HPLC assay was used. The reaction mixture (100 μl) contained 1.0 mm NAD+, 0.2 mm oxepin-CoA, 50 mm Tris-HCl (pH 8.0), 1.3 μg of PaaZ-E256Q, and 2.8 μg of PacL. Samples (30 μl) were taken after 25, 50, and 75 s, and the reaction was immediately stopped by adding 1% formic acid to obtain pH 4.5 before denatured protein was removed by centrifugation. The supernatants were analyzed by RP-HPLC. The formation of 3-oxo-5,6-dehydrosuberyl-CoA was quantified by determining the relative peak areas of oxepin-CoA and 3-oxo-5,6-dehydrosuberyl-CoA at different time points. The product formation was plotted against time, and the specific activity was calculated based on the best-fit line. The same assay was used for determination of PacL cofactor specificity with NADP+ instead of NAD+ and also to determine BoxD activity by replacing PacL.
Derivatization of Carbonyl Groups with Phenylhydrazine or Semicarbazide
To trap the reactive keto group of 3-oxo-5,6-dehydrosuberyl-CoA, a reaction mixture (0.4 ml) containing 1.5 mm NADPH, 1 mm NADP+, 100 mm Tris-HCl (pH 8.0), 0.03 mg of PaaG, 0.1 mg of PaaZ, 0.32 mg of PaaABC(D)E, 0.7 mm phenylacetyl-CoA, and 10 mm phenylhydrazine was incubated for up to 30 min at 22 °C. Samples were taken after different incubation times, the reactions were stopped by adding 1% formic acid to obtain pH 4.5, and denatured protein was removed by centrifugation. The samples were analyzed by RP-HPLC. A new product eluting at 11.8 min was formed over time that was collected and analyzed by MS. Similarly, assays containing either 10 mm phenylhydrazine or semicarbazide failed in trapping the labile ring-cleavage product 3-oxo-5,6-dehydrosuberyl-CoA semialdehyde or the derivative 2-hydroxycyclohepta-1,4,6-triene-1-formyl-CoA. Furthermore, aliquots of the enzymatic assay were quenched after different reaction times in MES-HCl (pH 5.5), which contained 10 mm semicarbazide or phenylhydrazine followed by additional incubation for 10 min. Again, no derivatization product was detected.
Labeling Experiments
For H218O-labeling assays, reaction mixtures (0.3 ml) containing 0.2 mg PaaABC(D)E, 0.01 mg of PaaG, 0.01 mg of PaaZ, 1 mm NADPH, 1 mm NADP+, 50 mm Tris-HCl (pH 8.0), and 0.4 mm phenylacetyl-CoA were incubated in 1.5-ml reaction tubes with 50% H218O and 50% H216O (v/v). Alternatively, ECH-Aa (0.12 mg) and PacL (0.08 mg) were used instead of PaaZ. For 18O-labeling experiments a closed tube (total volume 7.5 ml) with 50% 18O2, 50% 16O2 (v/v) gas phase was used. The reaction assay (2.5 ml) contained 0.1 mg of PaaZ, 1 mm NADP+, 50 mm Tris-HCl (pH 8.0), and 0.2 mm oxepin-CoA. The assay mixtures were stirred at 30 °C for 3 min before the reactions were stopped by adding 1% formic acid to obtain pH 4.5, and denatured protein was removed by centrifugation. The products were purified by RP-HPLC and frozen in liquid nitrogen until MS measurement.
Analysis of Oxygen Exchange by Mass Spectrometry
Unlabeled 3-oxo-5,6-dehydrosuberyl-CoA was enzymatically synthesized without 18O and purified by RP-HPLC. 200 μl of the unlabeled 3-oxo-5,6-dehydrosuberyl-CoA (containing 13% acetonitrile) was mixed with 174 μl of H218O (resulting in 50% H218O, 50% H216O) and analyzed by MS after different time intervals.
Spectroscopic Analysis of 3-Oxo-5,6-dehydrosuberyl-CoA
UV spectra of HPLC-purified 3-oxo-5,6-dehydrosuberyl-CoA were recorded in the range of 220–400 nm using a Cary 100 Bio spectrophotometer (Varian). After measurement of the sample, 80 mm Na2CO3 was added, which caused a pH shift from 6.8 to 10 before the sample was measured again. In a second sample 20 mm MgCl2 was added to purified 3-oxo-5,6-dehydrosuberyl-CoA before measurement.
Computational Analysis
The BLASTP searches were performed via the NCBI BLAST server (www.ncbi.nlm.nih.gov) (25, 26). The search was performed in June of 2010. All accession numbers are from GenBankTM. The amino acid sequences of PaaA and PaaC were used as queries for BLASTP searches against assembled bacterial genomes (www.ncbi.nlm.nih.gov). Unfinished bacterial genome projects were excluded from the BLASTP search. Occurrence and similarity of PaaA and PaaC were analyzed in a total number of 728 completely sequenced bacterial genomes. The percentage of organisms, which contained homologues for both key enzymes, was 16.6% (121 of 728 bacterial genomes). Homologues taken into account had an expectation value of <e−67 for PaaA or <e−11 for PaaC. The genomes of the organisms harboring the key genes were further analyzed for the presence of the different types of ALDHs involved in aerobic phenylacetate catabolism using the sequences of PaaZ (E. coli), PacL (A. aromaticum), and Bacillus ALDH (Bacillus subtilis) as queries for BLASTP searches. For construction of a phylogenetic tree, the amino acid sequences of a selection of the respective orthologues were aligned using ClustalW implemented within MEGA4 (27). The evolutionary history was inferred using the Neighbor-Joining method (28). The phylogenetic tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. Phylogenetic analyses were conducted in MEGA4. Other amino acid sequences were aligned using ClustalW implemented within BioEdit software.
RESULTS
Oxepin-CoA Ring Cleavage by the PaaZ-ECH Domain
The structure of oxepin-CoA (IV) suggests that the enoyl-CoA hydratase-like C-terminal domain (PaaZ-ECH) of the PaaZ fusion protein (accession number NP_415905) catalyzes the hydrolytic ring-cleavage, yielding an open-chain aldehyde (V, 3-oxo-5,6-dehydrosuberyl-CoA semialdehyde) (Fig. 1, A and B) (10). The N-terminal PaaZ-ALDH domain strongly resembles aldehyde dehydrogenases and may, thus, catalyze the subsequent oxidation of the aldehyde to the corresponding carboxylic acid (VI) (10). To identify the hydrolysis product, we incubated enzymatically synthesized and HPLC-purified (IV) with purified recombinant MBP-tagged PaaZ in the absence of NAD(P)+, as ALDH function strictly depends on NAD(P)+ in contrast to ECHs. HPLC analysis showed the rapid appearance of a new product that differed in retention time and UV spectrum from the PaaZ product (VI) formed in the presence of NADP+.
