Abstract
Prolylhydroxylase domain proteins (PHD) are cellular oxygen-sensing molecules that regulate the stability of the α-subunit of the transcription factor hypoxia inducible factor (HIF)-1. HIF-1 affects cardiac development as well as adaptation of the heart toward increased pressure overload or myocardial infarction. We have disrupted PHD2 in cardiomyocytes (cPhd −/−) using Phd2flox/flox mice in combination with MLCvCre mice, which resulted in HIF-1α stabilization and activation of HIF target genes in the heart. Although cPhd2−/− mice showed no gross abnormalities in cardiac filament structure or function, we observed a significant increased cardiac capillary area in those mice. cPhd2 −/− mice did not respond differently to increased mechanical load by transverse aortic constriction compared with their wild-type (wt) littermates. After ligation of the left anterior descending artery, however, the area at risk and area of necrosis were significantly smaller in the cPhd2−/− mice compared with Phd2 wt mice in line with the described pivotal role of HIF-1α for tissue protection in case of myocardial infarction. This correlated with a decreased number of apoptotic cells in the infarcted myocardium in the cPhd2−/− mice and significantly improved cardiac function 3 weeks after myocardial infarction.
Keywords: Cardiac Muscle, Cell Death, Enzymes, Heart, Hydroxylase, Hydroxyproline, Hypoxia, Signal Transduction
Introduction
When oxygen availability is impaired, the resulting hypoxia activates homeostatic mechanisms at the systemic and cellular level (1). Hypoxia-inducible factors (HIFs)2 are essential players in these responses because they regulate the transcription of a large number of genes that affect a myriad of cellular processes, including angiogenesis, metabolism, cell survival, and oxygen delivery (2). HIF is a heterodimeric protein comprising the oxygen-sensitive α-subunit HIF-1α or the more cell type-specifically expressed HIF-2α or HIF-3α and the oxygen-insensitive β-subunit (3). In the presence of oxygen, HIFα becomes hydroxylated at two critical proline residues by prolylhydroxylase domain (PHD) enzymes (4, 5). The PHD protein family responsible for HIFα regulation consists of three members called prolylhydroxylase domain (PHD)1, PHD2, and PHD3 (6, 7). Following prolyl-4-hydroxylation of the critical prolyl residues under normoxic conditions, the ubiquitin ligase von Hippel-Lindau tumor suppressor protein recognizes HIF-1α subunits and targets them for rapid ubiquitination and proteasomal degradation (8–10).
Based on the ubiquitous expression pattern and its dominant effect in normoxia, it had to be assumed that PHD2 is the most critical HIF-1α-regulating PHD isoform in most tissues (11–13). This notion, learned from in vitro studies, was confirmed by the up to now available genetically modified Phd2 mouse models (14). Phd2 knock-out embryos die between embryonic day (E) 12.5 and E14.5 (15). This time point coincides with the increased levels of PHD2 in wild-type (wt) mice starting from E9.0. A major role of PHD2 in regulating the HIF system is further underscored by mouse models with a somatic Phd2−/− knock out, which enable to analyze the in vivo function of PHD2 in the adult mice. Two independent inducible Phd2−/− mouse models were developed by Takeda et al. (16) and Minamishima et al. (17). The phenotype of these mice most obviously resembles the consequences of HIFα overexpression with increased angiogenesis, erythropoiesis, and extramedullar hematopoiesis (17, 18). Most interestingly, these mice also develop a cardiac phenotype with symptoms of dilated cardiomyopathy. In the heart, HIF-1α and thereby also the PHDs are known to influence key components of heart development, morphogenesis, and function (19, 20). Long term activation of HIF-1α in the heart seems to activate detrimental pathways resulting in the development of heart failure (21). Thus, it is tempting to speculate that loss of PHD2 in the heart is responsible for the dilated cardiomyopathy as observed in the inducible Phd2−/− mice. However, because these mice also develop an increased hematocrit, secondary blood hyperviscosity-dependent cardiac changes cannot be ruled out.
Therefore we have generated cardiomyocyte-specific PHD2 knock-out mice (cPhd2−/−) by crossing MLCvCre mice with Phd2flox/flox mice. While this work was ongoing Moslehi et al. have recently reported the generation of cardiac-specific PHD2 knock-out mice using αMHCCre expressing mice as deleter mice in combination with Phd2flox/flox mice (22). In line with our observation, Moslehi et al. observed no striking differences in morphology or function of resting young cPhd2−/− mice. Moslehi et al. additionally analyzed older mice and demonstrated that with increasing age cardiomyocyte-specific PHD2 knock-out mice developed changes, which phenocopy ischemic cardiomyopathy. At a young age these animals responded to increased afterload with fortified heart failure despite the successful development of cardiac hypertrophy.
In contrast to the results of Moslehi et al., we did not observe significant changes in the development of hypertrophy and heart failure of young PHD2-deficient mice toward increased afterload in our animal model. Moreover, we observed cardioprotective effects in the PHD2-lacking mice in a model of myocardial ischemia. Challenging the mice with permanent ligation of the left anterior descending artery (LAD) resulted in cardiac tissue protection and decreased myocardial infarct size in the cPhd2−/− mice compared with Phd2 wt littermates. Taken together, our results show for the first time that lack of PHD2 in cardiomyocytes protects the heart from an acute ischemic insult. This phenomenon is at least in part most likely due to an increased capillary area in the cPhd2−/− mice.
EXPERIMENTAL PROCEDURES
Mice
The generation and detailed characterization of Phd2flox/flox mice will be reported elsewhere.3 In brief, LoxP sites were cloned into the Phd2 gene via Red/ET recombination (23) flanking exons 2 and 3. To characterize the Phd2flox/flox mice, full inactivation of the Phd2 gene was achieved via an intercross with PGKCre mice. Consistent with earlier reports, no viable PHD2−/− mice were obtained (15, 17).
Cre-mediated excision of exons 2 and 3 results in a frameshift mutation from exon 1 to 4 leading to an early translational stop and subsequent Phd2 knock out. Exons 2 and 3 encode for almost the entire catalytic domain of Phd2. The successful deletion of Phd2 in the floxed mice was verified by Western blotting on protein extracts of embryonic hearts (E14.5) from PGKCre × Phd2flox/flox mice using a homemade Phd2 antibody against a C-terminal peptide (supplemental Fig. 1). No PHD2 protein could be detected in Phd2−/− hearts. Moreover, the fact that HIF-1α protein is stabilized in these samples verifies the loss of functionality of Phd2. To analyze, if a shorter PHD2 protein encoded by exon 1 is formed, we performed Western blotting using an antibody, which detects the N-terminal part of PHD2 (supplemental Fig. 2). There is indeed a shorter Phd2 protein detectable in the Phd2+/− and Phd2−/− mice, which in its apparent molecular mass matches the predicted number of 31.5 kDa. The putatively exon 1-encoded protein, however, was expressed compared with the wt PHD2 protein in wt or Phd2+/− embryos only at very low levels. Whether this shorter protein would have a dominant negative character and therefore would bind to its interaction partner (e.g. HIFα) and block its normal function has not been studied. However, it was suggested that in human PHD2 both Arg371 and Pro317 (in mice Arg348 and Pro294 located in exons 3 and 2, respectively) contribute to the HIF-1α or HIF-2α binding (24). In our mice these sites are not present anymore, and a dominant negative interaction with HIF-1α is therefore unlikely.
