Abstract
Biodegradation of plant biomass is a slow process in nature, and hydrolysis of cellulose is also widely considered to be a rate-limiting step in the proposed industrial process of converting lignocellulosic materials to biofuels. It is generally known that a team of enzymes including endo- and exocellulases as well as cellobiases are required to act synergistically to hydrolyze cellulose to glucose. The detailed molecular mechanisms of these enzymes have yet to be convincingly elucidated. In this report, atomic force microscopy (AFM) is used to image in real-time the structural changes in Valonia cellulose crystals acted upon by the exocellulase cellobiohydrolase I (CBH I) from Trichoderma reesei. Under AFM, single enzyme molecules could be observed binding only to one face of the cellulose crystal, apparently the hydrophobic face. The surface roughness of cellulose began increasing after adding CBH I, and the overall size of cellulose crystals decreased during an 11-h period. Interestingly, this size reduction apparently occurred only in the width of the crystal, whereas the height remained relatively constant. In addition, the measured cross-section shape of cellulose crystal changed from asymmetric to nearly symmetric. These observed changes brought about by CBH I action may constitute the first direct visualization supporting the idea that the exocellulase selectively hydrolyzes the hydrophobic faces of cellulose. The limited accessibility of the hydrophobic faces in native cellulose may contribute significantly to the rate-limiting slowness of cellulose hydrolysis.
Keywords: Atomic Force Microscopy, Enzyme Catalysis, Enzyme Mechanisms, Enzyme Processing, Microscopic Imaging, Cellobiohydrolase I, Cellulase, Cellulose, Hydrophobic Faces
Introduction
Natural cellulose is a bundle of linear β-1,4-linked glucan chains held tightly in a crystalline structure by the cumulative effect of many inter- and intrachain hydrogen bonds. Cellulose produced by higher plants is the most abundant biopolymer on Earth, accounting for 40–60% by dry weight of plant cell walls. In addition to the traditional uses of cellulose in the paper, food, and textile industries, the new concept of biofuels produced from lignocellulosic biomass is considered a promising route to sustainable energy production. Unfortunately, lignocellulosic material is intrinsically recalcitrant to chemical and enzymatic breakdown to simple sugars that can be fermented to liquid fuels. A deeper understanding of biomass recalcitrance will be required for the potential of lignocellulosic biofuels to be realized (1).
In native plant cell walls, cellulose exists as nanometer scale microfibril networks embedded in matrices of other biopolymers such as hemicelluloses, pectins, and lignins. Chemical pretreatment processes are often required to remove or relocate these “other” matrix polymers, thereby exposing the cellulose to a follow-up enzymatic hydrolysis to produce glucose. The cellulases are a class of enzymes, produced mainly by cellulolytic fungi and bacteria, that catalyze hydrolysis of the β-1,4-glucosidic bonds that link the glucosyl units of cellulose. Cellulases have been classified on the basis of their modes of action on the substrate into three distinct classes that react synergistically: (i) the endo-β-(1,4)-glucanases that cleave the cellulose chain at internal positions to produce new chain ends (2), (ii) the exo-β-(1,4)-d-glucanases that cleave successive cellobiosyl units from the ends of cellulose, and (iii) the β-d-glucosidases that hydrolyze cellobiose to glucose. Among these types of cellulases, the exoglucanases appear to catalyze most of the bond-cleavages in the saccharification of crystalline cellulose and are usually the major component of cellulase preparations, especially in the case of current fungus-derived commercial enzymes.
Cellulose substrates isolated from different sources, though all composed of linear β-1,4-linked glucose polymers, differ significantly in structure as measured in terms of amorphous content, crystallinity, and size and shape of crystallites (3); these structural differences may substantially affect susceptibility to cellulase action. In addition, depending on variations of the patterns of inter- and intrachain hydrogen bonds, cellulose may form different crystalline allomorphs (e.g. cellulose Iα, Iβ, II, III, and IV), upon which the binding and activities of cellulases may be different. Endoglucanases acting alone may attack the non-crystalline regions effectively, but effective attack on the crystalline portion of cellulose requires in addition the synergistic action of the exoglucanases. The fact that cellulose structure affects cellulase activities has long been documented (4), but experimental data presented in the literature have been subject to considerable uncertainty in interpretation of the relationships between cellulose structure and cellulase activities due to disagreements in identification and quantification of cellulose structure by different analytical methods (5). In the current study, cellulose crystals from the cell wall of a green alga, Valonia ventricosa, were intensively characterized by means of atomic force microscopy (AFM).2 Single, continuously monitored crystals of the characterized material were used as substrates in assays visualizing and quantifying the effects of catalysis by Trichoderma reesei CBH I (Cel7A) on the structure of cellulose crystals. The ultimate objective of this study is to elucidate the reaction mechanism of cellulase on true crystalline cellulose.