To corroborate that this represents the product of the PaaZ-ECH domain and to determine the reaction rate, we produced mutated MBP-tagged PaaZ in which we impaired ALDH function by exchanging an assumed catalytic glutamate of the ALDH domain for a glutamine (E256Q). As expected, the ALDH-deficient mutant protein PaaZ-E256Q formed the new product even in the presence of NADP+ (Figs. 2 and Fig. 3A). We then determined the rate of oxepin-CoA hydrolysis via a photometric test, by coupling PaaZ-E256Q-catalyzed ring cleavage with aldehyde oxidation by NAD+-dependent recombinant ALDH PacL from A. aromaticum (which catalyzes an analogous reaction to PaaZ-ALDH; see below). The specific hydratase activity amounted to 33 ± 1 units/mg (all activities were corrected for the size of untagged protein).
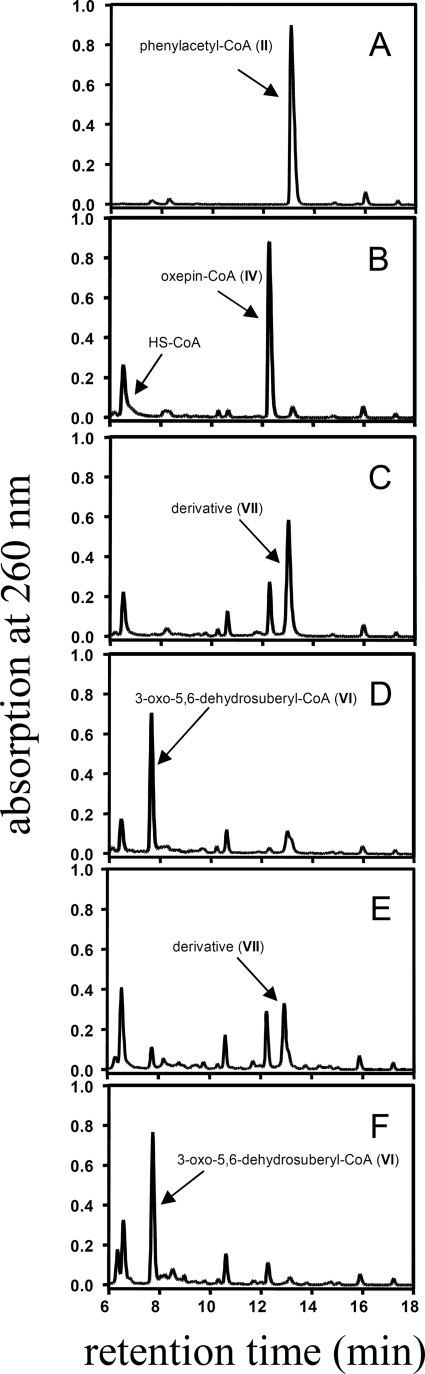
FIGURE 2.
Conversion of phenylacetyl-CoA to oxepin-CoA, 2-hydroxycyclohepta-1,4,6-triene-1-formyl-CoA, and 3-oxo-5,6-dehydrosuberyl-CoA by recombinant enzymes of the phenylacetate catabolic pathway from E. coli K12 (PaaZ, PaaZ-E256Q) and A. aromaticum (ECH-Aa, PacL). Products were separated by RP-HPLC in an acetonitrile gradient in 40 mm NH4Ac (pH 6.8) and detected at 260 nm. The initial reaction mixture contained 50 mm Tris-HCl (pH 8.0), 0.5 mm phenylacetyl-CoA, 2 mm NADPH, 1 mm NADP+, and 30 μg PaaG and was started by the addition of 160 μg of PaaABC(D)E to produce oxepin-CoA. The samples are before the addition of PaaABC(D)E (A) and 2 min after reaction start (B). Then the assay was split and incubated for another 1.5 min with 1.5 μg of PaaZ-E256Q (C) or 1.5 μg of PaaZ (D). A second initial reaction mixture, which contained 1 mm NAD+ instead of NADP+, was incubated for 2 min, split, and incubated for additional 1.5 min with 35 μg of ECH-Aa (E) or 35 μg of ECH-Aa and 41 μg PacL (F). CoA is released (B–F) through decomposition of the labile epoxide intermediate (PaaABC(D)E product). The roman numerals indicate the compounds in Figs. 1 and 4.
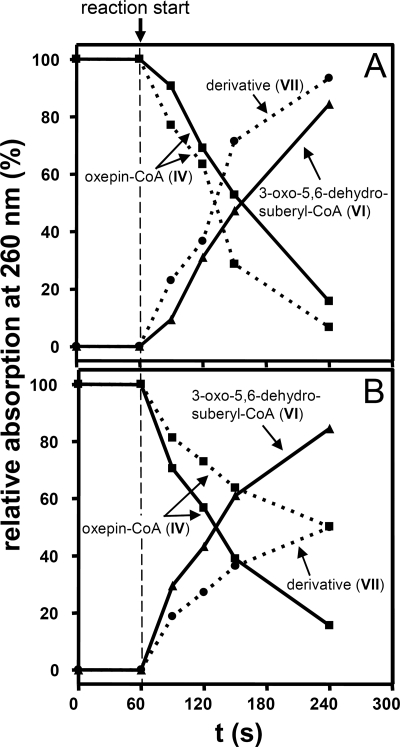
FIGURE 3.
Time course of substrate consumption and product formation by recombinant purified enzymes PaaZ or PaaZ-E256Q from E. coli K12 (A) and ECH-Aa and PacL from A. aromaticum (B). The reaction mixtures contained 50 mm Tris-HCl (pH 8.0), 0.25 mm oxepin-CoA, and 1.5 mm NADP+ (for A) or 1.5 mm NAD+ (for B). The reactions at 30 °C were started by the addition of PaaZ (0.5 μg), PaaZ-E256Q (0.8 μg), or ECH-Aa (46 μg). The continuous lines in A present the time course for PaaZ, and the dotted lines present the time course for PaaZ-E256Q. The continuous lines in B show the time course for ECH-Aa in the presence of 28 μg of PacL, and the dotted lines show the time course for ECH-Aa alone. The roman numerals indicate the compounds in Figs. 1 and 4.
Spontaneous Formation of a Stable Derivative from Labile 3-Oxo-5,6-dehydrosuberyl-CoA Semialdehyde
To test if the new product presents the expected hydrolysis product (V), we converted oxepin-CoA with the ALDH-deficient PaaZ-E256Q. We then tested by HPLC if further addition of unimpaired PaaZ produces (VI). Surprisingly, this was not the case. Therefore, the new product must present a rapidly generated derivative, as no other peaks were observed during the HPLC runs. Mass spectrometry showed that this non-physiological compound exhibited a mass of MH+ = 902 (supplemental Fig. S1) equal to oxepin-CoA, whereas the mass of 920 was expected for the hydrolytic ring cleavage product.
NMR analysis identified the enzymatically produced [U-13C]-labeled derivative as a seven-membered carbon-cycle, [U-13C]2-hydroxycyclohepta-1,4,6-triene-1-formyl-CoA (VII) (Table 1, Fig. 4). Apparently, after hydrolytic ring fission, the proposed open-chain aldehyde (V) rapidly forms a seven-membered carbon ring through Knoevenagel-type condensation. C2 deprotonates, which is facilitated by the electron-withdrawing forces of the adjacent thioester and keto group. It enables a nucleophilic attack at the reactive terminal C8 aldehyde group with subsequent water elimination, forming the seven-membered ring. This is followed by rearrangement of the keto group to the more stable enol-form of the compound, yielding the observed (VII) (Fig. 4). This compound likely inhibits PaaZ as the initial oxepin-CoA hydrolysis rate slowed down with increasing derivative concentration as observed by HPLC (data not shown).