All animals in this study were backcrossed to C57BL/6 mice at least five times. Phd2flox/flox mice were crossed with MLCvCre+/− mice (25) to generate Phd2flox/flox × MLCvCre+/− mice within two generations. Phd2flox/flox × MLCvCre+/− mice were then crossed with Phd2flox/flox mice to obtain cPhd2−/− (Phd2flox/flox × MLCvCre+/−) mice and littermate control wild-type mice (Phd2flox/flox).
Mice were genotyped by PCR using the following primers: Cre forward, 5′-CGTACTGACGGTGGGAGAAT-3′ and Cre reverse, 5′-CGGCAAAACAGGTAGTTA-3′; PHD2 forward, 5′-CTCACTGACCTACGCCGTGT-3′ and PHD2 reverse, 5′-CGCATCTTCCATCTCCATTT-3′.
The deletion of floxed exons in cDNAs isolated from ventricles or atria by PCRs was shown with the following primers: PHD2 forward, 5′-TACAGGATAAACGGCCGAAC-3′ and PHD2 reverse, 5′-GGCAACTGAGAGGCTGTAGG-3′. The forward primer binds in exon 1, the reverse primer binds in exon 5.
Animal Experimentation and Echocardiography
Animal experimentation was performed with cPhd2−/− mice and littermate Phd2 wt control mice. All protocols regarding animal experimentation were conducted according to the German animal protection laws and approved by the responsible governmental authority (Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit in Oldenburg; animal experimentation numbers 33.942502-04-10/0024 and 33.9-42502-04-10/0069). Pressure overload was induced by transverse aortic constriction (TAC) and performed essentially as described previously by our group with blunted 27-gauge needles as placeholders in 10-week-old female mice (26).
LAD ligations were performed by an investigator who was blinded regarding the genotypes of the mice. The measurement of infarct size was performed by an additional investigator, who was likewise blinded.
LAD ligation was performed on female mice having a minimum weight of 21 g (8–12 weeks of age). Mice were anesthetized (2% isoflurane). The trachea was surgically exposed, and tracheal intubation was performed. A blunt intubation cannula (intubation cannula, stainless steel with Y-adapter, 1.2-mm outer diameter, 30-mm length; Hugo Sachs Elektronik, Harvard Apparatus GmbH) was inserted into the trachea. Correct tube placement was confirmed by direct visualization of the cannula within the previously exposed trachea. The tracheal tube was connected to a mechanical ventilator (MiniVent; Hugo Sachs Elektronik, Harvard Apparatus GmbH), and the animals were ventilated by using a pressure-controlled ventilation mode (stroke volume 150 μl, 150 strokes/min fractional inspired O2 = 0.3). After exposing the heart, the pericardium was removed, and a 9-0 polyamide suture with a U-shaped needle was passed under the left anterior descending artery. The suture was tied to occlude the artery. The chest was closed and the mouse recovered.
Echocardiography and measurement of posterior wall thickness, septum thickness, left ventricular end systolic diameter, left ventricular end diastolic diameter, and fractional shortening (FS) were performed as described by Silter et al. (26).
Measurement of Infarct Size
Six hours after LAD ligation the mice were given heparin (250 units) and anesthetized, and the hearts were excised. Myocardial infarct size was determined by using Evans blue/2,3,5-triphenyltetrazolium chloride (TTC) staining as described by Bohl et al. (27). Briefly, the ascending aorta was cannulated with a 20-gauge tubing adapter, and 1% Evans blue was perfused into the aorta and coronary arteries to delineate the “area at risk” (AAR). The Evans blue dye was distributed uniformly to those areas of the myocardium, which were well perfused; hence, the area of the myocardium that was not stained with Evans blue was defined as the AAR. The left ventricle was separated from the rest of the heart and sectioned into three transverse slices. Sections were incubated in 2% TTC for 20 min at 37 °C to identify viable tissue. Infarct quantification was performed on digital photographs (SMZ 100; Nikon, Tokyo, Japan) using ImageJ (National Institutes of Health). The area of necrosis (AON) and AAR were determined as the average percent area per slice from the middle section and the lowest section. Also, myocardial samples were collected for cyrosectioning and assessment of apoptosis.
Protein Extraction and Immunoblot Analyses
Heart tissue was rapidly homogenized in a buffer containing 4 m urea, 140 mm Tris (pH 6.8), 1% SDS, 2% Nonidet P-40, and protease inhibitors (Roche Applied Science). Protein concentrations were quantified (DC Protein Assay; Bio-Rad). For immunoblot analysis protein samples were resolved by SDS-PAGE and transferred onto nitrocellulose membranes (Amersham Biosciences) by semidry blotting (PeqLab). Primary antibodies used were: anti-PHD2 (NB100-2219; Novus), anti-HIF-1α (NB100-479; Novus), and anti-vinculin (hVin-1, V9264; Sigma).
For detection of immunocomplexes, horseradish peroxidase-conjugated secondary goat anti-rabbit or goat anti-mouse antibodies (Santa Cruz Biotechnology) were used, and membranes were incubated with 100 mm Tris-HCl (pH 8.5), 2.65 mm H2O2, 0.45 mm luminol, and 0.625 mm coumaric acid for 1 min. Chemiluminescence signals were detected with the LAS3000 camera system (Fuji Film Europe, Düsseldorf, Germany).
