Abstract
We characterized the effect of histone acetylation on the structure of a nucleosome and the interactions between two nucleosomes. In this study, nucleosomes reconstituted with the Selex “Widom 601” sequence were acetylated with the Piccolo NuA4 complex, which acetylates mainly H4 N-terminal tail lysine residues and some H2A/H3 N-terminal tail lysine residues. Upon the acetylation, we observed directional unwrapping of nucleosomal DNA that accompanies topology change of the DNA. Interactions between two nucleosomes in solution were also monitored to discover multiple transient dinucleosomal states that can be categorized to short-lived and long-lived (∼1 s) states. The formation of dinucleosomes is strongly Mg2+-dependent, and unacetylated nucleosomes favor the formation of long-lived dinucleosomes 4-fold as much as the acetylated ones. These results suggest that the acetylation of histones by Piccolo NuA4 disturbs not only the structure of a nucleosome but also the interactions between two nucleosomes. Lastly, we suggest a structural model for a stable dinucleosomal state where the two nucleosomes are separated by ∼2 nm face-to-face and rotated by 34° with respect to each other.
Keywords: Chromatin Histone Modification, Chromatin Structure, Fluorescence Resonance Energy Transfer (FRET), Nucleosome, Single Molecule Biophysics
Introduction
A nucleosome core particle comprising ∼147-bp DNA wrapped in ∼1.65 superhelical turns around a histone octamer core is the basic subunit of chromatin (1). Individual nucleosome core particles are connected to its neighbors through a ∼10–50-bp linker DNA fragment forming beads-on-a-string nucleosomal arrays, which fold into 30-nm-thick fibers and can oligomerize to form extensively condensed chromatin (2, 3). It has been hypothesized that the structure and structural dynamics of nucleosomes and chromatin are tied to gene regulation.
A core histone octamer is composed of two H2A-H2B dimers and a (H3-H4)2 tetramer. Histones are targets for several post-translational modifications that are linked to various gene regulatory activities (4–6). Most of these modifications are found on the unstructured tail regions of histones that account for ∼28% of an octamer core and are extremely basic because of the high content of lysine and arginine residues (7). The N-terminal tails of histones are of particular interest because they may be in contact with DNA as well as with other histones across nucleosomes and consequently contribute to nucleosome packaging in chromatin (8–10).
All four histone tails contain highly conserved lysine residues serving as targets for various histone acetyltransferases, which transfer an acetyl group from acetyl-CoA to the nitrogen atom of the amine group in a lysine residue (11, 12). The Piccolo NuA4 is a subcomplex of nucleosome acetyltransferases of H4, which is the only histone acetyltransferase required for cell viability in yeast (13). The role of H4 acetylation has been under extensive study, and the prevailing consensus is that it is involved in gene activation mechanisms (14–16). NuA4 is also linked to transcription regulation and DNA damage repair and likely responsible for the global acetylation of H4 in yeast (17, 18). The Piccolo NuA4 complex consists of three subunits, Epl1, Yng2, and Esa1, and acetylates all four lysine residues of H4 N-terminal tail and one lysine residues of H2A and H3 tails in both naked histones and nucleosomes (17, 19). A mutagenesis study has found that all four lysine residues in the H4 tail are involved in maintaining the genome integrity (20). In addition, acetylation of H4 tails on newly synthesized histone H4 is conserved between Drosophila and humans (21), suggesting that it plays an important role in nucleosome assembly during replication. Acetylation of some H4 tail residues is associated with the regulation of chromatin structure. In particular, most of the genome in budding yeast showed decondensed chromatin state with over 80% of H4K16 acetylated (22, 23), and acetylation of H4K16 led to the disruption of H4 tail-mediated oligomerization of nucleosomes in vitro (9, 24). These reports suggest that the H4 tail is involved in internucleosomal interactions that can control the chromatin structure and eventually regulate the activity of genes.
There are two predominant hypotheses for the mechanism of gene activation by histone acetylation (25). First, acetylated residues serve as binding sites for regulatory proteins including histone chaperones, chromatin modification enzymes, and transcriptional regulatory proteins (12, 26). Second, histone acetylation alters the structure and/or structural dynamics of nucleosomes and/or chromatin, resulting in increased DNA accessibility (12, 27, 28). One of the proposed mechanisms to explain the structural changes induced by histone acetylation implicates the reduction of DNA-histone contacts by neutralizing the positive charges on histones upon acetylation of the lysine residues. Although multiple studies have reported that histone acetylation induces small changes in the structure of nucleosomes based on ensemble-averaging measurements, the nature of the structural changes has never been clearly addressed.
The basic N-terminal tails of histone H4 (residues 14–19) from one nucleosome interact with acidic patches of another nucleosome mainly on the H2A (1, 10, 19, 29), raising the possibility that acetylation of H4 tails will have an impact on internucleosomal interactions between two nucleosomes and consequently on the structure of chromatin. There are several models proposed for the structure of chromatin that can be categorized into two groups: one-start helices and two-start helices (30, 31). In both groups of models, interactions between two nucleosomes are the starting points of nucleosome packaging in chromatin. Thus, studying the interactions between two nucleosomes with and without H4 acetylation is an essential step toward the understanding of chromatin structure and changes in chromatin structure upon H4 acetylation. Mapping of internucleosomal protein-DNA interactions in dinucleosomes using a photoactivated cross-linker has been reported by Zheng and Hayes (32). However, the dynamics of interactions between two nucleosomes or the effect of histone acetylation on the dynamics of internucleosomal interactions has never been reported to date. This deficiency is largely due to the difficulty associated with constructing an experimental system with which one can monitor the dynamic and transient dinucleosomal states in solution.
In this article, we report the effect of histone acetylation by the Piccolo NuA4 complex on (i) the structure of nucleosomes and (ii) the dynamic interactions between two nucleosomes using single molecule FRET and ensemble fluorescence anisotropy. We also propose a structural model for a dinucleosomal state in solution. Our results make a unique and valuable contribution to the understanding of the structure and structural dynamics of chromatin and how they are altered by histone acetylation.
EXPERIMENTAL PROCEDURES
DNA Preparation and Reconstitution to Nucleosomes
A fluorescently labeled 147-bp dsDNA fragment with an additional 10-base linker sequence was generated by ligation using T4 DNA ligase. Several pieces of oligonucleotides (36–78-mer) were designed to contain biotin and Cy3 or Cy5 (Integrated DNA Technologies, Coraville, IA) near the 5′ terminus. The oligonucleotides were annealed to dsDNA by heating and cooling down from 95 to 5 °C for 18 min followed by purification using a PCR purification kit (Qiagen). The annealed dsDNA is treated with T4 ligase (New England Biolabs) for 16 h at 16 °C. The ligated 147-bp DNA fragment after purification with a PCR purification kit was used to assemble nucleosomes. A control 147-bp DNA fragment was prepared by enzyme digestion of a plasmid containing a Widom 601 sequence with EcoRV restriction sites at both ends (Integrated DNA Technologies) (supplemental materials). The restriction product 147-bp control DNA was purified before use. For DNA denaturing gel electrophoresis, DNAs were denatured in 7 m urea for 2 min at 90 °C and subsequently loaded on a polyacrylamide gel containing 7 m urea in 0.5× TBE buffer. DNAs were run on the gel for 60 min at 100 V. His6-yeast NAP1 histone chaperone (yNAP1) was expressed and purified by nickel affinity as described in Ref. 33, and Xenopus laevis histone octamer core particles were provided by Dr. Song Tan (Penn State University). Histone octamers and yNAP1 were incubated at 30 °C for 4 h before adding labeled DNA fragment and incubating them for an additional 4 h at 30 °C. The reactions were analyzed with 5% native polyacrylamide gel electrophoresis in 1× TBE buffer.