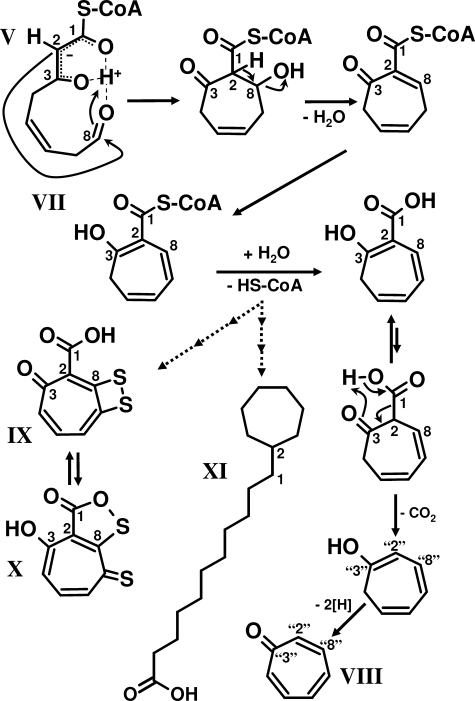
FIGURE 4.
Proposed mechanism of 2-hydroxycyclohepta-1,4,6-triene-1-formyl-CoA formation. The highly reactive 3-oxo-5,6-dehydrosuberyl-CoA semialdehyde (V, ring-cleavage product) undergoes spontaneous intramolecular Knoevenagel-type condensation forming the derivative 2-hydroxycyclohepta-1,4,6-triene-1-formyl-CoA (VII). The aromatic tropone (IX) is likely formed from VII, whereas the antibiotics tropodithietic acid (IX) and thiotropocin (X) as well as the ω-cycloheptyl fatty acids (XI) are derived either directly from VII or from the same compound after hydrolysis of the CoA thioester. Note that the numbering of C-atoms for the cyclic compounds is not according to nomenclature.
Aldehyde Oxidation by PaaZ-ALDH
PaaZ converted oxepin-CoA in the presence of NADP+ mainly to the final product (VI), whereas only trace amounts of (VII) were produced (Figs. 2 and Fig. 3A). To determine the ALDH reaction rate, we quantified the formation of NADPH in a photometric assay and measured a specific activity of 32 ± 3 units/mg, which is nearly identical to the enoyl-CoA hydratase activity (33 ± 1 units/mg). With NAD+, PaaZ exhibited 50% activity.
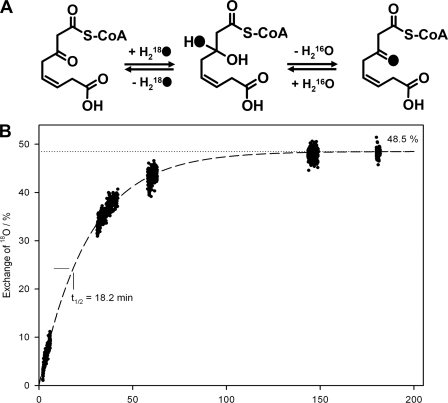
18O-Labeling of 3-Oxo-5,6-dehydrosuberyl-CoA after Transformation of Oxepin-CoA by PaaZ in the Presence of H218O
We applied 18O-labeling studies to verify the hydrolytic ring cleavage of (IV) and aldehyde oxidation of (V) by PaaZ. For this purpose, we enzymatically produced (VI) in the presence of 50% H218O, 50% H216O. In case of hydrolytic ring cleavage and subsequent aldehyde oxidation by PaaZ, we would expect the incorporation of up to two labeled oxygen from water (Fig. 1, A and B). The mass of the proposed PaaZ product (VI) is MH+ = 936 (10). When 50% H218O is used, one would expect a ratio of 1:2:1 of unlabeled (MH+ = 936), single-labeled (MH+ = 938), and double-labeled product (MH+ = 940). However, we observed a ratio of 4:5:1 (supplemental Fig. S2). This discrepancy can be explained as follows.
Proof of Oxygen Exchange between the Carbonyl Oxygen of 3-Oxo-5,6-dehydrosuberyl-CoA and Oxygen from Water
It is well known for aldehydes and ketones that the carbonyl oxygen rapidly exchanges with oxygen from water (29). This exchange is also expected for the free keto group of labeled (VI), which would result in an exchange of 18O with 16O from water via the ketone hydrate (Fig. 5A). This substitution can take place during sample preparation when the product was isolated in unlabeled water; double-labeled (VI) would then exchange 18O with 16O at the keto group leading to more single-labeled product. Moreover, the part of the single-labeled (VI) that harbors 18O at the keto group exchanges with the same rate, thus, forming more unlabeled product. Finally, after sufficient incubation time, one would expect a 1:1 ratio of unlabeled to single-labeled (VI), as labeled oxygen at the carboxyl group (i.e. from aldehyde oxidation) cannot exchange with water. Because the sample preparation via HPLC took about 10 min, we observed a yet incomplete exchange. To verify that oxygen does exchange between carbonyl 18O of the PaaZ product and 16O from water, we measured the exchange reaction in the opposite direction. Enzymatically synthesized and HPLC-purified 16O-(VI) was incubated with 50% H218O, 50% H216O and analyzed by MS after several time intervals. In fact, we could observe the formation of single-labeled 18O-(VI) with a rate that may well explain the initial labeling pattern (Fig. 5B, supplemental Fig. S2).
FIGURE 5.
Mechanism (A) and kinetic behavior (B) of oxygen exchange between the carbonyl oxygen of 3-oxo-5,6-dehydrosuberyl-CoA and oxygen from water in 50% H218O/50% H216O. Phenylacetyl-CoA was converted by PaaABC(D)E, PaaG, and PaaZ, and the product 3-oxo-5,6-dehydrosuberyl-CoA was purified by reverse-phase HPLC. After sample collection, 50% H218O (v/v) was added, and the first sample was immediately injected into the electrospray ionization source of a Finnigan LTQ-FT mass spectrometer and measured within 1.5 min. Further measurements were done in several time intervals. The curve demonstrates the incorporation of 18O from labeled water into the 3-oxo group via the keto hydrate and explains the reduced amount of double labeled 3-oxo-5,6-dehydrosuberyl-CoA in the performed H218O labeling experiments (see supplemental Fig. S2). 18O-atoms are illustrated as black dots in A.
As a control, we converted purified (IV) with PaaZ in the presence of 50% 18O2, 50% 16O2. As expected, we could not observe labeled (VI). Taken together, the data provides evidence for a hydrolytic rather than oxygenolytic nature of the ring fission.