The cellulase system produced by the filamentous fungus T. reesei is widely used as a commercial enzyme mixture in industry. This system contains two exoglucanases, cellobiohydrolases CBH I and CBH II, which belong to glycoside hydrolase families 7 and 6 respectively, along with five endoglucanases (I–V). It is generally believed that hydrolysis of crystalline cellulose to cellobiose is the rate-limiting step of cellulose degradation, which requires at least two type of synergistic reactions, endo-exo cooperation between endoglucanases and cellobiohydrolases (CBHs), and exo-exo cooperation between CBH I and II, which remove cellobiosyl residues from, respectively, the reducing and nonreducing ends of cellulose chains (6). Among the enzymes present in the cellulase preparation from T. reesei, CBH I is the most abundant component, accounting for >60% of the total protein (7). CBH I is structurally modular, consisting of a family 7 glycoside hydrolase catalytic module and a family 1 carbohydrate-binding module (TrCBM1), connected by a proline- and threonine-rich, highly glycosylated linker peptide. In a proposed (8) mechanism for the action of CBH I on crystalline cellulose, the surface-binding family 1 CBM first binds to the planar surface of cellulose. The reducing end of one cellulose chain is then threaded into the active site tunnel of the catalytic module. The cellulose chain is advanced “processively” through the tunnel, two glucosyl residues at a time, as catalytic residues in the tunnel catalyze the hydrolysis of every second β-1,4-glucosidic bond to depolymerize the chain to free cellobiose units. This last proposed feature is based primarily on the analytical finding that CBH I produces mainly cellobiose, as well as on the structure of the catalytic module (8) and the CBM (9). A recent study employing fast AFM of CBH I on a cellulose crystal appears supportive of this proposed mechanism (10).
During the past decade, researchers have investigated cellulase activity and cellulose-cellulase interaction through a combination of biochemical methods and wet chemistry, as well as imaging techniques such as fluorescence microscopy (11–14), transmission electron microscopy (TEM) (15, 16) and scanning electron microscopy (17). AFM has also been used to characterize the effects of different types of cellulases upon cellulose structure (10, 13, 18–20). Previously, we have studied the interactions with cellulose of both complete cellulase enzymes (13) and their binding modules (14, 21), by employing single-molecule spectroscopy techniques, such as total internal reflection fluorescence microscopy and AFM. It has been demonstrated that AFM is a powerful analytical tool that can be used to obtain highly accurate images of the cellulose surface under physiological conditions with nanometer resolution (22, 23). The sample can be imaged without physical (freezing) or chemical (fixation) treatments; the images thus obtained therefore reflect “native” structures. However, up until now, limited information has been obtained by real-time observation of biological events in time frames long enough to capture significant effects of cellulase action. In the present study, we use AFM to image in real-time the morphological changes occurring in single crystals of Valonia cellulose as a result of hydrolysis by T. reesei CBH I. The Valonia cellulose I has mostly an Iα structure and is a widely accepted native crystalline cellulose standard. Our objective was to learn how the enzyme affects the cellulose morphology at the nanometer scale.
EXPERIMENTAL PROCEDURES
T. reesei CBH I Purification and Valonia ventricosa Cell Culture
CBH I was purified from a seven-day broth of T. reesei grown on Sigmacell 50 (Sigma-Aldrich Corp.) as carbon source. The detailed protocol of CBH I purification has been described elsewhere (24). The cell culture of the green alga V. ventricosa was purchased from UTEX (The Culture Collection of Algae at The University of Texas at Austin) and was grown in enriched seawater medium (UTEX) at 23 °C under a photo regime of 16-h light at 80–100 foot candles of intensity alternated with 8-h periods in the dark. The cells were harvested by centrifugation and were stored at −20 °C.
Preparation of Valonia Cell Wall Cross-section for AFM
Frozen whole Valonia cells were fixed and embedded using the following protocol, with all steps performed in a microwave at 30 °C under vacuum except where noted. Samples were fixed in 2.5% (v/v) glutaraldehyde in PBS, washed 3× with PBS, fixed in 1% (w/v) OsO4 in PBS, washed 3× with distilled H2O, and dehydrated with increasing concentrations of ethanol (30%, 60%, 90%, 3 × 100%, diluted in distilled H2O). Fixed cells were then infiltrated with EMbed 812 resin (Electron Microscopy Sciences, Hatfield, PA) in increasing concentrations of resin (7%, 15%, 30%, 60%, 90%, 3 × 100% resin, diluted in ethanol), with the last two resin exchanges lasting for 48 h each on a rotator at room temperature. The samples were transferred to flat-bottomed BEEM-type capsules (Electron Microscopy Sciences), and the resin was polymerized by heating to 60 °C for 72 h in a nitrogen-purged vacuum oven. The samples were then sectioned to ∼240 nm thickness with a Diatome diamond knife (Electron Microscopy Sciences) on a Leica EM UTC ultramicrotome (Leica, Wetzlar, Germany). Sections were collected on freshly cleaved mica disks for AFM.