RT-PCR Analyses
After RNA extraction reverse transcription (RT) was performed with 2 μg of RNA and a first strand cDNA synthesis kit (Fermentas, St. Leon-Rot). mRNA levels were quantified by using 1 μl of the cDNA reaction and a SYBR Green qPCR reaction kit (Clontech) in combination with a MX3000P light cycler (Stratagene). The initial template concentration of each sample was calculated by comparison with serial dilutions of a calibrated standard. Primers were as follows: glucose transporter-1 (Glut-1) forward, 5′-TGGCCTTGCTGGAACGGCTG and Glut-1 reverse, 5′-TCCTTGGGCTGCAGGGAGCA-3′; mS12 forward, 5′-GAAGCTGCCAAGGCCTTAGA-3 and mS12 reverse, 5′-AACTGCAACCAACCACCTTC-3′; PHD2 forward, 5′-TTGCTGACATTGAACCCAAA-3′ and PHD2 reverse, 5′-GGCAACTGAGAGGCTGTAGG-3′; PHD1 forward, 5′-GCTAGGCTGAGGGAGGAAGT-3′ and PHD1 reverse, 5′-TCTACCCAGGCAATCTGGTC-3′; PHD3 forward, 5′-GGCCGCTGTATCACCTGTAT-3 and PHD3 reverse, 5′-TTCTGCCCTTTCTTCAGCAT-3′; brain natriuretic peptide (BNP) forward, 5′-AAGTCCTAGCCAGTCTCCAGA-3′ and BNP reverse, 5′-GAGCTGTCTCTGGGCCATTTC-3′; phosphofruktokinase 1 (Pfkl) forward, 5′-ACGAGGCCATCCAGCTCCGT-3′ and Pfkl reverse 5′-TGGGGCTTGGGCAGTGTCCT-3′; pyruvate dehydrogenase kinase 1 (PDK-1) forward, 5′-GTTCACGTCACGCTGGGCGA-3′ and PDK-1 reverse, 5′-CCAGGCGTCCCATGTGCGTT-3′; phosphoglycerate kinase (PGK) forward, 5′-ACGTCTGCCGCGCTGTTCTC-3′ and PGK reverse, 5′-ACCATGGAGCTATGGGCTCGGT-3′; VEGF forward, 5′-CGAACGTACTGCCGATTGAGA-3 and VEGF reverse, 5′-TGGTGAGGTTTGATCCGCATGATCTG-3′; angiopoietin (Ang)-1 forward, 5′-CGCTCTCATGCTAACAGGAGGTTGG-3′ and Ang-1 reverse, 5′-GCATTCTCTGGACCCAAGTGGCG-3′; Ang-2 forward, 5′-CACCCAACTCCAAGAGCTCGG-3′ and Ang-2 reverse, 5′-CACGTAGCGGTGCTGACCGG-3′; BCL2/adenovirus E1B 19-kDa protein-interacting protein 3 (Bnip3) forward, 5′-GTCCAGTGTCGCCTGGCCTC-3′ and Bnip3 reverse, 5′-TGGGAGCGAGGTGGGCTGTC-3′; iNOS, inducible nitric-oxide synthase; (iNOS) forward, 5′-GCTGCCTTCCTGCTGTCGCA-3′ and iNOS reverse, 5′-GGAGCCGCTGCTGCCAGAAA-3; hemoxygenase (Hox)-1 forward, 5′-TGGTGCAAGATACTGCCCCTGC-3′ and Hox-1 reverse, 5′-TGGGGGACAGCAGTCGTGGT-3′, apelin forward, 5′-TGCAGTTTGTGGAGTGCCACTG-3′ and apelin reverse, 5′-GCACCGGGAGGCACT-3′; adrenomedullin forward, 5′-TGGCCCCCTACAAGCCAGCAAT-3′ and adrenomedullin reverse, 5′-GCCAACGGGATACTCGCCCG-3′. The sets of primers for adenosine receptor A2b and CD73 were according to previously published sequences (28).
BNP Enzyme Immunoassay
Plasma BNP levels were measured using a commercially available enzyme immunoassay kit (catalog number EK-011-23; Phoenix Pharmaceuticals) according to the manufacturer's instructions.
Histological Analyses
For Trichrome staining, heart tissue was fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) and embedded in paraffin before sectioning.
Cryosectioning and Immunofluorescence Labeling
Freshly isolated hearts were treated as described before (29). Briefly, hearts were sectioned (5–10 μm) in a cryostat, mounted on glass slides, and dried for at least 30 min. After being washed with PBS, the sections were briefly incubated with 1% Triton X-100 and then fixed with 4% formaldehyde and permeabilized with 0.2% Triton X-100. Nonspecific binding of antibodies was blocked by incubation with 1% bovine serum albumin for 1 h. The sections were then incubated with the polyclonal anti-CD31 antibodies (DIA 310, diluted 1:20; Dianova) and anti-vinculin antibodies (hVin-1, diluted 1:100; Sigma) at room temperature for 1 h, washed three times in PBS, and then incubated with anti-mouse TR (diluted 1:200; Santa Cruz Biotechnology) and anti-rat FITC (1:200; Santa Cruz Biotechnology). Samples were counterstained for DNA (Hoechst). Finally, samples were washed extensively in PBS, mounted in Mowiol, and examined by fluorescence microscopy (Axio Observer D1; Carl Zeiss, Göttingen, Germany). The capillary area was determined by analyzing CD31-positive pixels using ImageJ. In addition, the number of capillaries were counted per view of field.
Immunohistochemistry
Paraffin-embedded sections were immunostained as described by others (22). The anti-HIF-1α primary antibody (Novus NB100-123) was used at a 1:1000 dilution.
Electron Microscopy
Standard procedures were applied for fixation (1.5% glutaraldehye and 1.5% paraformaldehyde in phosphate buffer) of hearts of cPhd2−/− and Phd2 wt mice. The hearts were then washed in PBS and fixed in 2% osmium tetroxide. The samples were dehydrated in a graded series of alcohol and embedded in Araldite. Ultrathin sections were stained with 2% methanolic uranyl acetate. Samples were then examined in a transmission electron microscope equipped with photodocumentation.
TUNEL Assay
Apoptotic cells were detected with a TUNEL assay according to the manufacturer's protocol (In Situ Cell Death Detection kit, catalog number 11 684 795 001; Roche Applied Science). Three hearts of each genotype were analyzed, and five random high power fields from each heart sample were quantified.
Statistical Analyses
Data were analyzed by two-tailed Student's t test or (in case of repeated echocardiography analysis after TAC treatment) as paired t test and presented as mean ± S.E. A p value < 0.05 was considered statistically significant.
RESULTS
Generation of Cardiac-specific PHD2 Knock-out Mice
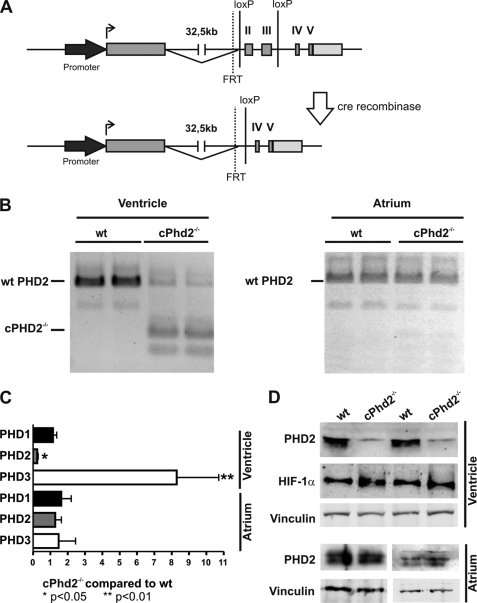
For generating cPhd2−/− mice, we crossed Phd2flox/flox mice with mice that express the Cre recombinase under the control of the MLCv promoter (25). The MLCv-driven Cre permits recombination of the floxed Phd2 exons 2 and 3 in ventricular cardiomyocytes (Fig. 1A). Isolation and PCR analysis of cDNAs from atria and ventricles of cPhd2−/− and littermate Phd2 wt control mice verified successful recombination in the ventricles but not in the atria of the cPhd2−/− mice (Fig. 1B). The additional bands seen in the PCR analysis of Phd2 wt mice most likely are the result of described PHD2 splice variants (30).