Substrate Specificity and Stereospecificity of the PaaZ-ECH Domain
We tested crotonyl-CoA, butyryl-CoA, β-methylcrotonyl-CoA, acryloyl-CoA, and methacryloyl-CoA as possible substrates for the PaaZ-ECH domain. Significant activity (0.23 ± 0.01 units/mg) was shown exclusively for crotonyl-CoA. The hydration product was identified as 3-hydroxybutyryl-CoA using HPLC (data not shown). It should be noted that earlier studies with PaaZ reported a specific activity of 48 units/mg for crotonyl-CoA (12). The assay conditions were similar, and the reasons for such a great discrepancy to our results are unclear.
To test the stereospecificity of the PaaZ-ECH hydration, we measured the backward dehydration reaction of 3-hydroxybutyryl-CoA to crotonyl-CoA in a coupled assay with crotonyl-CoA carboxylase/reductase. This enzyme converts crotonyl-CoA to ethylmalonyl-CoA under consumption of NADPH and CO2 (23). The reaction was only observed with (R)-3-hydroxybutyryl-CoA, but not with (S)-3-hydroxybutyryl-CoA, proving that the PaaZ-ECH domain acts as (R)-specific hydratase.
Derivatization with Phenylhydrazine and Semicarbazide
To test for reactive keto groups, we enzymatically synthesized (VI) in the presence of phenylhydrazine and observed the formation of a new product by HPLC (data not shown). We applied MS and detected a mass corresponding to 3-phenylhydrazono-5,6-dehydrosuberyl-CoA (see supplemental Fig. S3), which supports the reported structural data for the PaaZ product (10). To prove an aldehyde intermediate as the ring-cleavage product, we tried to trap (V) with phenylhydrazine or semicarbazide as a stable phenylhydrazone or semicarbazone under different conditions (up to 10 mm (pH 5.5–8.0)), by converting (IV) with ALDH-deficient PaaZ-E256Q. Apparently, formation of (VII) from (V) occurred too rapidly to permit derivatization, as no new peak was detected by HPLC. The 2-hydroxy group of (VII) may form a keto group through keto-enol tautomerism (Fig. 4), which should also become trapped. It is unclear why derivatization did not occur. Probably, the keto-enol-enolate equilibrium favors the enolate and may, thus, impede derivatization.
Spectroscopic Features of 3-Oxo-5,6-dehydrosuberyl-CoA
The UV spectrum of the PaaZ product (VI) showed a small shoulder between 290–330 nm beside the maximum at 260 nm caused by the nucleotide of the CoA ester. To test if this shoulder is caused by the enolate form that equilibrates with the β-keto group, we used Mg2+ or, alternatively, a pH increase to 10 to stabilize the enolate (16). As expected, the substance showed in both cases typical spectroscopic features of a 2,3-enolate group, with a significant absorption increase at 310 nm (supplemental Fig. S4).
Ring Cleavage in Organisms Lacking the PaaZ Fusion Protein
Many phenylacetate degrading organisms lack paaZ-like genes but instead harbor genes in their paa clusters that only encode predicted ALDHs without PaaZ-ECH-like domains. An example is pacL from A. aromaticum, which is currently misleadingly annotated as paaZ. Consequently, an obvious candidate for hydrolytic ring cleavage is missing in the clusters of many other phenylacetate-degrading bacteria (see supplemental Table S1). We, thus, performed a genome-wide BLASTP search in A. aromaticum for predicted PaaZ-ECH-like proteins ((R)-specific Hot dog-fold hydratases) using the PaaZ-ECH domain of E. coli as a query.
Two candidates were found; the MBP-tagged proteins were heterologically produced and purified. One of them (accession number CAI09349.1) showed no activity toward (IV), whereas the other candidate (ECH-Aa, accession number CAI08632.1) exhibited oxepin-CoA ring-cleavage activity. Ring fission with ECH-Aa yielded the same derivative as did cofactor-deprived PaaZ or ALDH-deficient PaaZ-E256Q, as shown by HPLC (Figs. 2 and 3B) and MS, however, with a much lower rate of 1.2 ± 0.1 units mg−1. It is noteworthy that ECH-Aa exhibited high activity for crotonyl-CoA (2300 ± 340 units/mg), indicating that oxepin-CoA hydrolysis might be a side reaction. As expected, ECH-Aa was also found to be (R)-specific, like PaaZ-ECH. We then tested by HPLC if ECH-Aa and purified recombinant MBP-tagged PacL (accession number CAI08120) are able to substitute for PaaZ. In fact, the concerted action of ECH-Aa and PacL lead to the formation of (VI), which was identified by retention time, UV spectrum, and MS (Figs. 2 and 3B). Furthermore, according 18O-labeling patterns were observed for (VI) when ECH-Aa and PacL were used instead of PaaZ (data not shown).
A photometric test to measure the rate of the PacL-catalyzed reaction was not possible because an excess of PaaZ-E256Q or ECH-Aa (which is needed to provide substrate (V)) for the determination of PacL activity inevitably led to accumulation of the derivative (VII). This interfered with detection of NADH formation due to overlapping spectra. We, therefore, used HPLC for quantifying the PacL-catalyzed formation of (VI) from (V) and determined a specific activity of 5.8 ± 0.1 units mg−1. The concurrent non-enzymatic formation of (VII) from (V) indicated the required excess of PaaZ-E256Q for determination of PacL activity. PacL preferred NAD+ as a cofactor with 60% activity with NADP+.
Determination of the Catalytic Properties of PaaZ, PacL, and ECH-Aa
A summary of some catalytic features of the three enzymes characterized in this work is shown in Table 2. The stoichiometry of the PaaZ-catalyzed reaction was 1 NADP+ reduced per 1 oxepin-CoA converted. No Km value could be determined for PacL due to the instability of the substrate (V).
TABLE 2.
Catalytic properties of recombinant PaaZ (Escherichia coli) and PacL and ECH-Aa (both from Aromatoleum aromaticum)
ND, not determined.
| Organism and enzymea | E. coli PaaZ | A. aromaticum PacL | A. aromaticum ECH |
|---|---|---|---|
| Substrates | Oxepin-CoA, crotonyl-CoA, NADP+ | 3-Oxo-5,6-dehydrosuberyl-CoA semialdehyde, NAD+ | Oxepin-CoA, crotonyl-CoA |
| Products | 3-Oxo-5,6-dehydrosuberyl-CoA, (R)-3-hydroxybutyryl-CoA, NADPH, H+ | 3-Oxo-5,6-dehydrosuberyl-CoA, NADH, H+ | 3-Oxo-5,6-dehydrosuberyl-CoA semialdehyde, (R)-3-hydroxybutyryl-CoA |
| Specific activityb (units/mg) | 32 (±3) | 5.8 (±0.1) | 1.2 (±0.1) |
| Apparent Km (μm) | Oxepin-CoA, 11 (±1); NADP+, 56 (±4) | ND | Oxepin-CoA 35 (±4) |
| Optimum pH | 8 | ND | 8 |
| Turnover (s−1) | 39 | 6 | 0.32 |
| Catalytic efficiency (l/mol·s) | 3.5 × 106 | ND | 0.01 × 106 |
| Specificity (%) | Oxepin-CoA 100, crotonyl-CoA 1 | Oxepin-CoA 0.02, crotonyl-CoA 100 |
a Accession numbers are NP_415905 for PaaZ, CAI08120 for PacL and CAI08632.1 for ECH-Aa.
b Activities were determined using photometric tests (PaaZ, ECH-Aa) or HPLC tests (PacL) and are corrected for the size of an untagged protein.