Acetylation of Nucleosomes or Histones by Piccolo NuA4 Complex
The Piccolo NuA4 complex composed of Esa1, Yng2, and Epl1 was coexpressed and purified using the polycistronic expression system developed by Tan et al. (34). Briefly, the complex was expressed in BL21(DE3)pLysS grown in LB medium to A600 = 0.7. The expression was induced with 0.4 mm isopropyl β-d-thiogalactopyranoside. After 4 h of induction at 37 °C, the cells were harvested by centrifugation and purified with a nickel affinity column in a buffer containing 50 mm sodium phosphate at pH 7.5, 300 mm NaCl, 5 mm β-mercaptoethanol, and 100 mm imidazole. Further purified proteins were dialyzed against 50 mm Tris-HCl at pH 7.5, 300 mm NaCl, and 5 mm β-mercaptoethanol to remove excess imidazole. The activity of the purified Piccolo NuA4 complex in the presence of radioactively labeled acetyl-CoA and histone octamers was confirmed using a filter binding assay as described (35). Briefly, the assays were performed either in 1× TBA (50 mm Tris, 50 mm Bis-Tris, and 100 mm sodium acetate) at pH 7.0 or in the imaging buffer, which is used for single molecule measurements of nucleosome structure (10 mm HEPES, pH 7.6, 50 mm NaCl, 50 mm KCl, and 3% glycerol). Both reactions contained 0.1–0.4 μCi of [3H]acetyl-CoA, 0.5 μm of Piccolo NuA4, and 1 μg of histone octamers and displayed the same level of activity as previously reported.
To observe the acetylation effect on the structure of nucleosomes, purified Piccolo NuA4 complexes were incubated (0.5 μm) with the surface immobilized nucleosomes for 30 min in the imaging buffer supplemented with 0.5 μm acetyl-CoA. On the other hand, to prepare the acetylated histones to reconstitute acetylated nucleosomes, 20 μg of histone octamers were acetylated with 5 μm of Piccolo NuA4 in 1× TBA buffer in the presence of 10 μm acetyl-CoA for 30 min incubation at 30 °C to be tetra-acetylated on H4 and mono-acetylated on either H3 or H2A as previously reported (17). This mixture was then loaded on nickel-nitrilotriacetic acid column and washed twice with the same buffer containing 250 mm NaCl. The acetylated histones were eluted with 2 m NaCl. We repeated the acetylation with [3H]acetyl-CoA instead of unlabeled acetyl-CoA and confirmed that eluted histones were acetylated based on the radioactive filter binding assay. The acetylated histones eluted with 2 m NaCl were diluted to 250 mm NaCl and concentrated using a centrifugal filter device (Amicon filter; Millipore) at 7500 × g for 80 min at 4 °C.
Fluorescence Anisotropy Measurements
The reconstituted 601A or 601B nucleosomes (∼40 nm) were incubated with or without Piccolo NuA4 complex (0.4 μm) and acetyl-CoA(4 μm) for 30 min at 30 °C in a buffer containing 10 mm HEPES (pH 7.6), 80 mm NaCl, 0.5 mm MgCl2, 1 mm DTT, and 3% glycerol. The fluorescence anisotropy was measured at excitation/emission 532 nm/570 nm for Cy3, 600/670 for Cy5, and 532/670 for Cy5 excited via FRET using a commercial fluorimeter (Fluoromax4; Horiba). The anisotropy r = (IVV − GIVH)/(IVV + 2GIVH) was calculated from the parallel IVV and perpendicular IVH polarized fluorescence intensities with the correction factor G for the setup.
Single Molecule FRET Measurements
The surface of a quartz microscope slide was silanized and subsequently coated with PEG (Laysan Bio, Arab, AL). One of 100 PEG molecules is biotinylated (Laysan Bio). We passivated the surface with BSA before immobilizing nucleosomes on the surface via streptavidin-biotin conjugation. To observe the acetylation effect on the structure of nucleosomes, FRET donor (Cy3)- and acceptor (Cy5)-labeled nucleosomes were immobilized on a surface-treated slide through streptavidin-biotin conjugation followed by incubation with purified Piccolo NuA4 complexes (0.5 μm) for 30 min in the imaging buffer, and 0.5 μm acetyl-CoA. To observe nucleosome-nucleosome interactions, Cy3-labeled nucleosomes (601C; see Fig. 5A) were immobilized on a surface-treated slide, and Cy5-labeled nucleosomes (601D or 601E; see Fig. 5, B and C) were injected onto the slide. To prevent Cy5-labeled nucleosomes from binding on the surface, we fully blocked biotin-binding sites of the streptavidin by incubating excess biotin on the surface before injecting Cy5-labeled nucleosomes. The fluorescence signals were taken within 1 h after nucleosome immobilization to avoid unfolding of nucleosomes. A protocatechuate dioxygenase (Sigma-Aldrich) and protocatechuic acid (Sigma-Aldrich) mixture in addition to Trolox (2 mm) was used to elongate the dye photobleaching lifetime and stabilize the emission.
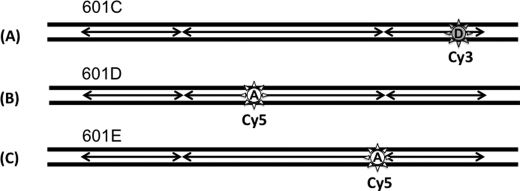
FIGURE 5.
Schematic representation of fluorophore positions on a 601 DNA fragment to investigate internucleosomal interactions. A, a 601C nucleosome was labeled with a donor Cy3 dye at the 129th base. B, a 601D nucleosome was labeled with an acceptor Cy5 dye at the 60th base from the left termini. C, an acceptor Cy5 dye was labeled at the 96th base of a 601E nucleosome.
Fluorescence signals from single Cy3 and Cy5 fluorophores were imaged on an electron multiplying CCD camera (EMCCD, iXon+897; Andor Technology, Belfast UK) in a prism coupled total internal reflection geometry based on a commercial microscope (TE2000; Nikon, Tokyo, Japan) with modifications. The excitation of the FRET donor Cy3 was achieved with a laser beam at 532 nm (Laser Quantum) that is perpendicularly polarized along the total internal reflection incidence. Fluorescence emission from FRET pairs (Cy3 and Cy5) were spectrally separated into two regions (550–645 and 645–750 nm) with a dichroic mirror, and the two spectrally separated images of Cy3 and Cy5 at single nucleosomes were projected on a single EMCCD chip to simultaneously collect fluorescence signals from the two fluorophores in a time-resolved manner by recording a series of fluorescence images at a frame rate of ms (i.e. a movie in which each frame is a fluorescence image with 35 ms of signal integration time). The intensities of Cy3 and Cy5 fluorescence at a given time point are the brightness of the corresponding pixels on the movie frame at the time point. The time series of fluorescence intensities of Cy3 and Cy5 were obtained from the series of fluorescence images contained in a movie and were plotted against the elapsed time (see Fig. 6A). FRET efficiency at each time point was calculated with a formula, Icy5/(Icy3 + Icy5). To construct the FRET efficiency distribution histogram of a FRET state, we manually selected regions in the FRET time traces that show non-zero FRET efficiencies as carefully as possible. The FRET efficiency values in the selected regions were combined together to build a histogram that shows the distribution of FRET efficiency values. To construct the lifetime histogram of a FRET state (a dinucleosomal state), we manually selected regions in a FRET time trace that represent a dinucleosomal state. We measured the durations of each selected region and counted one selected region as one event with the measured duration. These events with their measured durations were constructed as the lifetime histogram of a dinucleosomal state.