FIGURE 1.
Generation and analyses of cPhd2−/− mice. A, schematic description of the gene targeting strategy is shown. In the targeted Phd2 locus, exons 2 and 3 are flanked by two loxP sites. Exons 2 and 3 are deleted by Cre-mediated recombination by crossing Phd2flox/flox mice with MLCvCre mice. B, PCRs were performed with cDNAs isolated from left ventricles or atria of cPhd2−/− or Phd2 wt control mice to demonstrate the deletion of floxed exons in the ventricles but not the atria. Incomplete recombination in the ventricles is most likely due to the presence of noncardiomyocytes in the tissue samples. Additional bands resemble splice variants. C, RT-PCR analysis confirmed the significant reduction of PHD2 mRNA transcripts in the left ventricles but not the atria of 8-week-old cPhd2−/− mice compared with wt littermates. Transcripts of the HIF-target PHD3 were increased whereas PHD2 mRNA was unchanged. In total, 11 wild-type mice and 7 cPHD2−/− were analyzed. *, p < 0.05; **, p < 0.01. Data represent mean values ± S.E. (error bars). D, PHD2 protein was detected by Western blot analysis. Protein extracts prepared from left ventricles and atria of 8-week-old Phd2 wt and cPhd2−/− mice were analyzed with anti-PHD2, anti-HIF-1α, and anti-vinculin antibodies, confirming that PHD2 was successfully knocked out in the left ventricles but not in the atria.
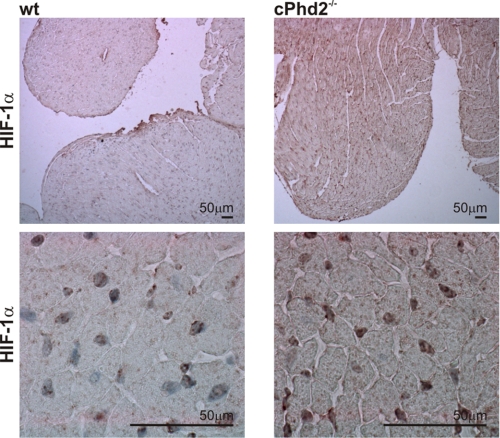
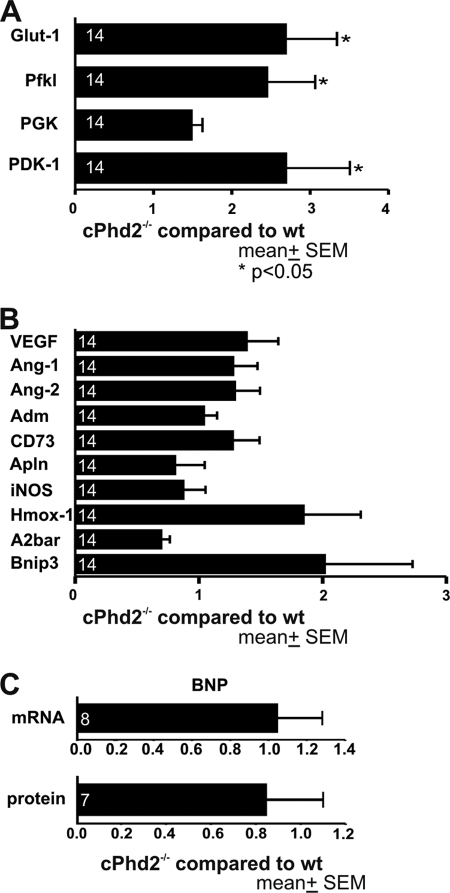
Quantification of PHD2 RNA levels in the left ventricles and atria of cPhd2−/− mice compared with Phd2 wt mice revealed a roughly 5-fold reduction in the knock-out ventricles but not the atria, confirming the MLCvCre-mediated knock out in the ventricular cardiomyocytes (Fig. 1C). Considering that the RNA was isolated from the whole left ventricles, which besides the cardiomyocytes also contain especially cardiac fibroblasts but also endothelial cells, smooth muscle cells etc., this reduction in PHD2 expression is in accordance with a successful recombination in the Cre-expressing cardiomyocytes. In line, cardiac PHD2 expression was also reduced at the protein level in left ventricles but not the atria of cPhd2−/− mice as determined by immunoblot analysis (Fig. 1D). As a consequence of the PHD2 knock out, HIF-1α protein levels were slightly increased (Fig. 1D). Immunohistochemical analysis confirmed the accumulation of HIF-1α protein in cell nuclei of the cardiomyocytes (Fig. 2). Detection of HIF-2α by immunohistochemistry was hampered by varying qualities of the batches of the commercially available polyclonal anti-HIF-2α antibodies. At least in vitro it was described that in contrast to PHD3, PHD2 seems to have higher activity toward HIF-1α compared with HIF-2α (30). Therefore the increased levels of HIF-1α in the cardiomyocytes are in line with an effective PHD2 knock out in our mouse model. A moderate activation of the HIF-signaling pathway in the hearts of cPhd2−/− mice was likewise detected by analyzing the expression of the HIF target genes PHD3 (Fig. 1C), Glut-1, Pfk1, and PDK1 (Fig. 3A). Whereas increased levels of Glut-1, Pfk1, and PDK1 reflect the impact of HIF on glucose transport and glucose metabolism, respectively (31), the elevated levels of PHD3 are indicative of the described negative HIF/PHD feedback loop, which involves the HIF-dependent transcription of PHD3 (32). In contrast, PHD1 RNA levels, which are not inducible by hypoxia (13), were not affected in the hearts of cPhd2−/− mice (Fig. 1C). HIF target genes involved in angiogenesis (VEGF, Ang-1, Ang-2), vasotonus regulation (adrenomedullin, apelin, iNOS, and Hox-1), purinergic signaling (CD73, and adenosine receptor A2b), and apoptosis (Bnip3) were found to be unchanged (Fig. 3B). BNP is produced in the ventricles and is regarded as surrogate marker for heart failure (33). Analyzing BNP RNA levels in the left ventricles or BNP plasma levels of Phd2 wt and cPhd2−/− mice revealed no significant differences, indicating that there were no obvious signs for heart failure in the cPhd2−/− mice (Fig. 3C).
FIGURE 2.
HIF-1α protein accumulates in cPhd2−/− hearts. Immunohistochemical analyses of hearts from 8-week-old cPhd2−/− and Phd2 wt mice are shown. HIF-1α protein accumulates in the nuclei of cPhd2−/− ventricles.
FIGURE 3.