Different ALDHs Involved in Phenylacetate Metabolism
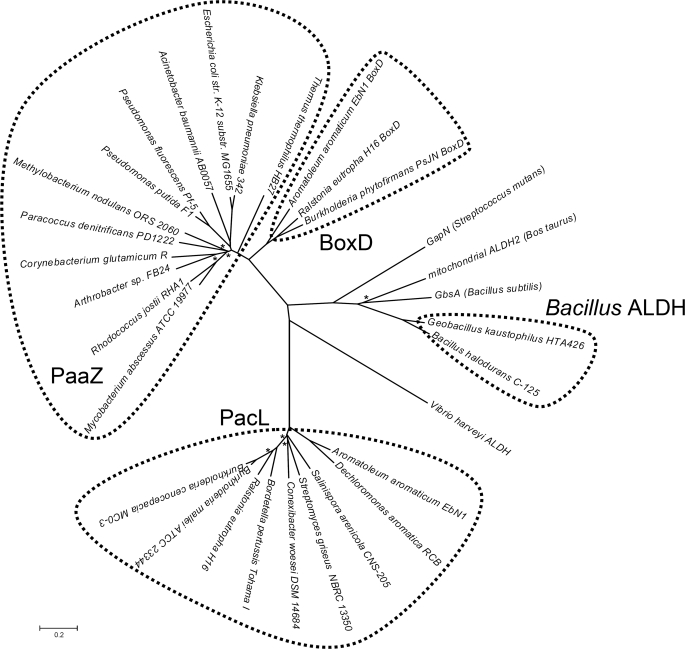
To gain insight into the PacL and PaaZ relation, we performed BLASTP searches against assembled bacterial genomes with the NCBI data base. Our results showed that 121 of 728 (>16%) bacterial species harbor key genes for the phenylacetate pathway (see supplemental Table S1). We searched the proteome of those bacteria for the presence of homologues of PaaZ from E. coli and of PacL from A. aromaticum using the respective amino acid sequences as queries. Most (56 organisms) showed homologues for PaaZ, 42 for PacL and 3 for both proteins (see supplemental Table S1). Strikingly, no homologues for either protein were found in Firmicutes. We checked their phenylacetate degradation cluster and found a further candidate gene located between paaG and paaH for the processing of the labile aldehyde intermediate in Bacillus halodurans (accession number BAB03922). All 11 members of the Firmicutes and one member of the Chloroflexi that make use of this route harbored homologues for this protein. We found no obvious (i.e. clustered) candidate gene in the remaining 10 bacterial species.
Phylogeny of PaaZ and Related Enzymes
PaaZ-ALDH of E. coli showed high similarity to aldehyde dehydrogenase BoxD of A. aromaticum (43% amino acid sequence identity, accession number CAI07676) and other bacteria. BoxD oxidizes 3,4-dehydroadipyl-CoA semialdehyde to 3,4-dehydroadipyl-CoA in the (micro)aerobic benzoate catabolic pathway (11). Despite the similarity of both enzymes, purified BoxD exhibited only minor activity toward (V) (≈0.02 units mg−1) as determined by HPLC.
The different types of ALDHs prompted us to construct a phylogenetic tree of the three types of aldehyde dehydrogenases involved in phenylacetate degradation, of BoxD and of several other ALDHs whose functions are known (see Fig. 6). The tree suggested that PaaZ-ALDH and BoxD very likely share a recent common ancestor as they cluster together. PacL and Bacillus ALDH formed different self-contained branches unrelated to PaaZ-ALDH, although they probably catalyze the same reaction. Bacillus-ALDH was highly similar to bovine mitochondrial ALDH2 (see Fig. 6) and to glycine betaine aldehyde dehydrogenase GbsA from B. subtilis (30) (42% amino acid identity), which may have been the ancestral function of the enzyme. PacL does not show significant homology toward any of the discussed ALDHs (≤28% amino acid identity).
FIGURE 6.
Phylogenetic unrooted tree of a selection of orthologues of PaaZ-ALDH, PacL, Bacillus ALDH, BoxD, and some ALDHs that are not involved in aromatic degradation. The tree was constructed using the Neighbor-Joining method (28) and is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. Asterisks indicate bootstrap values <70%. ALDHs that are not involved in aromatic catabolism are: GapN, glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans (64) (accession number Q59931); mitochondrial bovine ALDH2 (53) (accession number P20000); GbsA: glycine betaine aldehyde dehydrogenase from B. subtilis (30) (accession number NP_390984); Vibrio harveyi ALDH (60) (accession number AAA89078).
DISCUSSION
Hydrolytic Ring Cleavage by PaaZ-ECH
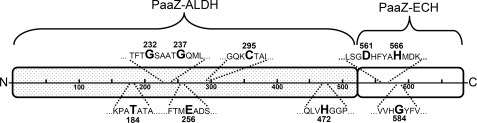
The predicted Hot dog-fold PaaZ-ECH domain acts as an (R)-specific hydratase that exhibits high catalytic efficiency for oxepin-CoA (Table 2). This enables efficient ring cleavage and removal of toxic pathway intermediates. Catalytic residues of (R)-specific Hot dog-fold hydratases (EC 4.2.1.119) have been identified by structural and mutational analysis of the hydratase 2 domain of eukaryotic multifunctional enzyme type 2 (MFE-2) and of Aeromonas caviae hydratase, suggesting an acid-base mechanism with strictly conserved catalytic aspartate and histidine residues (31–33). A similar mechanism also applies to (S)-specific hydratases despite lacking amino acid sequence and structural similarity. In this case, two glutamate residues serve as catalytic acid and base (34). Hydration by both types of hydratases, hence, occurs with reciprocal stereochemistry due to mirror-image active centers (31). Multiple sequence alignments of the hydratase 2 domains from MFE-2 and eukaryotic and prokaryotic fatty acid synthases revealed the conserved consensus sequence (Y/F)X1,2(L/V/I/G)(S/T/G/C)GDXNP(L/I/V)HX5(A/S) (35) with the catalytic residues shown in bold. The remaining conserved amino acids likely stabilize the structure with hydrophobic and hydrogen bond interactions (33). Alignment of all 56 PaaZ revealed the consensus sequence F-(A/G/S)X2(S/T)(G/W)DXFY(A/M)HX(D/N)X3(A/L/T) for the PaaZ-ECH domain. Consistently, this consensus sequence also shows the strictly conserved Asp and His (shown in bold) with a spacer of four amino acids, suggesting that those residues are responsible for oxepin-CoA hydrolysis (Fig. 7, see also supplemental Fig. S5).
FIGURE 7.
Schematic view of fusion protein PaaZ from E. coli K12. The N-terminal ALDH domain and the C-terminal ECH domain of PaaZ are illustrated as separate parts. The highlighted amino acids are: Thr-184, responsible for cofactor specificity; Gly-232 and Gly-237, important for cofactor binding; Glu-256, likely acting as catalytic general base; Cys-295, catalytic residue responsible for the initial formation of a covalent thiohemiacetal intermediate; His-472, possible candidate for the catalytic general base; Asp-561 and His-566, proposed catalytic residues for hydrolytic ring-cleavage; Gly-584, likely responsible for the formation of an oxyanion hole.