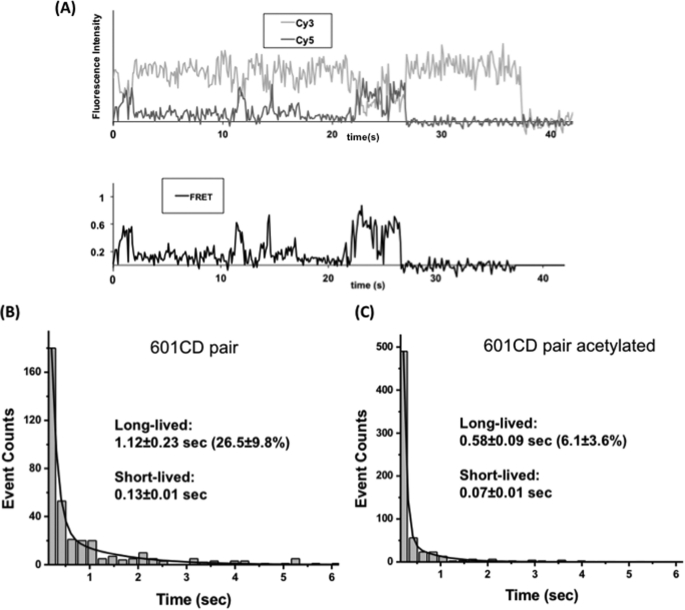
FIGURE 6.
Internucleosomal interactions between 601C and 601D nucleosomes. A, a typical FRET trace displaying the internucleosomal interactions between 601C and 601D nucleosomes. B, a histogram of the lifetimes of dinucleosomal states formed by unacetylated 601CD pairs. C, a histogram of the lifetimes of dinucleosomal states formed by 601CD pairs acetylated by Piccolo NuA4.
RESULTS
DNA Fragment Preparation and Reconstitution of Nucleosomes Using yNAP1 Protein
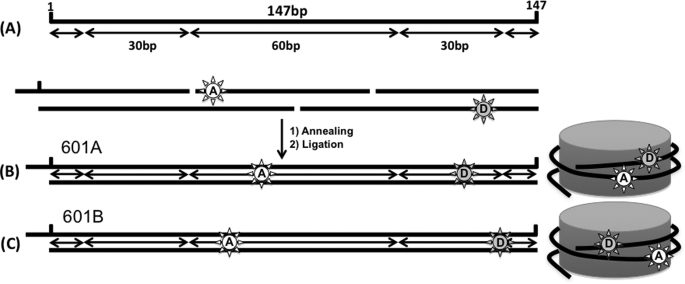
A 147-bp DNA fragment was labeled with a FRET pair: a Cy3 (donor fluorophore) around where the DNA contacts H2A/H2B and a Cy5 (acceptor fluorophore) around where the DNA contacts H3/H4. The two dyes are 65–83 bp apart within the DNA fragment. We designed two nucleosomal DNA fragments based on the Widom 601 sequence: 601A DNA fragment labeled at the 60th and 22th bases from each termini, respectively, and 601B DNA fragment labeled at the 53th and 11th bases from each termini, respectively (Fig. 1). Based on a crystal structure (Protein Data Bank code 1AOI), their intrinsic FRET efficiencies are approximately in the range of 0.5–0.7, which is appropriate for detecting DNA motion.
FIGURE 1.
Nucleosomal DNA constructs based on the Widom 601 sequence to study the effect of histone acetylation by Piccolo NuA4. The filled stars represent Cy3 (FRET donor, D), and the empty stars represent Cy5 (FRET acceptor, A). A, the 147-bp DNA fragment is comprised of ∼60 bp of H3/H4 tetramer-binding site in the center and ∼30 bp of H2A/H2B dimer-binding sites near the termini. B, 601A DNA fragment with an additional 10-base single-stranded DNA at one end is labeled with a FRET pair (Cy3 at the 125th base and Cy5 at the 60th). The base numbers are counted from the left end of the 147-bp nucleosome positioning sequence. C, 601B DNA fragment with the same DNA sequence as 601A is labeled with a FRET pair at different locations (Cy3 at the 136th base and Cy5 at the 53th base).
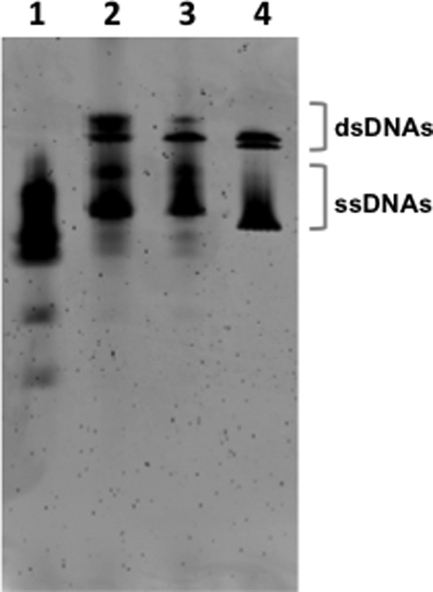
To construct the nucleosomal DNA, we ligated several 40–80 base oligonucleotides, two of which are labeled with Cy3 or Cy5 (Integrated DNA Technologies). The ligation was confirmed with denaturing DNA-PAGE (Fig. 2). Fig. 2 shows one major single-stranded DNA for the ligated DNAs and the control DNA (lanes 2–4). Multiple major bands were observed for the annealed but unligated DNA (lane 1). Some of the single-stranded DNAs in the ligated DNAs and the control DNA were renatured during electrophoresis because the running buffer did not contain urea. The double band for the dsDNA region of the control DNA (lane 4) might be due to two different conformations of dsDNA created during the renaturation process in the gel. The slightly slow migration of ligated DNAs as compared with the control DNA is due to the extra 10 oligonucleotides at the end and the fluorophores attached to the DNAs. The ligation products were purified with a PCR purification kit before nucleosome reconstitution.
FIGURE 2.
Denaturing DNA polyacrylamide gel image. Denaturing polyacrylamide gel electrophoresis was performed to monitor the ligation steps. Several pieces of oligonucleotides were first annealed to make a 147-bp dsDNA fragment with an additional 10-base single strand at the N terminus (lane 1: before ligation) and then ligated with Taq DNA ligase (lane 2) or T4 DNA ligase (lane 3) to seal the nicks on the DNA construct. Ligated DNA fragments were compared with the 147-bp 601 DNA, which was restricted from a plasmid containing a 601 sequence (see “Experimental Procedures” for detailed preparation) (lane 4). ssDNA, single-stranded DNA.