Preparation of Valonia Cellulose Crystals for AFM Imaging in Buffer and CBH I Solution
The cellulose crystals were isolated from Valonia cells using the previously described method (25). Purified Valonia cellulose crystals were stored at −20 °C. Freshly cleaved mica coated with an amine polymer, poly-l-lysine, provided a flat, smooth, and stationary substrate on which the cellulose crystals were firmly immobilized to overcome the forces applied by the AFM probe and to permit high resolution of the sample structure. Five μl of poly-l-lysine solution (0.1% w/v in water, Sigma-Aldrich) was added to freshly cleaved mica. After 5 min of incubation, the excess poly-l-lysine solution was removed using a spin coater (Model KW-4A, Chemat Technology, Northridge, CA) operated at 500 rpm for 30 s followed by 4000 rpm for 30 s. After that, 5 μl of a suspension of cellulose crystals (1 mg/ml in water) was added, followed by incubation and spin coating as in the above procedure. The coated mica was then dried in an oven at gentle heat (45 °C) overnight. AFM was operated in tapping mode in buffer and in air. We found that application of a 150-μl liquid droplet (enzyme solution or buffer) to the cellulose-coated mica provided a sample in aqueous environment that was sufficient for imaging for at least 10 h without significant artifacts caused by water evaporation. CBH I (1 mg/ml) was applied to the cellulose-coated mica in acetate buffer (20 mm acetate, 0.02% NaN3, 100 mm NaCl, pH 5.0): the same buffer, without CBHI, was used for buffer-control imaging.
AFM and Image Analysis
AFM measurements were conducted using a Multi-ModeTM scanning probe microscope with NanoScope V controller (Veeco, Santa Barbara, CA). To ensure absolute stability, the AFM was located in a specially designed laboratory with acoustic and vibration isolation. A customized Nikon optical microscope with deep focus (800× magnification) was used to aid in positioning of the AFM tip to the desired location. Silicon nitride probes and sharp nitride lever probes (Veeco) were used in tapping mode to image cellulose fibers in liquid, and DP18/HI'RES/Al BS probes (MikroMasch, San Jose, CA) were used in air. Images of cross section and roughness were analyzed using Nanoscope software (version 7.30). Surface roughness was evaluated by flattening the images (second order) and calculating mean roughness (Ra) and root mean squared roughness (Rq). Ra is the arithmetic average of the absolute values of the surface height deviations measured from the mean plane, expressed as follows in Equation 1.
 |
In contrast, Rq is root mean square average of height deviations taken from the mean image data plane, expressed as follows in Equation 2.
 |
RESULTS AND DISCUSSION
Size and Shape of Valonia Cellulose Crystals
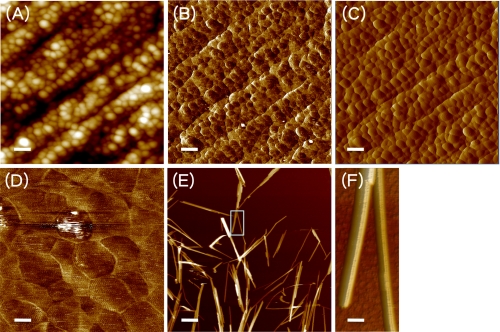
The green alga V. ventricosa produces large cellulose fibrils. Cross-sections of Valonia cellulose crystallites appear square with sizes ranging from 10 to 20 nm as observed by TEM (26, 27). The negative staining method used in those studies measured the crystallites as nonstaining objects, and it was expected that the measured size of the crystallite could be smaller than that of the actual microfibril due to staining effects and resolution limitation. In the current study, the cross-section of the Valonia cell wall was imaged by AFM, with height (Fig. 1A), phase (Fig. 1B), and amplitude (Fig. 1C) images being taken simultaneously. By analyzing these images, the contours of each microfibril could be seen clearly. The shape of each microfibril is hexagonal (Fig. 1D); because the microfibrils are mostly irregular with two narrow sides, they can appear pentagonal in low resolution images (Fig. 1, A–C). The sizes of individual microfibrils range from 10 to 50 nm, with maximum frequency near 35 nm. The isolated cellulose crystals were also imaged (Fig. 1, E and F). Several crystals often form small bundles, and single crystals were measured in the cross-section profile of height images as being 15 to 40 nm in height with maximum frequency of 25 nm, and the length was measured as 200 nm to 2 μm with maximum frequency of 1 μm. The reason that the measured size of single isolated crystals was slightly smaller than that in native cell wall could be one of the following: 1) the measured cross-sections may not be perfectly perpendicular to the long axis of the microfibril, which would result in larger size measured than the actual one; 2) although we used a sharp tip (1 nm in apex) for the AFM imaging to minimize the tip broadening effect, it would be difficult to deconvolute such effects; or 3) the preparation process used to isolate the cellulose crystal could also be expected to cause surface damage and peeling. Nevertheless, the shape of Valonia cellulose observed is an irregular hexagon with two narrow sides. Incidentally, subunits corresponding to the 3–5 nm elementary fibril reported previously (28) were not observed in this study.