Proposed Catalytic Mechanism of PaaZ-ECH
The structure of the PaaZ substrate (IV) allows two possible sites for hydrolytic cleavage, at C3 or C8 (see Fig. 1B). PaaZ adds water to crotonyl-CoA, which harbors a double bond between C2 and C3 adjacent to the CoA-ester like oxepin-CoA, thus indicating that the PaaZ-ECH domain likely introduces water at C3 of the side chain. In the case of hydrolytic ring cleavage at C8, one would not expect 18O-exchange with water, as observed. The keto-group of (VI) should then be derived from the former epoxy-oxygen, which in turn derives from (unlabeled) O2 introduced during enzymatic synthesis of (IV) by monooxygenase PaaABC(D)E (Fig. 1A).
We suggest a reaction mechanism that requires a positively charged ether O-atom and an oxyanion of the CoA-thioester (residue numbers for E. coli enzyme). A strictly conserved Gly-584 may form an oxyanion hole stabilizing this enolate anion intermediate (34) (Figs. 1B and 7 and supplemental Fig. S5), similar to other (R)-specific hydratases (31, 33). This state is also favored through mesomerism. It enables the addition of a hydroxyl group to C3 and a proton to C2, likely mediated by Asp-561 and His-566 (Fig. 1B and 7 and supplemental Fig. S5). The proton of the attached hydroxyl group then dissociates, forming a keto group. Simultaneously, ring cleavage between C3 and the ether O-atom occurs. Finally, the resulting ring-opened C8 alcohol rearranges through keto-enol tautomerism to (V), the substrate of the ALDH domain of PaaZ (Fig. 1B). PaaZ, hence, exploits the structure of the oxepin for an attack that resembles conventional hydration of α,β unsaturated acyl-CoA esters (like crotonyl-CoA) to achieve ring cleavage. The need of CoA esters for the formation of an enolate intermediate in this reaction and also in the PaaG-catalyzed epoxide-oxepin (III-IV) isomerization may explain why the pathway intermediates are processed as CoA-esters.
Ring Cleavage in Organisms Lacking PaaZ
A PaaZ-ECH-like protein from A. aromaticum ECH-Aa encoded outside the paa gene cluster is able to cleave oxepin-CoA (IV), yet with low catalytic efficiency. Possibly, a similar gene may code for the true enzyme. Here we searched only for homologues of (R)-specific Hot dog-fold hydratases. However, despite the stereoselectivity of the PaaZ-ECH domain for crotonyl-CoA hydration, the hydrolytic cleavage semialdehyde product (V) does not harbor a stereocenter. Therefore, an (S)-specific enoyl-CoA hydratase may well catalyze oxepin-CoA (IV) ring fission via an acid-base mechanism. This possibility is supported by the fact that some bacteria contain neither genes for PaaZ nor for (R)-specific Hot dog-fold hydratases. In A. evansii lacking a fusion protein, a pacL knock-out mutant was unable to grow aerobically on phenylacetate but accumulated the aromatic tropone (VIII) (2,4,6-cycloheptatriene-1-one; Fig. 4) during growth in the presence of phenylacetate (36). Recently, (VIII) was also shown to be derived from phenylacetate (37). It resembles the derivative (VII) which is formed during oxepin-CoA hydrolysis by PaaZ in trace amounts along with the main product (VI) or exclusively if PaaZ-ALDH function is impaired through mutation or by lack of NAD(P)+. Moreover, ECH-Aa produces the same derivative in the absence of PacL (Fig. 2). The unstable hydrolysis product (V) may undergo spontaneous Knoevenagel-type condensation forming (VII) (tropone precursor, Fig. 4). Hydrolysis of the CoA-thioester, subsequent decarboxylation, and oxidation then likely leads to tropone (VIII).
Bacterial Primary and Secondary Metabolites with the Derivative (VII) as Precursor
The antibiotics tropodithietic acid (IX) and its tautomers, e.g. thiotropocin (X) from Roseobacter, Pseudomonas, and Caulobacter species (37–40) are also highly similar to (VII). Their formation proceeds via phenylacetate and depends on early phenylacetate catabolic pathway enzymes (37, 41). Tropodithietic acid may even act as a quorum signal in symbiotic Roseobacter species (42). Moreover, a very similar sulfur-bridged dicyclic tropolone with broad antibacterial activity spectrum was isolated from Burkholderia cenocepacia (43, 44), which can grow on phenylacetate (45). Astonishingly, the derivative (VII) resembles parts of the unusual predominant eponymous ω-cycloheptyl fatty acids (XI) from Alicyclobacillus cycloheptanicus (46). The terminal cycloheptane ring is derived from phenylacetate and formed through a so far unknown ring expansion mechanism similar to the production of tropodithietic acid/thiotropocin (46). Possibly all tropone/tropolone-related compounds found in bacteria may be formed this way either from proffered phenylalanine/phenylacetate (catabolic) or from a C9 Shikimate pathway intermediate (anabolic). Analogous formation may also apply to some tropone/tropolone compounds from plants/fungi, as there is limited knowledge at present (44).
We postulate that the derivative (VII) is the sought precursor of the discussed secondary metabolites and ω-cycloheptyl fatty acids. The proposed spontaneous intramolecular Knoevenagel-type condensation of the labile ring-cleavage product (V) is, thus, pivotal for the formation of such heptacyclic compounds. The required minimum set of enzymes for an efficient production of (XII) from phenylacetate are, thus, PaaK, PaaABC(D)E, PaaG (Fig. 1A), and a ring-cleaving hydratase that lacks ALDH function (e.g. PaaZ-E256Q). This knowledge may set the stage for antibiotics-producing bacterial systems when combined with enzymes for the final conversions of (XII) into tropodithietic acid or related bioactive compounds.
Oxidation of the Labile 3-Oxo-5,6-dehydrosuberyl-CoA Semialdehyde
The resulting semialdehyde ring-cleavage product (V) is further oxidized to the respective carboxylic acid (VI) by PaaZ-ALDH (or PacL) under formation of NADPH (Fig. 1A). This fits well with the NADPH consumption through the multicomponent oxygenase/reductase PaaABC(D)E (10), thus, creating a short NADPH/NADP+ cycle for the initial steps of phenylacetate degradation. PaaZ-ALDH, PacL, Bacillus ALDH, and BoxD all belong to the large enzyme family of NAD(P)+-dependent ALDHs (EC 1.2.1.x) (47). They contain a characteristic Rossman-fold for cofactor binding with the conserved fingerprint motive GXXXXG (17, 48, 49). Only the first glycine is mandatory for cofactor binding (49) as observed in the BoxD orthologues and in some PaaZ orthologues, whereas both glycines are conserved in PaaZ from E. coli (Fig. 7), PacL, and Bacillus ALDH (supplemental Fig. S6).