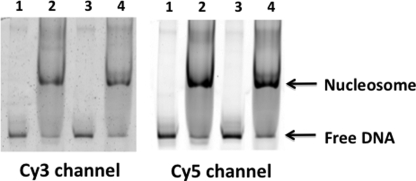
The purified DNA ligation products were reconstituted into nucleosomes using yNAP1 histone chaperone and analyzed in a 5% native-PAGE (Fig. 3) (36). Nucleosomes containing the 147-bp DNA displayed one major band, some minor amounts of higher molecular weight species, and free DNAs. The existence of only one major band indicates no significant positioning heterogeneity. Free DNA without histones does not yield any FRET because the distance between Cy3 and Cy5 is longer than 10 nm for each DNA construct. To filter out signal from oligomer nucleosomes, we analyzed data only from nucleosomes with single photobleaching steps of the two fluorophores (i.e. Cy3 and Cy5). A single photobleaching step in a fluorescence intensity time trace is a signature of a single fluorophore. Similar intensities of the major bands were observed for both constructs, indicating that the reconstitution efficiencies in the two sequences were similar.
FIGURE 3.
5% native polyacrylamide gel images of nucleosomes. The nucleosomes were reconstituted with yeast nucleosome assembly protein 1 (yNAP1). Lane 1, 601A DNA; lane 2, 601A nucleosome; lane 3, 601B DNA; lane 4, 601B nucleosome.
Histone Acetylation by Piccolo NuA4 Results in Unwrapping of Nucleosomal DNA through Unwinding
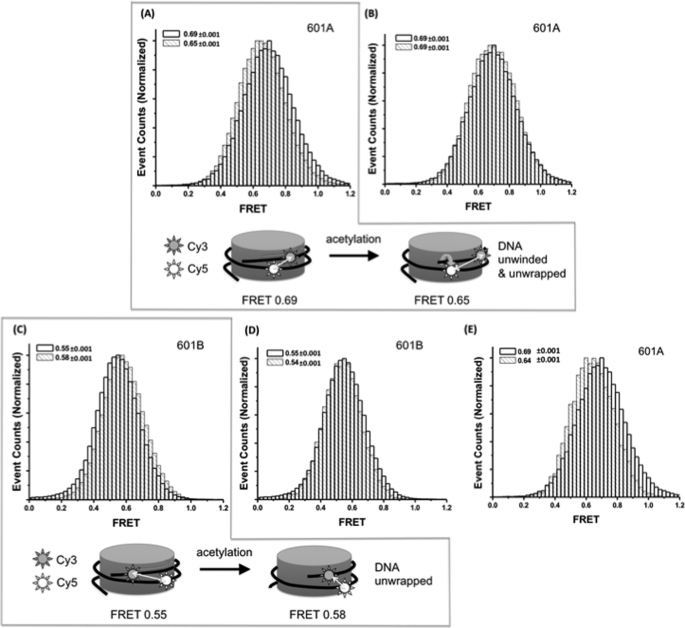
Fig. 4 (A and C) shows the FRET efficiency distributions of the two nucleosomes (601A and 601B). Consistent with the native PAGE analysis, both 601A and 601B nucleosomes showed one major intrinsic FRET state, whose average FRET efficiency is 0.69 and 0.55 for 601A and 601B, respectively. We observed some other minor FRET states whose FRET efficiencies are far from the major FRET state. These minor states may represent noncanonical nucleosomal states or nonstandard translational settings. Data from these populations and data with a low signal-to-noise ratio (lower than 5.0) were excluded from the analysis. To examine whether histone acetylation by Piccolo NuA4 modulates the nucleosome structure, nucleosomes were acetylated by incubating the nucleosomes with the Piccolo NuA4 complex in the presence of acetyl-CoA for 30 min. The efficiency of acetylation reaction was verified separately by an independent radioactivity assay (35). After the acetylation, the FRET efficiency of 601A decreased from 0.69 to 0.65, whereas the FRET efficiency of 601B increased from 0.55 to 0.58. There are two possible scenarios to account for these FRET changes. The first is directional unwrapping of DNA without changes in DNA topology, and the second scenario is partial detachment of DNA from the histone core that likely accompanies DNA unwrapping and/or DNA topology changes.
A major difference between the two scenarios is the change in DNA topology. The first scenario does not yield any changes in the DNA topology, whereas the second scenario does. Any topology changes of DNA would be seen as the relative dipole orientation changes of the FRET pair in our system. This is because the cyanine dyes in our system do not rotate freely (experimental justification is following) and consequently maintain a fixed dipole orientation with respect to DNA. To monitor the dipole orientation of the fluorophores, we measured fluorescence anisotropy values of Cy3 and Cy5 in 601A and 601B nucleosomes before and after the acetylation. The results are listed in Table 1. Cy3 and Cy5 in 601A or 601B showed much higher anisotropy than free dyes, and their anisotropy values are close to the maximum possible value 0.4, both of which strongly suggest that the dyes do not rotate on DNA and maintain fixed dipole orientation to the DNA (more discussion in supplemental materials). The anisotropy of Cy3 or Cy5 in 601A or 601B does not change upon the acetylation within the precision of our measurements. However, acetylated 601A nucleosomes displayed decreased anisotropy of Cy5 excited via FRET compared with the control 601A nucleosomes, which are unacetylated nucleosomes or nucleosomes treated with Piccolo NuA4 in the absence of acetyl-CoA (CTL-NUC and CTL-PCL). Based on the anisotropy change, the change in the angle between Cy3 and Cy5 dipoles upon the acetylation was calculated to be 4.6° (42.4–37.8 = 4.6; Table 1 and supplemental materials for the detailed explanation and calculation procedure). This result indicates that the acetylation induces a change in the DNA topology such as DNA twist or writhe. The relative dipole orientation of Cy3 and Cy5 with each other (β in Table 1) is related to the orientation factor κ2 in FRET. Taking this factor into account, we can calculate how much FRET change we can expect solely from the DNA topology change induced by the acetylation (detailed calculation procedure in supplemental materials). By comparing the observed FRET change to the calculated FRET change induced by DNA topology change, we can deduce whether the observed FRET change is solely from DNA topology change or whether it also includes effect of FRET distance change that can be attributed to DNA unwrapping. Because we have too many unknown parameters to calculate the absolute distances between FRET pairs, we calculated only the ratio between FRET distances (r1/r2) in the 601A nucleosomes before (r1) and after (r2) acetylation. As shown in Table 2, r1/r2 for 601A was below 1, indicating that the distance between Cy3 and Cy5 increased upon the acetylation. This change suggests DNA unwrapping upon the acetylation.
TABLE 1.