FIGURE 1.
Atomic force micrographs of Valonia cell wall microfibrils. A layered structure is shown in height (A), phase (B), and amplitude (C) images taken simultaneously in 1 × 1 μm scan size of native the Valonia cell wall cross-section. Microfibrils are nearly aligned in parallel, and each five to eight layers of microfibrils form a lamella. Zoomed-in phase image (200 × 200 nm, D) shows the cross-section shape of individual microfibrils are mostly anisotropic hexagonal. Isolated Valonia cellulose crystals were also imaged in acetate buffer. Height images of 5 × 5 μm scan size (E) show small bundles formed by several crystals, and a close-up on the selected area from E shows individual crystals (F), the size of which is ∼25 nm based on height measurement. Clear ridges can be seen on the cellulose crystal. Scale bar, 100 nm (A–C), 20 nm (D), 1 μm (E), and 30 nm (F).
CBH I Binds to Cellulose and Moves
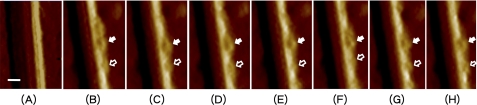
Fig. 2 shows a single cellulose crystal imaged in acetate buffer (as “zero-time control,” Fig. 2A) and at 171, 179, 188, 196, 205, 214, and 222 min, respectively, (Fig. 2, B–H) after addition of CBH I. With the same scanning size and area, we observed that cellulose fibers had well defined ridges and relatively clean surfaces in buffer. After addition of CBH I, there were some new features (right side of cellulose) showing up on the cellulose crystal. These new features could be explained as CBH I enzymes binding to the cellulose surface. These new features were observed over a period of time (222 min), as shown in Fig. 2 (B–H). The apparent size of these features is approximately a quarter of the AFM measured height (25 nm) of the cellulose crystal. Based on published crystal structures for the CBH I catalytic domain (8), and on small-angle x-ray scattering structures for the intact enzyme (29), projection of a bound CBH I molecule away from the cellulose surface may be inferred to be between 5 and 10 nm. A productively bound molecule (bound through both CBM and a catalytic module “threaded” upon a cellulose chain) will have a “projection height” closer to the smaller size, whereas an enzyme molecule bound to the cellulose only by the CBM might have projection heights ranging up to ∼10 nm. Therefore, we propose that the new features appearing on cellulose are most likely to be CBH I enzymes. Although the AFM time-resolution (frame rate ∼ 1 frame/(8 min)) used in this study is not capable of demonstrating processive motion of specific, individual CBH I molecules, the observed new features were similar to those tracked in a recent high speed AFM report (10). Fig. 2 also shows that these features change relative locations on the cellulose surface over the time imaged. Because there are apparently two different sizes of features, with their relative distances changing during the observed time frames (Fig. 2, C and D), we further speculate that the two size categories represent two different kinds of modules (i.e. catalytic and carbohydrate-binding modules) and that the observed changes in intermodule distance reflect conformational changes in the entire CBH I molecule as CBH I catalyzes bond cleavage and moves along on a cellulose chain. Rigorous statistical analysis of continuous real-time imaging using AFM will be required to confirm this speculation.
FIGURE 2.
Three-dimensional rendering of time lapse AFM amplitude images of an individual isolated Valonia cellulose crystal in acetate buffer (A) and at 171, 179, 188, 196, 205, 214, and 222 min after addition of CBH I enzyme (B–H). Features observed attached to one side of the cellulose crystal are believed to be CBH I enzyme molecules. Features observed attached to one side of the cellulose crystal are believed to be CBH I enzyme molecules. Features appearing to have two different sizes may represent the two modules of CBH I (i.e. CD and CBM indicated by solid and open arrows, respectively). The location of CBH I on cellulose was shown to change, which indicates movement of the enzyme. The measured relative position of putative CDs and CBMs was also observed to change between ∼5–10 nm, which is consistent with the modeled linker length, indicating that CBH I may be a dynamic structure during the catalytic process. Scale bar, 10 nm.