NAD+ specificity of ALDHs is largely determined by a glutamate residue (50), whereas polar amino acid residues (e.g. threonine, as observed in PaaZ, Thr-184, see Fig. 7) at this position favor the binding of NADP+ (51, 52) (for sequence alignments of PaaZ, PacL, Bacillus ALDH, BoxD (27), and NAD+-specific bovine mitochondrial ALDH2 (53) see supplemental Fig. S6). Some other PaaZ orthologues harbor Ser, Pro, Gly, or His instead of Thr. PacL, which has an isoleucine (Ile-236) at this location, prefers NAD+; this may apply to all PacL orthologues, with some harboring a valine instead of isoleucine. Bacillus ALDH orthologues harbor a serine or a glutamate at that position (Ser/Glu-177) with yet no available experimental data for cofactor specificity.
Aldehyde oxidation is achieved through initial formation of a covalent thiohemiacetal intermediate of the substrate with a cysteine residue of the ALDH-NAD(P)+ (54–57). Homologues to this catalytic cysteine (Cys-295 in PaaZ E. coli; see Fig. 7 and supplemental Fig. S6) as well as two strictly conserved glutamate residues were identified in the discussed ALDHs (in PaaZ E. coli Glu-256 and Glu-395; see Fig. 7 and supplemental Fig. S6). A conserved histidine or glutamate residue, which is close to the catalytic cysteine, may act as a general base (58–60). This conserved histidine is not present in all ALDHs but can be found in PaaZ (Fig. 7) and BoxD (supplemental Fig. S6). The crystal structure of BoxD from Burkholderia xenovorans LB400 indicates that the glutamate (Glu-255 in BoxD A. aromaticum) is the most likely candidate for the general base (61). This suggests that the corresponding Glu-256 in PaaZ from E. coli has the same essential function that is underlined by the ALDH-deficient PaaZ-E256Q mutant.
Consequences for Nomenclature of MaoC-like Proteins
It is noteworthy that the paaZ gene was originally identified as monoamine oxidase C (maoC) without knowledge of the encoded proteins function, as it is flanked upstream by maoA, padA, and maoB in Klebsiella aerogenes (62), whereas the paa genes are located downstream. MaoA is an amine oxidase that accepts various substrates like tyramine or phenylethylamine, the latter yielding phenylacetaldehyde. This is the substrate for phenylacetaldehyde dehydrogenase PadA, producing phenylacetate (63). Therefore, two related pathways are clustered, but PaaZ itself is not involved in the processing of amines. However, mainly the ECH domain of PaaZ and similar proteins and their genes are currently misleadingly referred to as MaoC-like. We, therefore, suggest the renaming of these proteins and the respective genes and propose the correct term (R)-specific enoyl-CoA hydratase.
Evolutionary Considerations
Formation and accumulation of (VII) is likely inevitable during growth on phenylacetate, and apparently some bacteria found use for this “dead-end product,” making secondary metabolites and even unusual fatty acids. Likely, the PaaZ fusion protein does not release the semialdehyde intermediate but immediately oxidizes it to the more stable carboxylic acid to largely prevent formation of dead-end products. These products are not only inhibitory but also trap CoA. Metabolic channeling achieved by fusion of two genes thus was positively selected. At least three different ALDHs have been recruited for the processing of the labile semialdehyde (V). They do not share a recent common ancestor and evolved independently as illustrated by a phylogenetic tree (Fig. 6). The fact that BoxD is homologous to PaaZ-ALDH tempts to speculate whether one is derived from the other or if the two proteins just share a common ancestor.
Supplementary Material
Acknowledgments
We thank Bettina Knapp for mass spectrometry measurements, Tobias Erb for kindly providing some CoA esters and purified crotonyl-CoA carboxylase/reductase, Liv Rather for providing BoxD, and Ivan Berg and Michael Carter for critical reading. We also thank the Hans-Fischer Gesellschaft.
This work was supported by the Deutsche Forschungsgemeinschaft (to G. F., W. E., and W. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S2 and Figs. S1–S6.
- Paa
- phenylacetic acid
- Box
- benzoate oxidation
- ECH
- enoyl-CoA hydratase
- ALDH
- aldehyde dehydrogenase
- MBP
- maltose-binding protein
- RP
- reverse phase
- LTQ-FT
- linear ion trap with Fourier-transform ion cyclotron resonance mass spectrometry
- HSQC
- heteronuclear single quantum correlation
- TOCSY
- two-dimensional total correlation spectroscopy
- MaoC
- monoamine oxidase C.
REFERENCES
- 1. Fuchs G. (2008) Ann. N.Y. Acad. Sci. 1125, 82–99 [DOI] [PubMed] [Google Scholar]
- 2. Butler C. S., Mason J. R. (1997) Adv. Microb. Physiol. 38, 47–84 [DOI] [PubMed] [Google Scholar]
- 3. Gibson D. T., Parales R. E. (2000) Curr. Opin. Biotechnol. 11, 236–243 [DOI] [PubMed] [Google Scholar]
- 4. Boll M., Fuchs G. (1995) Eur. J. Biochem. 234, 921–933 [DOI] [PubMed] [Google Scholar]
- 5. Luengo J. M., García J. L., Olivera E. R. (2001) Mol. Microbiol. 39, 1434–1442 [DOI] [PubMed] [Google Scholar]
- 6. Erb T. J., Ismail W., Fuchs G. (2008) Curr. Microbiol. 57, 27–32 [DOI] [PubMed] [Google Scholar]
- 7. Martínez-Blanco H., Reglero A., Rodriguez-Aparicio L. B., Luengo J. M. (1990) J. Biol. Chem. 265, 7084–7090 [PubMed] [Google Scholar]
- 8. Vitovski S. (1993) FEMS Microbiol. Lett. 108, 1–5 [DOI] [PubMed] [Google Scholar]
- 9. Olivera E. R., Miñambres B., García B., Muñiz C., Moreno M. A., Ferrández A., Díaz E., García J. L., Luengo J. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6419–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Teufel R., Mascaraque V., Ismail W., Voss M., Perera J., Eisenreich W., Haehnel W., Fuchs G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14390–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rather L. J., Knapp B., Haehnel W., Fuchs G. (2010) J. Biol. Chem. 285, 20615–20624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park S. J., Lee S. Y. (2003) J. Bacteriol. 185, 5391–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ismail W., El-Said Mohamed M., Wanner B. L., Datsenko K. A., Eisenreich W., Rohdich F., Bacher A., Fuchs G. (2003) Eur. J. Biochem. 270, 3047–3054 [DOI] [PubMed] [Google Scholar]