Fluorescence anisotropy of Cy3 and Cy5 labeled at the 601A and 601B nucleosomes
Anisotropy was measured with the excitation wavelength of 532 nm (for Cy3) or 600 nm (for Cy5) and emission wavelength of 570 nm (for Cy3) or 670 nm (for Cy5). For FRET anisotropy (i.e. Cy5 emission anisotropy when it was excited via FRET) was measured at 670-nm emission with 532-nm excitation. The wavelength range was ±2.5 nm for excitations (Ex) and ±14 nm for emissions (Em). β is the average angle between the emission dipole of Cy3 and the absorption dipole of Cy5 (see the supplemental materials). β was calculated based on the measured anisotropy (r) using the formula r = 2/5{(3cos2 β − 1)/2}. The detailed calculation procedure is described in the supplemental materials. CTL-NUC, unacetylated nucleosomes; CTL-PCL, unacetylated nucleosomes incubated with Piccolo NuA4 complex in the absence of acetyl-CoA; PCL, acetylated nucleosomes incubated with Piccolo NuA4 complex in the presence of acetyl-CoA.
| Ex/Em | 601A |
601B |
||||||
|---|---|---|---|---|---|---|---|---|
| 532/570 (Cy3) | 600/670 (Cy5) | 532/670 (Cy5 via FRET) | β | 532/570 (Cy3) | 600/670 (Cy5) | 532/670 (Cy5 via FRET) | β | |
| Free | 0.200 ± 0.003 | 0.130 ± 0.004 | 0.200 ± 0.003 | 0.130 ± 0.004 | ||||
| CTL-NUC | 0.314 ± 0.011 | 0.329 ± 0.009 | 0.140 ± 0.011 | 37.8 ± 1.0 | 0.338 ± 0.010 | 0.328 ± 0.015 | 0.078 ± 0.008 | 45.7 ± 0.5 |
| CTL-PCL | 0.321 ± 0.008 | 0.343 ± 0.013 | 0.144 ± 0.011 | 37.9 ± 1.0 | 0.337 ± 0.014 | 0.337 ± 0.013 | 0.078 ± 0.011 | 45.7 ± 0.9 |
| PCL | 0.319 ± 0.010 | 0.347 ± 0.012 | 0.105 ± 0.010 | 42.4 ± 0.9 | 0.347 ± 0.015 | 0.331 ± 0.014 | 0.078 ± 0.012 | 45.8 ± 1.0 |
TABLE 2.
FRET efficiencies of acetylated and unacetylated nucleosomes and the FRET distance ratio (r1/r2) of unacetylated nucleosomes (r1) to acetylated nucleosomes (r2)
E2 is the FRET efficiency of 601A or 601B nucleosomes upon incubation with Piccolo NuA4 complex in the presence of acetyl-CoA (Fig. 4, A and C). E1 is the FRET efficiency without Piccolo NuA4 complex in the same condition as in E2 (Fig. 4, B and D). The detailed calculation procedure is described in the supplemental materials.
| E2 | E1 | E2 − E1 | r1/r2 | |
|---|---|---|---|---|
| 601A | 0.65 ± 0.001 | 0.69 ± 0.001 | −0.04 ± 0.001 | 0.992 ± 0.006 |
| 601B | 0.58 ± 0.001 | 0.55 ± 0.001 | 0.03 ± 0.001 | 1.021 ± 0.007 |
These results suggest partial detachment of DNA from histones through unwinding that results in DNA unwrapping and DNA topology change. This suggestion is supported by the acetylation-induced writhe of DNA resulting in the release of negative supercoils of the nucleosomal DNA (37, 38). On the other hand, in case of 601B, no change upon the acetylation was observed in the emission anisotropy of Cy5 excited via FRET, although the FRET efficiency increased. This lack of DNA topology change upon the acetylation in 601B may be because the Cy3 in 601B is near the end of DNA fragment, and consequently the extent of supercoiling of the Cy3 region of DNA was too low to be detected within the precision of our measurements. The r1/r2 for 601B is above 1, indicating that the distance between Cy3 and Cy5 was decreased upon the acetylation. Although there has not been any evidence of yNAP1 participating in the mechanism of gene activation by histone acetylation, we cannot completely rule out the possibility of yNAP1 contributing to the structural changes of nucleosomes we observed.
Next, to rule out the simple binding of Piccolo NuA4 to nucleosomes as a source of the DNA structural changes, control measurements in the absence of acetyl-CoA were performed. The results are shown in Fig. 4 (B and D), which demonstrates no or negligible changes. In addition, Fig. 4E shows the difference in the structure of nucleosomal DNA before and after the acetylation by Piccolo NuA4 without the Piccolo NuA4 bound to the nucleosome. To obtain the acetylated nucleosomes without the Piccolo NuA4 complex bound, we reconstituted the nucleosomes with the histones that had already been acetylated with Piccolo NuA4. The complex bound to the histones was removed from the histones by salt wash as described under “Experimental Procedures.” As shown in Fig. 4E, the acetylation effect without Piccolo NuA4 bound to the nucleosome was nearly identical to those in Fig. 4A where the Piccolo NuA4 complex is likely still bound to the nucleosome, suggesting that the changes in FRET upon histone acetylation by Piccolo NuA4 are due to directional unwrapping of nucleosomal DNA around the histone core.
FIGURE 4.
Effect of histone acetylation by Piccolo NuA4 on the structure of nucleosomes. Dye-labeled nucleosomes were incubated without (empty bar with thicker boarder) or with (light striped bar) Piccolo NuA4 for 30 min at 25 °C. A, FRET efficiency histograms of 601A nucleosomes upon incubation with or without Piccolo NuA4 in the presence of acetyl-CoA. B, FRET efficiency of 601A nucleosomes was decreased from 0.69 to 0.65 upon incubation, whereas there was no change in FRET before and after the incubation with the Piccolo NuA4 complex in the absence of acetyl-CoA. C, 601B nucleosomes displayed increased FRET efficiency from 0.55 to 0.58 upon incubation with Piccolo NuA4 in the presence of acetyl-CoA. D, only a minimal change in FRET was observed when nucleosomes were incubated with Piccolo NuA4 in the absence of acetyl-CoA. E, the same level of FRET change as in A was observed from 601A nucleosomes reconstituted with histones preacetylated with Piccolo NuA4 (i.e. no complex bound to the nucleosomes).
Interactions between Two Nucleosomes Lead to the Formation of Transient Dinucleosomal States
We tested whether two nucleosomes can form a dinucleosome without any linker DNA and accessory proteins. We constructed two sets of identical 601 sequence nucleosomes with one set labeled with Cy3 at the 129th base of the nucleosomal DNA (601C) and the other labeled with Cy5 at the 60th base (601D) (Fig. 5). The Cy3-labeled nucleosomes were immobilized on the surface of a microscope slide, and Cy5-labeled nucleosomes were injected onto the slide. In the absence of Mg2+ ions (10 mm Tris-HCl, pH 7.5, and 10 mm KCl), no interaction between two nucleosomes was observed. When we added Mg2+ ions (3 mm MgCl2), we observed various FRET states that are mostly transient (Fig. 6A). The lifetime analysis of these dinucleosomal states revealed that there are short-lived and long-lived (several hundred milliseconds or longer) states. The first column of a lifetime histogram was excluded from data analysis because there are always undetectable shortest-lived FRET events, which makes the lifetime measurements inaccurate. Fig. 6B shows a double exponential decay of the lifetimes of dinucleosomes, where 26.5 ± 9.8% of the total events belong to the long-lived (1.12-s lifetime) component.
Histone Acetylation by Piccolo NuA4 Inhibits the Formation of the Long-lived Dinucleosomal States and Reduces the Stability of Dinucleosomal States
We proceeded to study the effect of histone acetylation by the Piccolo NuA4 complex on the interactions between two nucleosomes. Fig. 6C shows the double exponentially decaying lifetime of the dinucleosomal states upon the acetylation, where only 6.1 ± 3.6% of the total events belong to the long-lived (0.58 s) component. As the result indicates, the stability of the long-lived states is reduced (1.12 s versus 0.58 s), and the formation of the long-lived states is inhibited (26.5% versus 6.1%) upon acetylation.