Cellulose Surface Roughness Increases with CBH I Reaction
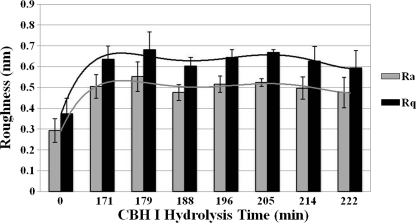
AFM is capable of measuring the surface structure at atomic resolution. We analyzed the structural changes in the cellulose surface concomitant with the action of CBH I. One of the features obtained from AFM measurements is the degree of roughness, which can be used to analyze changes in the surface brought about by friction, adhesion, and catalytic activity. There are many mathematical approaches to calculating surface roughness from AFM images, including probability height distribution, fractal analysis, mean roughness (Ra), and root-mean-squared roughness (Rq). Ra and Rq are the most commonly reported of the measures of surface roughness that can be extracted from AFM images. Fig. 3 represents roughness measurement of the cellulose crystal shown in Fig. 2. We found that cellulose surface roughness increased (in both Ra and Rq cases) by ∼0.3 nm after addition of CBH I, which further confirms that the apparent morphological changes of cellulose observed by AFM were the results of CBH I reaction. The roughness remained almost constant during a subsequent 274 min of measurement in the presence of CBH I (Fig. 3). Valonia celluloses are known to be primarily Iα in crystalline form (30). The distance between Iα cellulose sheets (d110) is 0.39 nm, which is similar to the measured surface roughness increases resulting from CBH I action, indicating that CBH I enzymes affect only the surface layer of cellulose crystals. Previously, it has been demonstrated that the TrCBM1 binds to the hydrophobic faces (1 1 0) of cellulose (21, 31, 32), and these faces are as narrow as 2–4 nm (14, 32). Presumably the hydrophobic faces consist of more than one cellulose chain, thus the roughness change may indicate that the cellulose chains are hydrolyzed individually.
FIGURE 3.
Cellulose surface roughness increases after addition of CBH I. Time lapse roughness analysis (based on Fig. 2) of cellulose fibers in buffer (0 min, control experiment) and in CBH I solution (171, 179, 188, 196, 205, 214, and 222 min). Roughness increases by 0.3 nm shortly after addition of CBH I and then remains at about the same level over 3 h measurement. Trend lines are used to guide the eye. Ra, mean roughness; Rq, root mean squared roughness. Error bars indicate S.D.
Cellulose Crystal Size Decreases during CBH I Hydrolysis
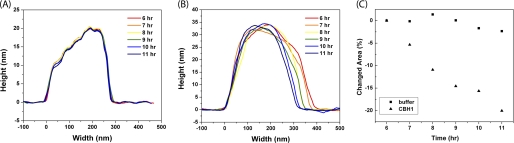
Continuous images of cellulose were taken in real-time during incubation with either buffer or CBH I solution. The size and shape of the cellulose crystal were analyzed by plotting the cross-section profiles of each image frame (Fig. 4, A and B). No change was observed during an 11-h incubation in buffer (Fig. 4A). After addition of CBH I, on the other hand, Fig. 4B shows size reduction of the observed area taken from the average of two successive frames with 50 cross-section lines in each frame (based on 512 × 512 scan lines), which demonstrated that the average width of the cellulose crystal decreased, but the average height remained relatively constant. The apparent reduction of cellulose specifically on one side of the crystal may imply that the CBH I tends to hydrolyze cellulose from certain surfaces. It has been previously reported that the family 1 CBM from CBH I binds only to certain faces of cellulose crystal, i.e. the (1 1 0) faces (or the hydrophobic faces) in the case of Valonia Iα cellulose (28). It could be further speculated that CBH I hydrolysis also occurs only on the hydrophobic faces as discussed below.
FIGURE 4.
Shown are selected cross-section profiles of Valonia cellulose analyzed by AFM height image in acetate buffer (A) and CBH I solution (B) during 11 h of continuous imaging. Averages of 100 cross-section profiles (two successive frames with 50 cross-section lines in each frame) are plotted. In buffer, the cellulose profile remains unchanged. In CBH I solution, cellulose surface morphology changes and size decreases from one side of the crystal resulting cellulose in width changes, whereas height remains approximately the same. C, shown is the percentage change of integrated areas relative to the first point of the sequence (the sixth hour after CBHI addition). Squares: cellulose in acetate buffer showing almost no changes; triangles: cellulose in CBH I solution showing decreases of ∼20%. Note that the cellulose crystals measured in buffer and in CBH I solution are two independent but identically conducted experiments.