PHD2-deficient ventricles exhibit increased expression of HIF target genes. A and B, quantitative real-time RT-PCR (qPCR) analysis of left ventricles from 8-week-old cPhd2−/− or Phd2 wt mice was performed. Transcript levels of genes involved in glucose transport and glucose metabolism (A) as well as HIF-target genes related to angiogenesis, vasotonus regulation, and apoptosis (B) were analyzed. The numbers in the bars indicate the number of animals analyzed. *, p < 0.05. Data represent mean values ± S.E. (error bars). C, changes of BNP mRNA in the left ventricles or in serum protein levels of cPhd2−/− compared with Phd2 wt mice were not detectable by qPCR and ELISA, respectively. Data represent mean values ± S.E.
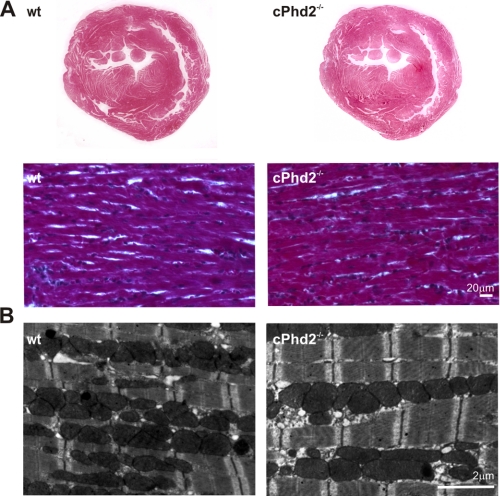
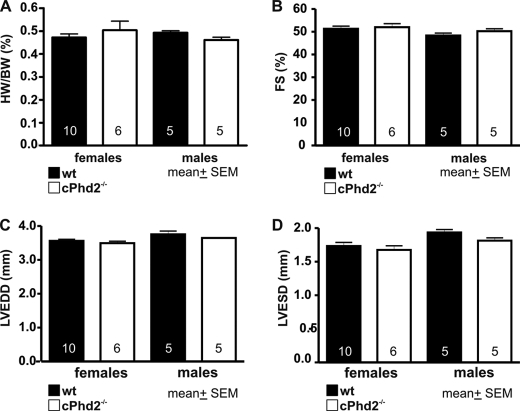
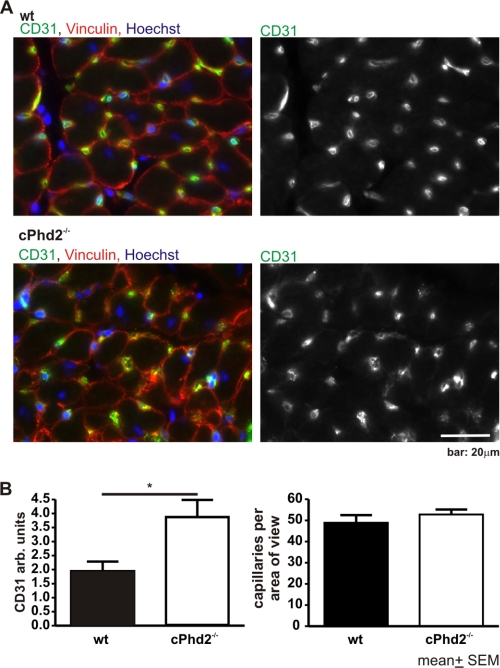
Histological assessment of the heart revealed no gross abnormalities in the cPhd2−/− mice (Fig. 4A). Furthermore, electron microscopy analysis displayed no significant changes in ultrastructure or filament architecture (Fig. 4B). Heart weight (Fig. 5A) and heart function as determined by echocardiography of the cPhd2−/− mice were found to be not different compared with Phd2 wt mice (Fig. 5, B–D). Interestingly, a significant difference between Phd2 wt and cPhd2−/− mice, however, could be observed when analyzing capillaries in the heart via CD31 immunostaining (Fig. 6). In the cPhd2−/− hearts significantly more CD31-positive staining was detected. This was due to an increased capillary area but not to an increase in the number of capillaries (Fig. 6B). Taken together, basal heart structure and function are, besides an increased diameter of the capillaries, not changed in 8 weeks old mice as a consequence of a lack of PHD2 in ventricular cardiomyocytes. In this regard it should be noted that MLCvCre × HIF-1αflox/flox mice similarly do not present obvious changes in resting heart function (34). In contrast to resting conditions, however, HIF-1α indeed seems to play a role in the adaptation of the heart to increased mechanical load or ischemia (20, 26). Therefore, we analyzed the cPhd2−/− mice under stressed conditions, i.e. TAC or LAD.
FIGURE 4.
cPhd2−/− hearts show no gross structural abnormalities. A, paraffin-embedded tissue sections of Phd2 wt and cPhd2−/− hearts stained with Trichrome. B, transmission electron microscopy of the Phd2 wt and cPhd2−/− hearts.
FIGURE 5.
Cardiac PHD2 deficiency does not affect heart function in 8-week-old mice. A, hearts of Phd2 wt or cPhd2−/− mice were excised, and the ratios of heart weight (HW) compared with body weight (BW) were determined. Data represent mean values ± S.E. (error bars). The numbers in the bars indicate the number of animals analyzed. B–D, fractional shortening (FS) (B), left ventricular end diastolic diameter (LVEDD) (C), and left ventricular end systolic diameter (LVESD) (D) were analyzed by echocardiography and showed no differences between indicated genotypes. The numbers in the bars indicate the number of animals analyzed. Data represent mean values ± S.E.
FIGURE 6.
Increased capillary area in PHD2-deficient hearts. A, hearts of cPhd2−/− mice (n = 4) and Phd2 wt (n = 4) littermates were excised and analyzed for angiogenesis as determined by anti-CD31 staining. Sections were co-stained for vinculin and DNA (Hoechst). B, the area of capillaries, which were determined in A, were quantified by determining the extent of CD31-positive staining, and the number of capillaries were counted. *, p < 0.05. Data represent mean values ± S.E. (error bars).
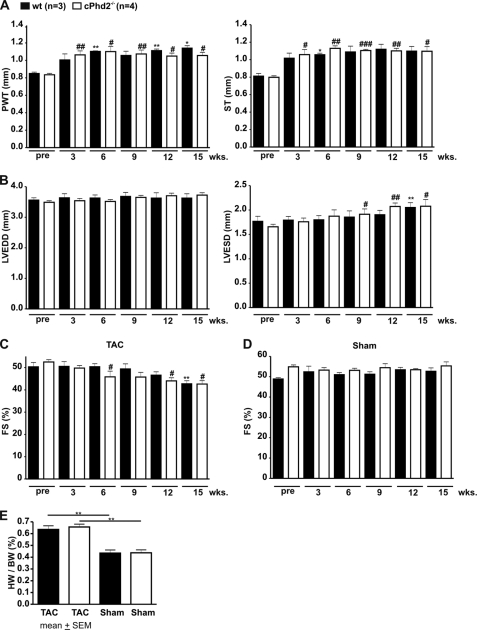
Lack of PHD2 in the Heart Does Not Affect the Cardiac Response to Increased Afterload
Increased mechanical load by TAC stimulates an adaptation program, which results in hypertrophy and if the load sustains in cardiac failure. We applied the aortic constriction between the branches of the truncus brachiocephalicus and the arteria carotis communis and followed the mice up to 15 weeks by echocardiography. As expected TAC-treated Phd2 wt and cPhd2−/− mice developed cardiac hypertrophy as a consequence of the aortic constriction (Fig. 7A) whereas sham-treated animal did not (data not shown). The load-induced increase in posterior wall thickness and septum thickness, however, was not different in Phd2 wt compared with cPhd2−/− mice. Likewise, both, i.e. TAC-treated Phd2 wt and cPhd2−/− mice, but not the sham-treated animals (Fig. 7D), developed an increased left ventricular end systolic diameter (Fig. 7B) and a drop in FS (Fig. 7C) to a similar extent at around 12–15 weeks after the intervention. Furthermore, no change in the heart to body weight ratio was seen in TAC-treated Phd2 wt mice compared with cPhd2−/− mice (Fig. 7E).