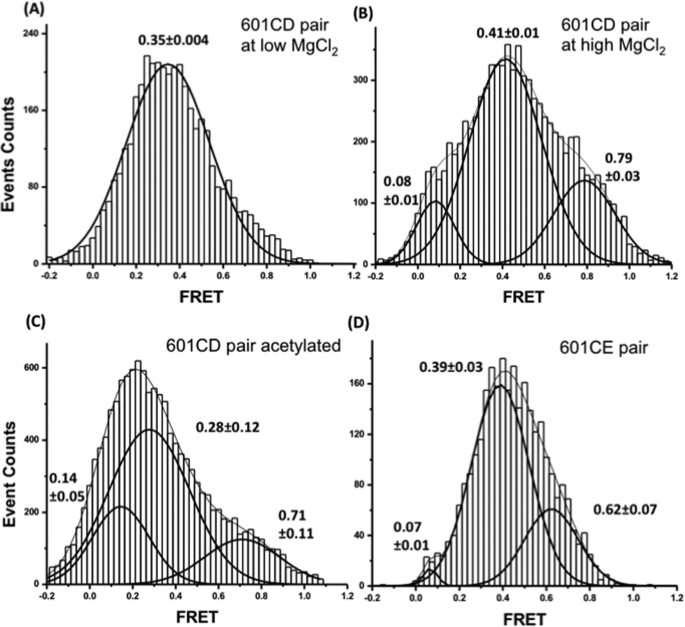
Next, we determined the FRET efficiency of the long-lived dinucleosomal states. We collected fragments of FRET traces only from dinucleosomal states that lived longer than the lifetime of the long-lived state (∼1 s), and constructed a histogram of their FRET efficiencies. As shown in Fig. 7A, at 1–2 mm Mg2+ concentrations, only one FRET state was observed around FRET efficiency 0.35. As we increased the Mg2+ concentration to 3 mm, FRET efficiencies of the long-lived unacetylated dinucleosomes had at least two distinct non-zero FRET peaks, which we designated as the low FRET peak (0.41) and the high FRET peak (0.79) (Fig. 7B). These results suggest that two nucleosomes in solution form multiple long-lived dinucleosomal states, and the high FRET state corresponds to the most compact dinucleosomal structure. Nucleosomes with their histones acetylated by Piccolo NuA4 also show two non-zero FRET states (Fig. 7C). The FRET efficiency peak positions of the low and high FRET states of the acetylated dinucleosomes are lower than those of the unacetylated nucleosomes, although the difference is within error. Nonetheless, the overall shape of the FRET efficiency histogram from the acetylated nucleosomes is shifted significantly toward lower FRET levels as compared with the unacetylated nucleosomes, which is evident in Fig. 7. These results suggest that acetylation of histones by Piccolo NuA4 reduces the compaction of the long-lived dinucleosomal states.
FIGURE 7.
FRET efficiency distributions of long-lived dinucleosomal states. A, unacetylated 601CD pairs show only one dinucleosomal state at 1 and 2 mm [MgCl2] (results from the two concentrations were combined together to construct one histogram). B, unacetylated 601CD pairs form at least two dinucleosomal states (0.41 and 0.79 FRET states, respectively) at 3 mm [MgCl2]. C, 601CD pairs acetylated with Piccolo NuA4 complex also form at least two different dinucleosomal states at 3 mm [MgCl2] (0.28 and 0.71 FRET states, respectively). D, a FRET efficiency distribution of long-lived FRET events from 601CE pairs revealed at least two different dinucleosomal states at 3 mm [MgCl2] (0.39 and 0.62 FRET states, respectively).
Nucleosomes Are Rotated with Respect to Each Other in a Long-lived Dinucleosome
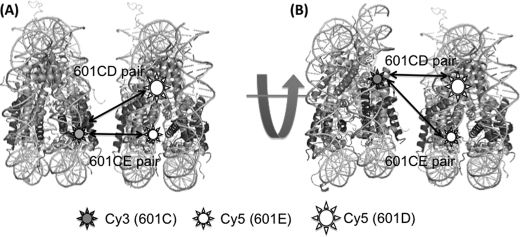
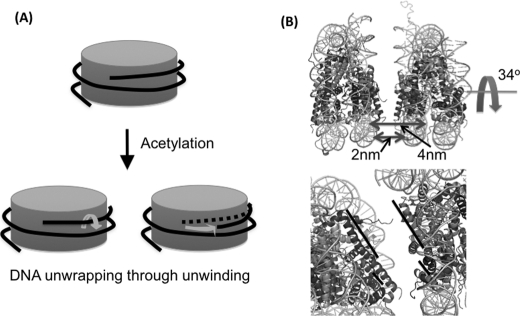
We studied how two nucleosomes stack onto each other when they form a long-lived dinucleosome. We made a nucleosome with the same DNA sequence that has an acceptor dye labeled at the 96th base (601E; Fig. 5C). The FRET efficiency between 601C and 601E must be higher than that between 601C and 601D if the two nucleosomes stack onto each other without any rotation (Fig. 8A). However, when we analyzed the FRET efficiencies of long-lived dinucleosomal states (Fig. 7C), we found that the FRET efficiency of the high FRET state is decreased from 0.79 of the 601CD pair to 0.62 of the 601CE pair. This result strongly suggests that the two nucleosomes in this long-lived dinucleosomal structure are rotated with respect to each other (Fig. 8B). Based on the two FRET efficiencies of the high FRET state from the two dinucleosomes, we calculated the rotation angle and the distance between the nucleosomes to be 34 ± 4° and 4.0 ± 0.1 nm, respectively (Fig. 9B, supplemental materials on the detailed calculation). The face-to-face distance between the two nucleosomes is ∼2 nm.
FIGURE 8.
Fluorophore locations in 601CD and 601CE nucleosome pairs. The two nucleosomes are stacked without rotation (A) and with rotation (B). 601CE pair must have a higher FRET efficiency than 601CD according to A.
FIGURE 9.
A, schematic representation of the directional unwrapping of nucleosomal DNA through DNA unwinding observed from this study. B, schematic representation of the most compact dinucleosomal state in our experimental condition (mononucleosome structure from Protein Data Bank code 1AOI). The distance between the two nucleosomes is measured between the dye locations in the phosphate backbone of nucleosomal DNA, which is close to the sum of the shortest distance between the two nucleosomal DNA and the thickness of dsDNA, i.e. the face-to-face distance between the two nucleosomes is ∼2 nm. It is apparent from B that H2A and H4 helices are better aligned in the suggested model (black bars in the lower panel indicate the axes of the helices).
DISCUSSION
Gansen et al. (39) recently examined the single pair FRET of mononucleosomes under quasi-bulk conditions, showing that the major effect of histone acetylation occurred only on the linker of the 601 DNA not in the internal regions, proposing that histone acetylation opens the nucleosomal DNA starting from the linker DNA. Previous work showed that histone acetylation reduced the linking number change of nucleosome core particle (38). In addition, considering that the helical repeat of nucleosome bound DNA is not altered by histone acetylation (40), we suggest that the change in the anisotropy of the 601A nucleosome upon histone acetylation is due to the reduction in the number of times the DNA winds around the nucleosome central axis (40). This unwinding would result in DNA topology change and directional unwrapping of DNA that may result in an enhanced accessibility to DNA. This inference is in line with a prevailing hypothesis of histone acetylation enhancing the accessibility of genes, which is supported by an early seminal study reporting that hyperacetylation of nucleosomes increases the susceptibility of nucleosomal DNA to DNase I digestion and enhances thermal denaturation of nucleosomes (41). Based on our observation, we concluded that histone acetylation by Piccolo NuA4 complex induces directional unwrapping of nucleosomal DNA at least in the region contacting H2A/H2B through unwinding of DNA (Fig. 9A). The reason why the internally labeled nucleosomes in the previous report did not show FRET changes may be because it is difficult to detect very small FRET changes in the quasibulk FRET experiment (39). Because we used only 147-bp DNA fragments without linker DNA, which display no heterogeneity in translational positioning of nucleosomes, our system may have a high sensitivity to small FRET changes.