The cross-section area of each single cellulose crystal was further integrated based on AFM image section profiles, and a size reduction of 20% was estimated based on 11 h of measurement (Fig. 4C), whereas there was no change observed for the same period of time in buffer (Fig. 4A). The hydrolysis rate of CBH I could not be calculated, because 1) it is not known how many CBH I molecules are involved in the period of 11 h hydrolysis, and 2) it is difficult to estimate how many cellulose chains are hydrolyzed in the 20% size reduction.
CBH I Hydrolysis Mechanism
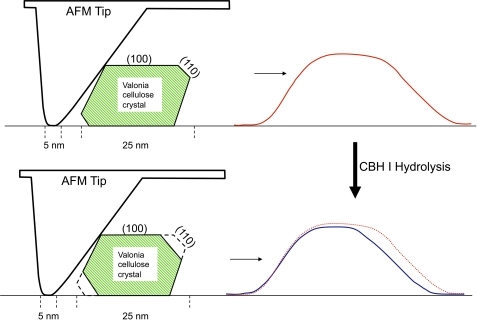
Fig. 5 shows a schematic diagram of an anisotropic hexagonal cellulose crystal and predictions of scanning profiles produced during AFM imaging. In this diagram, the cellulose crystal is initially 25 nm in width and is shaped based on the cellulose Iα structure with two hydrophobic faces (1 1 0) both 3-nm wide. Most likely, the broader hydrophilic faces will attach to the mica surface when a single cellulose crystal makes contact with the mica surface during AFM sample preparation. The schematic of the AFM tip is generated based on a 1-nm radius for the tip apex and the general shape provided by the manufacturer. According to this proposed orientation of the crystal on the mica surface, both hydrophobic (1 1 0) and (−1 −1 0) faces, are exposed and susceptible to enzymatic hydrolysis. AFM, however, can probe only the surfaces geometrically accessible to the tip, so for the situation shown in Fig. 5, only the hydrolytic removal of material from the (1 1 0) face will show up in scan profiles as a difference between scans taken before and after CBH I hydrolysis. The schematic diagram shown in Fig. 5 therefore provides a quite likely explanation for the one-sided reduction of the cellulose crystal width in Fig. 4B. In addition, Fig. 5 illustrates a reasonable explanation for the observation that no significant changes in measured crystal height are detected during enzyme hydrolysis, contrasted with substantial changes in crystal width.
FIGURE 5.
A schematic diagram of AFM measurement of a cellulose crystal, in which the cross-section of a single 25-nm cellulose crystal is represented based on cellulose Iα structure with narrow 3-nm hydrophobic faces (1 1 0). In this schematic representation, the crystal attaches to the mica surface by its broader hydrophilic surface. If the CBH I hydrolyzes both (1 1 0) and (−1 −1 0) faces, the AFM can only measure the changes in the (1 1 0) face, resulting in cross-section profiles of AFM height imaging reduced in width from one side and unchanged in height (Fig. 4). Tip scanning profiles in red and blue lines represent cellulose before and after hydrolysis, respectively. The tip and the cellulose are not in scale.
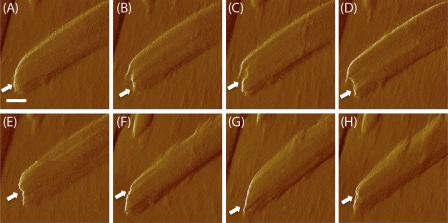
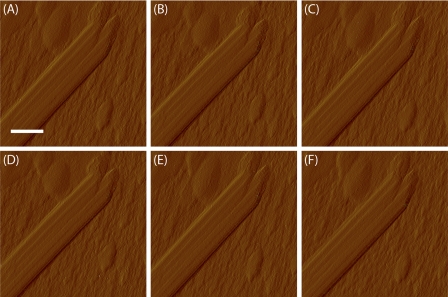
It has been documented that CBH I carries out exoattack on cellulose from its reducing end generating sharpened tips (16). Fig. 6 shows a succession of real-time AFM amplitude images (9 min per scan) of the end of the same cellulose crystal, beginning (”zero time” in the figure legend) 6 h after the addition of CBH I. The amplitude mode yields clear images with high contrast. During the course of the enzyme hydrolysis, we observed various shape changes at the end of the cellulose crystal, such as from smooth (Fig. 6A, 0 min), to sawtoothed (Fig. 6B, 9 min), and then to nicked (Fig. 6C, 26 min). The fiber end appeared smooth again in Fig. 6G (249 min), but with a sharper angle than that in Fig. 6A. The observed tip sharpening is similar to that reported previously (16) where CBH I erodes bacterial cellulose and renders the reducing end pointed in TEM images. We extend the results of the earlier work by providing real-time images of irregularities produced in cellulose-crystal tips by CBH I-catalyzed hydrolytic erosion. For the same reason illustrated in Fig. 5, AFM only detects one side of sharpening, whereas TEM image can show sharpening from both sides. In this study, operating AFM in an aqueous environment allows us to image the process under physiological conditions without intervening chemical/physical treatments. The observed phenomenon is thus closer to that occurring in nature. A control experiment was done by imaging cellulose fiber ends exposed to acetate buffer only. A series of real-time AFM amplitude images were taken starting at 6 h after adding acetate buffer solution rather than CBH I solution. No observable changes were found in either a short time (0 to 34 min with ∼9 min frame rate) or a long time scale (360 min) (Fig. 7). The ends of isolated cellulose crystals sometimes appear to be nicked branches (Fig. 7), which could have resulted from the harsh preparation conditions (strong acid/base or high temperature) of cellulose isolation.