FIGURE 7.
PHD2 does not affect cardiac hypertrophy and cardiac function after TAC in female mice. Sustained pressure overload was induced in 10-week-old female Phd2 wt and cPhd2−/− mice by TAC. Subsequently (A) posterior wall thickness (PWT) and septum thickness (ST) as well as (B) left ventricular end diastolic diameter (LVEDD) and left ventricular end systolic diameter (LVESD) were analyzed before (pre) and up to 15 weeks after TAC by echocardiography. Based on the left ventricular end diastolic diameter and left ventricular end systolic diameter, fractional shortenings (FS) of TAC-treated (C) and sham-operated animals (D) were determined. In addition, heart weight (HW) to body weight (BW) ratios were determined 15 weeks after TAC (E). *, p < 0.05 TAC-treated Phd2 wt versus nontreated/pre Phd2 wt mice; **, p < 0.01 TAC-treated Phd2 wt versus nontreated/pre Phd2 wt mice; #, p < 0.05 TAC-treated cPhd2−/− versus nontreated/pre cPhd2−/− mice; ##, p < 0.01 TAC-treated cPhd2−/− versus nontreated/pre cPhd2−/− mice; ###, p < 0.001 TAC-treated cPhd2−/− versus nontreated/pre cPhd2−/− mice. Data represent mean values ± S.E. (error bars).
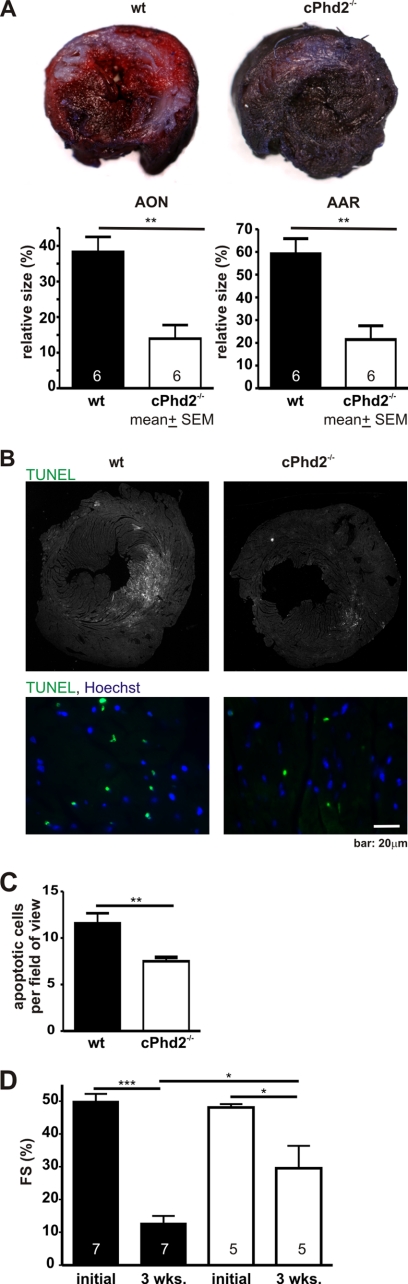
Cardiac PHD2 Knock-out Mice Are Protected from Acute Myocardial Ischemia
Ligation of the LAD allows studying the acute and chronic pathophysiological processes in myocardial ischemia and was applied to Phd2 wt and cPhd2−/− mice. The extent of tissue, which was well supplied by blood 6 h after the intervention, was analyzed by Evans blue perfusion. Myocardial slices were additionally stained with TTC to discriminate between nonperfused dead and vital tissue. This technique relies on the ability of dehydrogenase enzymes and co-factors in the vital tissue to react with tetrazolium salts to form a formazan pigment. Vital tissue appears red and reflects the AAR, whereas dead/nonvital tissue appears white, indicating the AON. The AAR and the AON were significantly smaller in the cPhd2−/− mice compared with littermate Phd2 wt control mice (Fig. 8A). This correlated with a significantly smaller number of apoptotic cardiomyocytes in the infarcted area in the cPhd2−/− mice as determined by TUNEL assays (Fig. 8, B and C).
FIGURE 8.
cPHD2−/− mice are protected from myocardial infarction. A, hearts of female Phd2 wt and cPhd2−/− mice were harvested 6 h after LAD ligation, and the AON (white area) and the AAR (non-blue red area) were determined with Evans blue perfusion and TTC staining. Representative mid-myocardial cross-sections of stained hearts are shown. AAR and AON were significantly smaller in cPHD2−/− hearts. The numbers in the bars indicate the number of animals analyzed. **, p < 0.01. Data represent mean values ± S.E. (error bars). B, heart sections of cPhd2−/− mice (n = 3) and wt mice (n = 3) were analyzed for apoptotic cells by TUNEL assay after LAD ligation. Nuclei were stained with Hoechst (blue), apoptotic nuclei were stained green by the TUNEL assay. Representative images are shown. C, apoptotic cells per field of view within the AAR after myocardial infarction of cPhd2 −/− and Phd2 wt mice were quantified. **, p < 0.01. Data represent mean values ± S.E. (error bars). D, cardiac function is preserved in cPHD2−/− mice 3 weeks after myocardial infarction. FS was analyzed before (initial) and 3 weeks after LAD ligation by echocardiography. *, p < 0.05; ***, p < 0.001. Data represent mean values ± S.E. The numbers in the bars represent the number of animals analyzed.
Echocardiography was performed at base line and 3 weeks after myocardial infarction in cPhd2−/− mice compared with Phd2 wt control mice (Fig. 8D). Before LAD ligation the FS was not different between cPhd2−/− and control mice and was reduced in both groups after myocardial infarction. In line with the smaller AAR and AON, myocardial function was better preserved in the cPhd2−/− mice as indicated by a significant higher FS 3 weeks after surgery in cPhd2−/− mice than in Phd2 wt control mice.