This result suggests that the stability of a nucleosome decreases upon histone acetylation by Piccolo NuA4, and consequently the disassembly of a nucleosome may become facilitated. A higher chance of disassembly upon histone acetylation would enhance gene accessibility on an ensemble average and thereby contribute to gene activation. Although our system is incapable of probing unwrapping of H3/H4 contacting region of DNA, the unwrapping of H2A/H2B contacting region alone may be sufficient to facilitate at least a partial disassembly of a nucleosome because H2A/H2B dimers have been reported to dissociate separately prior to H3/H4 during nucleosome disassembly (42–45). Because the structural changes upon histone acetylation in a heterogeneous population of various nucleosomal states are very difficult to detect in an ensemble averaging measurement, these observations demonstrate a unique benefit of our single molecule approach.
A very low nucleosome concentration results in the depletion of a H2A/H2B dimer in nucleosomes (46), and adding excess unlabeled nucleosomes could improve the structural integrity of nucleosomes in single molecule experiments (39). These results suggest that nucleosomes interact with each other in solution to increase their stability. We observed transient interactions between two nucleosomes in solution in the presence of Mg2+ ions (1–3 mm). The lack of this interaction in the absence of the magnesium ion strongly supports that the internucleosomal interactions we observed are related to the strong Mg2+-dependent folding of nucleosomal arrays. In particular, the internucleosomal interactions we observed may suggest the initial steps of the Mg2+-dependent folding of nucleosomal arrays. Although our system lacks linker DNA, linker histones, and other factors that play important roles in nucleosome packaging, the inherent interaction between two nucleosomes is likely an essential variable to control the thermodynamics of chromatin structure. Furthermore, salt-dependent folding and oligomerization of nucleosomal arrays suggested multi-step internucleosomal interactions during nucleosome packaging (27, 47). Interestingly, Poirier et al. (47) reported that Mg2+-dependent compaction of trinucleosomes using the 601 nucleosome positioning sequence displayed multiple dinucleosomal states based on FRET analysis. They labeled Cy3 and Cy5 dye at the first and the third nucleosomes, respectively, based on the crystal structure of tetranucleosome (48), with which they detected FRET between the two nucleosomes in face-to-face interactions. Consistent with this result, we observed Mg2+-dependent transient dinucleosomal interactions, which are no FRET in the absence of Mg2+, a low FRET state at a low [Mg2+] (1–2 mm), and the low and the high FRET states at a high [Mg2+] (3 mm). Based on this result, we suggest that the Mg2+-dependent dinucleosomal states observed in our system support the multi-step nucleosome compaction in chromatin. The similarity of the FRET values of the low FRET state at 3 mm [Mg2+] and the low FRET state at low [Mg2+] (0.41 versus 0.35) suggest that they belong to one dinucleosomal state. The high FRET state may represent the most compact dinucleosomal state as in a maximally folded nucleosome array, whereas the low FRET state may represent a less compact dinucleosomal state as in a moderately folded nucleosome array (e.g. an early intermediate state in the folding process).
Histone tails have been reported to play a central role in mediating internucleosomal contacts (49, 50). For example, dramatic unfolding of chromatin was observed upon the removal of histone tails even at a high Mg2+ concentration (8). Importantly, maximally condensed chromatin fibers were obtained with any one of the histone tails deleted with the exception of the H4 tails (10). The H4 tail is known to interact with nucleosome surface such as H2A acidic patch, which is crucial for the folding of nucleosomal arrays into the 30-nm chromatin fiber (51, 52). Most recently, Sinha and Shogren-Knaak (29) observed the direct interactions between the H4 tail and H2A core and the long range nucleosome contacts undergoing intermolecular self-association. These results indicate that histone tails, particularly the H4 tails, play an important role in internucleosomal interactions, thereby regulating the folding or oligomerization of nucleosome arrays. Assuming that the interaction between H4 tail and H2A acidic patch is an important driving force for nucleosome packaging, acetylation of H4 tails is likely to disturb the formation of internucleosomal contacts, which is consistent with our results. Based on the shortened lifetime of dinucleosomes after the acetylation, we suggest that the acetylated histone tails reduce the stability and the formation efficiency of dinucleosomal states. Acetylated tails seem to affect the stability and the structure of both the moderately compact and the most compact dinucleosomal structure based on the broadened FRET efficiency distribution and lower average FRET efficiency upon the acetylation (Fig. 7).
The H3 and the H4 tails contain >50% of random coils or <50% α-helical content in a nucleosome core particle (53), and the α-helical content of the tail domains was increased by the acetylation (54). From this structural point of view, it may be that the acetylation induced stability of tails with more ordered conformations that inhibit the interaction between the tails and other nucleosomes.
Finally, the high FRET efficiency of 601CE pair is significantly lower than that of the 601CD pair, and the low FRET efficiencies are the same in the two cases within error. Based on an assumption that the two nucleosomes stack onto each other to form a dinucleosome, we suggest a structural model of the most compact and stable dinucleosomal state in our experimental condition (10 mm Tris-HCl, pH 7.5, 10 mm KCl, and 3 mm MgCl2). The face-to-face interactions between two nucleosomes are supported by the compacted trinucleosome and the crystal structure of a tetranucleosome that represents a crossed-linker conformation forming a two-start helix without the linker histone (47, 48). According to the model, the two nucleosomes are rotated with respect to each other by 34° and the internucleosome distance is 4.0 nm, which is close to the ∼3 nm obtained from the crystal structure of a tetranucleosome (48) (Fig. 9B; the face-to-face distance between the two nucleosomes is ∼2 nm). Notably, in the most compact and stable dinucleosomal configuration, the rotation aligns the major helix of H4 parallel to that of H2A, suggesting that the interaction between H4 and H2A helices may play a role in nucleosome packaging.
Supplementary Material
Acknowledgments
We thank Dr. Song Tan (Penn State University) for providing the Piccolo NuA4 overexpressing bacterial strain, the scheme to prepare the control 147-bp 601 DNA fragment, and X. laevis histone octamer core particles and Joon Huh and Robert Dutnall (University of California, San Diego) for providing yNAP1 plasmid. We are also grateful for Dr. Tan's advice as well as comments on the manuscript and Dr. John Choy (National Cancer Institute) for comments on the manuscript.
Footnotes
This work was supported by a Searle Scholar Award (to T.-H. L.) and a Henry and Dreyfus New Faculty Award (to T.-H. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental materials.