- 14. Schachter D., Taggart J. V. (1953) J. Biol. Chem. 203, 925–934 [PubMed] [Google Scholar]
- 15. Stadtman E. R. (1957) Methods Enzymol. 3, 931–941 [Google Scholar]
- 16. Decker K. (1959) Die aktivierte Essigsäure, pp. 84–89, Ferdinand Enke Publisher, Stuttgart, Germany [Google Scholar]
- 17. Gescher J., Ismail W., Olgeschläger E., Eisenreich W., Wörth J., Fuchs G. (2006) J. Bacteriol. 188, 2919–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shenoy A. R., Visweswariah S. S. (2003) Anal. Biochem. 319, 335–336 [DOI] [PubMed] [Google Scholar]
- 19. Bertani G. (1951) J. Bacteriol. 62, 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 21. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 22. Ellis K. J., Morrison J. F. (1982) Methods Enzymol. 87, 405–426 [DOI] [PubMed] [Google Scholar]
- 23. Erb T. J., Berg I. A., Brecht V., Müller M., Fuchs G., Alber B. E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10631–10636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Binstock J. F., Schulz H. (1981) Methods Enzymol. 71, 403–411 [DOI] [PubMed] [Google Scholar]
- 25. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 26. Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Sayers E. W. (2009) Nucleic Acids. Res. 37, D26–D31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 28. Saitou N., Nei M. (1987) Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 29. Rétey J., Umani-Ronchi A., Arigoni D. (1966) Experientia 22, 72–73 [DOI] [PubMed] [Google Scholar]
- 30. Boch J., Nau-Wagner G., Kneip S., Bremer E. (1997) Arch. Microbiol. 168, 282–289 [DOI] [PubMed] [Google Scholar]
- 31. Koski M. K., Haapalainen A. M., Hiltunen J. K., Glumoff T. (2004) J. Biol. Chem. 279, 24666–24672 [DOI] [PubMed] [Google Scholar]
- 32. Koski M. K., Haapalainen A. M., Hiltunen J. K., Glumoff T. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1302–1305 [DOI] [PubMed] [Google Scholar]
- 33. Hisano T., Tsuge T., Fukui T., Iwata T., Miki K., Doi Y. (2003) J. Biol. Chem. 278, 617–624 [DOI] [PubMed] [Google Scholar]
- 34. Holden H. M., Benning M. M., Haller T., Gerlt J. A. (2001) Acc. Chem. Res. 34, 145–157 [DOI] [PubMed] [Google Scholar]
- 35. Qin Y. M., Haapalainen A. M., Kilpeläinen S. H., Marttila M. S., Koski M. K., Glumoff T., Novikov D. K., Hiltunen J. K. (2000) J. Biol. Chem. 275, 4965–4972 [DOI] [PubMed] [Google Scholar]
- 36. Rost R., Haas S., Hammer E., Herrmann H., Burchhardt G. (2002) Mol. Genet. Genomics 267, 656–663 [DOI] [PubMed] [Google Scholar]
- 37. Thiel V., Brinkhoff T., Dickschat J. S., Wickel S., Grunenberg J., Wagner-Döbler I., Simon M., Schulz S. (2010) Org. Biomol. Chem. 8, 234–246 [DOI] [PubMed] [Google Scholar]
- 38. Cane D. E., Wu Z., Van Epp J. E. (1992) J. Am. Chem. Soc. 114, 8479–8483 [Google Scholar]
- 39. D'Alvise P. W., Melchiorsen J., Porsby C. H., Nielsen K. F., Gram L. (2010) Appl. Environ. Microbiol. 76, 2366–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brinkhoff T., Bach G., Heidorn T., Liang L., Schlingloff A., Simon M. (2004) Appl. Environ. Microbiol. 70, 2560–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Geng H., Bruhn J. B., Nielsen K. F., Gram L., Belas R. (2008) Appl. Environ. Microbiol. 74, 1535–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Geng H., Belas R. (2010) J. Bacteriol. 192, 4377–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Korth H., Brüsewitz G., Pulverer G. (1982) Zentralbl. Bakteriol. Mikrobiol. Hyg. A 252, 83–86 [PubMed] [Google Scholar]
- 44. Bentley R. (2008) Nat. Prod. Rep. 25, 118–138 [DOI] [PubMed] [Google Scholar]
- 45. Law R. J., Hamlin J. N., Sivro A., McCorrister S. J., Cardama G. A., Cardona S. T. (2008) J. Bacteriol. 190, 7209–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moore B. S., Walker K., Tornus I., Handa S., Poralla K., Floss H. G. (1997) J. Org. Chem. 62, 2173–2185 [DOI] [PubMed] [Google Scholar]
- 47. Perozich J., Nicholas H., Lindahl R., Hempel J. (1999) Adv. Exp. Med. Biol. 463, 1–7 [DOI] [PubMed] [Google Scholar]
- 48. Liu Z. J., Sun Y. J., Rose J., Chung Y. J., Hsiao C. D., Chang W. R., Kuo I., Perozich J., Lindahl R., Hempel J., Wang B. C. (1997) Nat. Struct. Biol. 4, 317–326 [DOI] [PubMed] [Google Scholar]
- 49. Vedadi M., Vrielink A., Meighen E. (1997) Biochem. Biophys. Res. Commun. 238, 448–451 [DOI] [PubMed] [Google Scholar]
- 50. Perozich J., Kuo I., Lindahl R., Hempel J. (2001) Chem. Biol. Interact. 130–132, 115–124 [DOI] [PubMed] [Google Scholar]
- 51. Perozich J., Kuo I., Wang B. C., Boesch J. S., Lindahl R., Hempel J. (2000) Eur. J. Biochem. 267, 6197–6203 [DOI] [PubMed] [Google Scholar]
- 52. Zhang L., Ahvazi B., Szittner R., Vrielink A., Meighen E. (1999) Biochemistry 38, 11440–11447 [DOI] [PubMed] [Google Scholar]
- 53. Steinmetz C. G., Xie P., Weiner H., Hurley T. D. (1997) Structure 5, 701–711 [DOI] [PubMed] [Google Scholar]
- 54. Vedadi M., Szittner R., Smillie L., Meighen E. (1995) Biochemistry 34, 16725–16732 [DOI] [PubMed] [Google Scholar]
- 55. Weiner H., Farrés J., Rout U. J., Wang X., Zheng C. F. (1995) Adv. Exp. Med. Biol. 372, 1–7 [DOI] [PubMed] [Google Scholar]
- 56. Farrés J., Wang T. T., Cunningham S. J., Weiner H. (1995) Biochemistry 34, 2592–2598 [DOI] [PubMed] [Google Scholar]
- 57. Hempel J., Nicholas H., Lindahl R. (1993) Protein Sci. 2, 1890–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang L., Ahvazi B., Szittner R., Vrielink A., Meighen E. (2000) Biochemistry 39, 14409–14418 [DOI] [PubMed] [Google Scholar]
- 59. Zhang L., Ahvazi B., Szittner R., Vrielink A., Meighen E. (2001) Chem. Biol. Interact. 130–132, 29–38 [DOI] [PubMed] [Google Scholar]
- 60. Ahvazi B., Coulombe R., Delarge M., Vedadi M., Zhang L., Meighen E., Vrielink A. (2000) Biochem. J. 349, 853–861 [PMC free article] [PubMed] [Google Scholar]
- 61. Bains J., Boulanger M. J. (2008) J. Mol. Biol. 379, 597–608 [DOI] [PubMed] [Google Scholar]
- 62. Sugino H., Sasaki M., Azakami H., Yamashita M., Murooka Y. (1992) J. Bacteriol. 174, 2485–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Díaz E., Ferrández A., Prieto M. A., García J. L. (2001) Microbiol. Mol. Biol. Rev. 65, 523–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cobessi D., Tête-Favier F., Marchal S., Branlant G., Aubry A. (2000) J. Mol. Biol. 300, 141–152 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.