FIGURE 6.
Detailed structure of the end of a single cellulose crystal measured by AFM amplitude images. The cellulose crystal was treated in CBH I solution, with imaging begun after 6 h treatment (A) and continued for an additional 274 min. Representative images are shown at 9 (B), 26 (C), 43 (D), 86 (E), 240 (F), 249 (G), and 274 (H) additional minutes of CBH I treatment, respectively. At successive time points, the crystal ends appeared smooth (A), sawtoothed (B), and then nicked (C), indicating that CBH I erodes cellulose from the end of the crystal irregularly. White arrows highlight the crystal end change regions. Apparent overall end-sharpening (A–G) is also observed only on one side, due to the effects of AFM tip geometry illustrated in Fig. 5. These observations support previously reported cellulose tip sharpening caused by CBH I and imaged by TEM. The AFM imaging provides real-time measurement and more details of morphological changes in crystal ends during CBH I hydrolysis. Scale bar, 20 nm.
FIGURE 7.
“Buffer only” control for the experiment in Fig. 6. The cellulose crystal end shows no change in acetate buffer either in the short time scale (A–E) or in the long time scale (F). AFM amplitude images were taken 0 min (A), 9 min (B), 17 min (C), 26 min (D), 34 min (E), and 360 (F) min after the end of 6 h of initial exposure to acetate buffer. Scale bar, 20 nm.
Conclusions
Atomic force microscopy has been used to make real-time measurements, both qualitative and quantitative, of the morphological changes in single crystals of cellulose during hydrolysis by T. reesei CBH I. We demonstrated structural changes including decrease in width, increases of surface roughness, and changes in shape of fiber ends. We have observed that the cross-section of Valonia cellulose crystal is apparently an irregular hexagon with two narrow hydrophobic faces that are binding surfaces for TrCBM1, and are likely to be the surfaces from which CBH I-catalyzed reactions remove cellulose chains. To the best of our knowledge, this is the first direct measurement of CBH I hydrolysis of crystalline cellulose that supports the hypothesis that CBH I hydrolyzes only the hydrophobic faces of cellulose. We further speculate that development of pretreatment approaches aimed at modification of cellulose to increase the accessibility of the hydrophobic planes to enzyme catalyst may lead to further improvements in the efficiency of enzymatic saccharification of biomass.
This work was supported by the United States Department of Energy (DOE), Office of Energy Efficiency and Renewable Energy, the Office of the Biomass Program for the work to develop enzymes, and the United States DOE Office of Science, Office of Biological and Environmental Research through the BioEnergy Science Center, a DOE Bioenergy Research Center, for the work on AFM visualization and analysis.
- AFM
- atomic force microscopy
- CBH I
- cellobiohydrolase I
- CBM
- cellulose-binding module
- TEM
- transmission electron microscopy.