REFERENCES
- 1. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2. Robinson P. J., Fairall L., Huynh V. A., Rhodes D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6506–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Finch J. T., Klug A. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 1897–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cosgrove M. S., Boeke J. D., Wolberger C. (2004) Nat. Struct. Mol. Biol. 11, 1037–1043 [DOI] [PubMed] [Google Scholar]
- 5. Felsenfeld G., Groudine M. (2003) Nature 421, 448–453 [DOI] [PubMed] [Google Scholar]
- 6. Fischle W., Wang Y., Allis C. D. (2003) Curr Opin Cell Biol. 15, 172–183 [DOI] [PubMed] [Google Scholar]
- 7. Luger K., Richmond T. J. (1998) Curr. Opin. Genet. Dev. 8, 140–146 [DOI] [PubMed] [Google Scholar]
- 8. Arya G., Schlick T. (2009) J. Phys. Chem. A 113, 4045–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bertin A., Renouard M., Pedersen J. S., Livolant F., Durand D. (2007) Biophys. J. 92, 2633–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dorigo B., Schalch T., Bystricky K., Richmond T. J. (2003) J. Mol. Biol. 327, 85–96 [DOI] [PubMed] [Google Scholar]
- 11. Shahbazian M. D., Grunstein M. (2007) Annu. Rev. Biochem. 76, 75–100 [DOI] [PubMed] [Google Scholar]
- 12. Hansen J. C., Nyborg J. K., Luger K., Stargell L. A. (2010) J. Cell Physiol. 224, 289–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boudreault A. A., Cronier D., Selleck W., Lacoste N., Utley R. T., Allard S., Savard J., Lane W. S., Tan S., Côté J. (2003) Genes Dev. 17, 1415–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allfrey V. G., Faulkner R., Mirsky A. E. (1964) Proc. Natl. Acad. Sci. U.S.A. 51, 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pogo B. G., Allfrey V. G., Mirsky A. E. (1966) Proc. Natl. Acad. Sci. U.S.A. 55, 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hebbes T. R., Thorne A. W., Crane-Robinson C. (1988) EMBO J. 7, 1395–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Berndsen C. E., Selleck W., McBryant S. J., Hansen J. C., Tan S., Denu J. M. (2007) Biochemistry 46, 2091–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Selleck W., Fortin I., Sermwittayawong D., Côté J., Tan S. (2005) Mol. Cell Biol. 25, 5535–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kan P. Y., Caterino T. L., Hayes J. J. (2009) Mol. Cell Biol. 29, 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Megee P. C., Morgan B. A., Smith M. M. (1995) Genes Dev. 9, 1716–1727 [DOI] [PubMed] [Google Scholar]
- 21. Sobel R. E., Cook R. G., Perry C. A., Annunziato A. T., Allis C. D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1237–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suka N., Luo K., Grunstein M. (2002) Nat. Genet. 32, 378–383 [DOI] [PubMed] [Google Scholar]
- 23. Durrin L. K., Mann R. K., Kayne P. S., Grunstein M. (1991) Cell 65, 1023–1031 [DOI] [PubMed] [Google Scholar]
- 24. Robinson P. J., An W., Routh A., Martino F., Chapman L., Roeder R. G., Rhodes D. (2008) J. Mol. Biol. 381, 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Choi J. K., Howe L. J. (2009) Biochem. Cell Biol. 87, 139–150 [DOI] [PubMed] [Google Scholar]
- 26. Cosgrove M. S., Wolberger C. (2005) Biochem. Cell Biol. 83, 468–476 [DOI] [PubMed] [Google Scholar]
- 27. Hansen J. C. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 361–392 [DOI] [PubMed] [Google Scholar]
- 28. Anderson J. D., Lowary P. T., Widom J. (2001) J. Mol. Biol. 307, 977–985 [DOI] [PubMed] [Google Scholar]
- 29. Sinha D., Shogren-Knaak M. A. (2010) J. Biol. Chem. 285, 16572–16581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Staynov D. Z. (2008) Bioessays 30, 1003–1009 [DOI] [PubMed] [Google Scholar]
- 31. Tremethick D. J. (2007) Cell 128, 651–654 [DOI] [PubMed] [Google Scholar]
- 32. Zheng C., Hayes J. J. (2003) J. Biol. Chem. 278, 24217–24224 [DOI] [PubMed] [Google Scholar]
- 33. Tóth K. F., Mazurkiewicz J., Rippe K. (2005) J. Biol. Chem. 280, 15690–15699 [DOI] [PubMed] [Google Scholar]
- 34. Tan S., Kern R. C., Selleck W. (2005) Protein Expression Purif. 40, 385–395 [DOI] [PubMed] [Google Scholar]
- 35. Berndsen C. E., Denu J. M. (2005) Methods 36, 321–331 [DOI] [PubMed] [Google Scholar]
- 36. Choy J. S., Wei S., Lee J. Y., Tan S., Chu S., Lee T. H. (2010) J. Am. Chem. Soc. 132, 1782–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Norton V. G., Imai B. S., Yau P., Bradbury E. M. (1989) Cell 57, 449–457 [DOI] [PubMed] [Google Scholar]
- 38. Norton V. G., Marvin K. W., Yau P., Bradbury E. M. (1990) J. Biol. Chem. 265, 19848–19852 [PubMed] [Google Scholar]
- 39. Gansen A., Tóth K., Schwarz N., Langowski J. (2009) J. Phys. Chem. B 113, 2604–2613 [DOI] [PubMed] [Google Scholar]
- 40. Bauer W. R., Hayes J. J., White J. H., Wolffe A. P. (1994) J. Mol. Biol. 236, 685–690 [DOI] [PubMed] [Google Scholar]
- 41. Ausio J., van Holde K. E. (1986) Biochemistry 25, 1421–1428 [DOI] [PubMed] [Google Scholar]
- 42. Kimura H., Cook P. R. (2001) J. Cell Biol. 153, 1341–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bruno M., Flaus A., Stockdale C., Rencurel C., Ferreira H., Owen-Hughes T. (2003) Mol. Cell 12, 1599–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park Y. J., Chodaparambil J. V., Bao Y., McBryant S. J., Luger K. (2005) J. Biol. Chem. 280, 1817–1825 [DOI] [PubMed] [Google Scholar]
- 45. Gansen A., Valeri A., Hauger F., Felekyan S., Kalinin S., Tóth K., Langowski J., Seidel C. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15308–15313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Claudet C., Angelov D., Bouvet P., Dimitrov S., Bednar J. (2005) J. Biol. Chem. 280, 19958–19965 [DOI] [PubMed] [Google Scholar]
- 47. Poirier M. G., Oh E., Tims H. S., Widom J. (2009) Nat. Struct. Mol. Biol. 16, 938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schalch T., Duda S., Sargent D. F., Richmond T. J. (2005) Nature 436, 138–141 [DOI] [PubMed] [Google Scholar]
- 49. Gordon F., Luger K., Hansen J. C. (2005) J. Biol. Chem. 280, 33701–33706 [DOI] [PubMed] [Google Scholar]
- 50. Fletcher T. M., Hansen J. C. (1995) J. Biol. Chem. 270, 25359–25362 [DOI] [PubMed] [Google Scholar]
- 51. Dorigo B., Schalch T., Kulangara A., Duda S., Schroeder R. R., Richmond T. J. (2004) Science 306, 1571–1573 [DOI] [PubMed] [Google Scholar]
- 52. Zhou J., Fan J. Y., Rangasamy D., Tremethick D. J. (2007) Nat. Struct. Mol. Biol. 14, 1070–1076 [DOI] [PubMed] [Google Scholar]
- 53. Banères J. L., Martin A., Parello J. (1997) J. Mol. Biol. 273, 503–508 [DOI] [PubMed] [Google Scholar]
- 54. Wang X., Moore S. C., Laszckzak M., Ausió J. (2000) J. Biol. Chem. 275, 35013–35020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.