DISCUSSION
Small molecule PHD inhibitors have been demonstrated to be promising in strategies for treating diseases related to hypoxic adaptation like anemia, stroke, or myocardial infarction (14, 35, 36). The concept of cardiac tissue protection is delineated from several recent preclinical studies. Intramyocardial injection of small hairpin RNA targeting PHD2 following LAD ligation results in improved neovascularization in the peri-infarct region and improved cardiac function as indicated by determining FS (37). Treatment of mice with the PHD inhibitors dimethyloxaloylglycine or GSK60 had similar protective effects in mouse or rat myocardial infarction models (28, 38). Furthermore, a recent report by Hyvärinen et al. has underscored the cardiac protective effect of HIF-1α in a PHD2 hypomorphic mouse model (39). Taken collectively, these studies point to a cardiac tissue protective mechanism by inhibiting PHD activity in ischemia; however, they do not explicitly answer whether inhibiting the activities of PHDs in cardiomyocytes is involved in this process. In our study, in which the surgeon and the investigator analyzing the myocardial infarct size were blinded, both, i.e. AAR and AON, were significantly decreased after LAD ligation in cPhd2−/− mice, which lack PHD2 just in ventricular cardiomyocytes compared with Phd2 wt littermates. This correlated with a decreased number of apoptotic cells. Moreover, 3 weeks after LAD ligation cPhd2−/− mice demonstrated better heart function compared with Phd2 wt control animals. To this end the present study adds to the understanding of the consequences of loss of PHD2 specifically in cardiomyocytes.
In line with recently published results obtained with a mouse model similar to the one developed in our study, filament structure and cardiac function were not affected by the cardiomyocyte-specific loss of PHD2 in young mice (22). However, we did not observe any change in the response of cPhd2−/− mice compared with Phd2 wt mice to sustained pressure overload in regard to cardiac hypertrophy or development of heart failure. This is in contrast to the results of Moslehi et al. (22), who have described a decompensation in PHD2-deficient hearts in response to increased afterload. TAC in the mouse is a commonly used experimental model for pressure overload-induced cardiac hypertrophy and heart failure. The discrepancy in the TAC model comparing the study by Moslehi et al. and our study may be in part explained by the different deleter Cre mice applied. Furthermore, the development of cardiac failure after TAC intervention is highly depending on the severity of aortic constriction. The severity is mainly determined by the ratio of the basal diameter of the aorta and the diameter of the needle, which is used as placeholder during the constriction procedure. Whereas the needle size was the same in both studies, the severity of the constriction still seems to be more severe in the study of Moslehi et al. because the fractional shortening in the challenged mice was reduced already after 4–8 weeks, whereas our mice responded just after 12–15 weeks with a diminished FS.
Interestingly, we observed a significant increase in myocardial capillary size as defined by CD31 staining but not in the number of capillaries. This is consistent with the observation that genes involved in angiogenesis were not found to be up-regulated in the cPhd2−/− mice presented in this study. Still, it is conceivable that the increased capillary diameter results in an improved cardiac blood supply which might contribute to the cardiac tissue protection in the cPhd2−/− mice 6 h after ligation of the LAD.
A HIF-1α-mediated cardioprotection has also been observed in cardiac-specific HIF-1 transgenic mice after myocardial infarction (40) and in ischemia-reperfusion in mice carrying a hypomorphic Phd2 allele (39) and, very recently, in Phd1−/− mice (41). The cause of cardioprotection is likely multifactoral and due to the activation of several HIF target genes and the subsequent modulation of pathways involved in, for example, β-catenin signaling (41), the purinergic signaling pathways (28), and the glucose metabolism (39). However, in the cPhd2−/− mice presented in our study purinergic signaling was not affected as no significant changes in adenosine 2B receptor and CD73 transcriptional levels were observed. Yet, similar to the study by Hyvärinen et al. (39), we noticed significantly increased mRNA levels for Glut-1 and several enzymes of glycolysis which are all known HIF targets (42). A shift from oxidative to glycolytic metabolism in the heart confers an advantage in surviving ischemic insults. This might, together with the increased capillary vessel area in cPhd2−/− hearts, explain the cardioprotection after acute myocardial infarction.
Taken together, our data indicate that deficiency of PHD2 in the heart does not affect the response toward increased mechanical load but induces an acute tissue protective effect in case of myocardial ischemia. Long term inhibition of PHD2 as well as long term stabilization of HIF-1α in the heart seem to impair heart function as demonstrated in old (8 months) cPhd2−/− mice (22) (and own observations) and heart-specific HIF-1α transgenic mice (21). Further studies, which clarify for how long and when in relation to the myocardial insult PHD activity needs to be diminished, are therefore needed to delineate whether the application of small molecule PHD inhibitors are feasible and useful for inducing tissue protective effects in case of myocardial infarction.
Supplementary Material
Acknowledgments
We thank Gudrun Federkeil for help with the cryosections and Annette Hillemann for expert technical assistance.
This work was supported by Kröner Fresenius Stiftung Grant P25/10//A12/10 (to A. Z.) and Deutsche Forschungsgemeinschaft Grants Ka1269/11-1 (to D. M. K.), Br 1336/2-3 and SFB 655 A8 (to G. B.), and Wi 3291/1-1 (to B. W.). The work was performed as collaborative project within the COST Action TD0901 “HypoxiaNet.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
B. Wielockx, K. Anastassiadis, A. F. Stewart, and G. Breier, unpublished observations.
- HIF
- hypoxia-inducible factor
- AAR
- area at risk
- Ang
- angiopoietin
- AON
- area of necrosis
- Bnip3
- BCL2/adenovirus E1B 19-kDa protein-interacting protein 3
- BNP
- brain natriuretic peptide
- cPhd2−/−
- cardiac-specific PHD2 knock out
- E
- embryonic day
- FS
- fractional shortening
- Glut-1
- glucose transporter-1
- Hox-1
- hemoxygenase
- iNOS
- inducible nitric-oxide synthase
- LAD
- left anterior descending artery
- PDK1
- pyruvate dehydrogenase kinase 1
- pfk1
- phosphofruktokinase 1
- PGK
- phosphoglycerate kinase
- PHD
- prolylhydroxylase domain
- TAC
- transverse aortic constriction
- TTC
- 2,3,5-triphenyltetrazolium chloride
- wt
- wild type.