REFERENCES
- 1. Himmel M. E., Ding S. Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., Foust T. D. (2007) Science 315, 804–807 [DOI] [PubMed] [Google Scholar]
- 2. Reese E. T., Siu R. G., Levinson H. S. (1950) J. Bacteriol. 59, 485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Montanari S., Rountani M., Heux L., Vignon M. R. (2005) Macromolecules 38, 1665–1671 [Google Scholar]
- 4. Fan L. T., Lee Y. H., Beardmore D. H. (1980) Biotechnol. Bioeng. 22, 177–199 [Google Scholar]
- 5. Park S., Baker J. O., Himmel M. E., Parilla P. A., Johnson D. K. (2010) Biotechnol. Biofuels 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henrissat B., Driguez H., Viet C., Schülein M. (1985) Bio/Technology 3, 722–726 [Google Scholar]
- 7. Enari T. M., Niku-Paavola M. L. (1987) Crit. Rev. Biotechnol. 5, 67–87 [DOI] [PubMed] [Google Scholar]
- 8. Divne C., Ståhlberg J., Reinikainen T., Ruohonen L., Pettersson G., Knowles J. K., Teeri T. T., Jones T. A. (1994) Science 265, 524–528 [DOI] [PubMed] [Google Scholar]
- 9. Kraulis J., Clore G. M., Nilges M., Jones T. A., Pettersson G., Knowles J., Gronenborn A. M. (1989) Biochemistry 28, 7241–7257 [DOI] [PubMed] [Google Scholar]
- 10. Igarashi K., Koivula A., Wada M., Kimura S., Penttilä M., Samejima M. (2009) J. Biol. Chem. 284, 36186–36190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Helbert W., Chanzy H., Husum T. L., Schülein M., Ernst S. (2003) Biomacromolecules 4, 481–487 [DOI] [PubMed] [Google Scholar]
- 12. Moran-Mirabal J. M., Santhanam N., Corgie S. C., Craighead H. G., Walker L. P. (2008) Biotechnol. Bioeng. 101, 1129–1141 [DOI] [PubMed] [Google Scholar]
- 13. Liu Y. S., Luo Y. H., Baker J. O., Zeng Y. N., Himmel M. E., Smith S. J., Ding S. Y. (2010) Proc. SPIE 7571, 757103 [Google Scholar]
- 14. Ding S. Y., Xu Q., Ali M. K., Baker J. O., Bayer E. A., Barak Y., Lamed R., Sugiyama J., Rumbles G., Himmel M. E. (2006) BioTechniques 41, 435–446 [DOI] [PubMed] [Google Scholar]
- 15. Bayer E. A., Chanzy H., Lamed R., Shoham Y. (1998) Curr. Opin. Struct. Biol. 8, 548–557 [DOI] [PubMed] [Google Scholar]
- 16. Boisset C., Fraschini C., Schülein M., Henrissat B., Chanzy H. (2000) Appl. Environ. Microbiol. 66, 1444–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bohn A., Fink H. P., Ganster J., Pinnow M. (2000) Macromol. Chem. Phys. 201, 1913–1921 [Google Scholar]
- 18. Lee I., Evans B. R., Woodward J. (2000) Ultramicroscopy 82, 213–221 [DOI] [PubMed] [Google Scholar]
- 19. Liu H., Fu S. Y., Zhu J. Y., Li H., Zhan H. Y. (2009) Enzyme Microb. Technol. 45, 274–281 [Google Scholar]
- 20. Quirk A., Lipkowski J., Vandenende C., Cockburn D., Clarke A. J., Dutcher J. R., Roscoe S. G. (2010) Langmuir 26, 5007–5013 [DOI] [PubMed] [Google Scholar]
- 21. Liu Y. S., Zeng Y., Luo Y., Xu Q., Himmel M. E., Smith S. J., Ding S. Y. (2009) Cellulose 16, 587–597 [Google Scholar]
- 22. Baker A. A., Helbert W., Sugiyama J., Miles M. J. (2000) Biophys. J. 79, 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ding S. Y., Himmel M. E. (2006) J. Agric. Food Chem. 54, 597–606 [DOI] [PubMed] [Google Scholar]
- 24. Baker J. O., Mitchell D. J., Grohmann K., Himmel M. E. (1991) in Enzymes in Biomass Conversion (Leatham G. F., Himmel M. E. eds.) pp. 313–330, American Chemical Society, Washington D. C [Google Scholar]
- 25. Imai T., Putaux J. L., Sugiyama J. (2003) Polymer 44, 1871–1879 [Google Scholar]
- 26. Chanzy H., Henrissat B., Vuong R., Revol J. F. (1986) Holzforschung 40 (Suppl.), 25–30 [Google Scholar]
- 27. Revol J. F. (1982) Carb. Polymers 2, 123–134 [Google Scholar]
- 28. Gardner K. H., Blackwell J. (1971) J. Polymer Sci. Part C: Polymer Symposia 36, 327–340 [Google Scholar]
- 29. Abuja P. M., Schmuck M., Pilz I., Tomme P., Claeyssens M., Esterbauer H. (1988) Eur. Biophys. J. 15, 339–342 [Google Scholar]
- 30. Sugiyama J., Vuong R., Chanzy H. (1991) Macromolecules 24, 4168–4175 [Google Scholar]
- 31. Lehtiö J., Sugiyama J., Gustavsson M., Fransson L., Linder M., Teeri T. T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu Q., Tucker M. P., Arenkiel P., Ai X., Rumbles G., Sugiyama J., Himmel M. E., Ding S. Y. (2009) Cellulose 16, 19–26 [Google Scholar]