REFERENCES
- 1. Schofield C. J., Ratcliffe P. J. (2005) Biochem. Biophys. Res. Commun. 30, 617–626 [DOI] [PubMed] [Google Scholar]
- 2. Wenger R. H. (2002) FASEB J. 16, 1151–1162 [DOI] [PubMed] [Google Scholar]
- 3. Wang G. L., Semenza G. L. (1995) J. Biol. Chem. 270, 1230–1237 [DOI] [PubMed] [Google Scholar]
- 4. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 5. Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., von Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 6. Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 7. Bruick R. K., McKnight S. L. (2001) Science 294, 1337–1340 [DOI] [PubMed] [Google Scholar]
- 8. Hon W. C., Wilson M. I., Harlos K., Claridge T. D., Schofield C. J., Pugh C. W., Maxwell P. H., Ratcliffe P. J., Stuart D. I., Jones E. Y. (2002) Nature 417, 975–978 [DOI] [PubMed] [Google Scholar]
- 9. Min J. H., Yang H., Ivan M., Gertler F., Kaelin W. G., Jr., Pavletich N. P. (2002) Science 296, 1886–1889 [DOI] [PubMed] [Google Scholar]
- 10. Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. (1999) Nature 399, 271–275 [DOI] [PubMed] [Google Scholar]
- 11. Berra E., Benizri E., Ginouvès A., Volmat V., Roux D., Pouysségur J. (2003) EMBO J. 22, 4082–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lieb M. E., Menzies K., Moschella M. C., Ni R., Taubman M. B. (2002) Biochem. Cell Biol. 80, 421–426 [DOI] [PubMed] [Google Scholar]
- 13. Appelhoff R. J., Tian Y. M., Raval R. R., Turley H., Harris A. L., Pugh C. W., Ratcliffe P. J., Gleadle J. M. (2004) J. Biol. Chem. 279, 38458–38465 [DOI] [PubMed] [Google Scholar]
- 14. Katschinski D. M. (2009) Acta Physiol. 195, 407–414 [DOI] [PubMed] [Google Scholar]
- 15. Takeda K., Ho V. C., Takeda H., Duan L. J., Nagy A., Fong G. H. (2006) Mol. Cell. Biol. 26, 8336–8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takeda K., Cowan A., Fong G. H. (2007) Circulation 116, 774–781 [DOI] [PubMed] [Google Scholar]
- 17. Minamishima Y. A., Moslehi J., Bardeesy N., Cullen D., Bronson R. T., Kaelin W. G., Jr. (2008) Blood 111, 3236–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takeda K., Aguila H. L., Parikh N. S., Li X., Lamothe K., Duan L. J., Takeda H., Lee F. S., Fong G. H. (2008) Blood 111, 3229–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krishnan J., Ahuja P., Bodenmann S., Knapik D., Perriard E., Krek W., Perriard J. C. (2008) Circ. Res. 103, 1139–1146 [DOI] [PubMed] [Google Scholar]
- 20. Krishnan J., Suter M., Windak R., Krebs T., Felley A., Montessuit C., Tokarska- Schlattner M., Aasum E., Bogdanova A., Perriard E., Perriard J. C., Larsen T., Pedrazzini T., Krek W. (2009) Cell Metab. 9, 512–524 [DOI] [PubMed] [Google Scholar]
- 21. Bekeredjian R., Walton C. B., MacCannell K. A., Ecker J., Kruse F., Outten J. T., Sutcliffe D., Gerard R. D., Bruick R. K., Shohet R. V. (2010) PLoS One 5, e11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moslehi J., Minamishima Y. A., Shi J., Neuberg D., Charytan D. M., Padera R. F., Signoretti S., Liao R., Kaelin W. G., Jr. (2010) Circulation 122, 1004–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y., Muyrers J. P., Testa G., Stewart A. F. (2000) Nat. Biotechnol. 18, 1314–1317 [DOI] [PubMed] [Google Scholar]
- 24. Percy M. J., Furlow P. W., Beer P. A., Lappin T. R., McMullin M. F., Lee F. S. (2007) Blood 110, 2193–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Minamisawa S., Gu Y., Ross J., Jr., Chien K. R., Chen J. (1999) J. Biol. Chem. 274, 10066–10070 [DOI] [PubMed] [Google Scholar]
- 26. Silter M., Kögler H., Zieseniss A., Wilting J., Schafer K., Toischer K., Rokita A. G., Breves G., Maier L. S., Katschinski D. M. (2010) Eur. J. Physiol. 459, 569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bohl S., Medway D. J., Schulz-Menger J., Schneider J. E., Neubauer S., Lygate C. A. (2009) Am. J. Physiol. Heart Circ. Physiol. 297, H2054–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eckle T., Köhler D., Lehmann R., El Kasmi K., Eltzschig H. K. (2008) Circulation 118, 166–175 [DOI] [PubMed] [Google Scholar]
- 29. Zieseniss A., Schroeder U., Buchmeier S., Schoenenberger C. A., van den Heuvel J., Jockusch B. M., Illenberger S. (2007) Cell Tissue Res. 327, 583–594 [DOI] [PubMed] [Google Scholar]
- 30. Hirsilä M., Koivunen P., Günzler V., Kivirikko K. I., Myllyharju J. (2003) J. Biol. Chem. 278, 30772–30780 [DOI] [PubMed] [Google Scholar]
- 31. Wenger R. H., Stiehl D. P., Camenisch G. (2005) Sci. STKE 2005, re12. [DOI] [PubMed] [Google Scholar]
- 32. Stiehl D. P., Wirthner R., Köditz J., Spielmann P., Camenisch G., Wenger R. H. (2006) J. Biol. Chem. 281, 23482–23491 [DOI] [PubMed] [Google Scholar]
- 33. Palazzuoli A., Gallotta M., Quatrini I., Nuti R. (2010) Vasc. Health Risk Manag. 6, 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang Y., Hickey R. P., Yeh J. L., Liu D., Dadak A., Young L. H., Johnson R. S., Giordano F. J. (2004) FASEB J. 18, 1138–1140 [DOI] [PubMed] [Google Scholar]
- 35. Myllyharju J. (2009) Curr. Pharm. Des. 15, 3878–3885 [DOI] [PubMed] [Google Scholar]
- 36. Yan L., Colandrea V. J., Hale J. J. (2010) Expert Opin. Ther. Pat. 20, 1219–1245 [DOI] [PubMed] [Google Scholar]
- 37. Huang M., Chan D. A., Jia F., Xie X., Li Z., Hoyt G., Robbins R. C., Chen X., Giaccia A. J., Wu J. C. (2008) Circulation 118, S226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bao W., Qin P., Needle S., Erickson-Miller C. L., Duffy K. J., Ariazi J. L., Zhao S., Olzinski A. R., Behm D. J., Pipes G. C., Jucker B. M., Hu E., Lepore J. J., Willette R. N. (2010) J. Cardiovasc. Pharmacol. 56, 147–155 [DOI] [PubMed] [Google Scholar]
- 39. Hyvärinen J., Hassinen I. E., Sormunen R., Mäki J. M., Kivirikko K. I., Koivunen P., Myllyharju J. (2010) J. Biol. Chem. 285, 13646–13657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kido M., Du L., Sullivan C. C., Li X., Deutsch R., Jamieson S. W., Thistlethwaite P. A. (2005) J. Am. Coll. Cardiol. 46, 2116–2124 [DOI] [PubMed] [Google Scholar]
- 41. Adluri R. S., Thirunavukkarasu M., Dunna N. R., Zhan L., Oriowo B., Takeda K., Sanchez J., Otani H., Maulik G., Fong G. H., Maulik N. (2010) Antioxid. Redox Signal., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Semenza G. L. (2007) J. Bioenerg. Biomembr. 39, 231–234 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.