Abstract
Phylogenetic analysis has previously shown that plant receptor-like kinases (RLKs) are monophyletic with respect to the kinase domain and share an evolutionary origin with the animal interleukin-1 receptor-associated kinase/Pelle-soluble kinases. The lysin motif domain-containing receptor-like kinase-3 (LYK3) of the legume Medicago truncatula shows 33% amino acid sequence identity with human IRAK-4 over the kinase domain. Using the structure of this animal kinase as a template, homology modeling revealed that the plant RLK contains structural features particular to this group of kinases, including the tyrosine gatekeeper and the N-terminal extension α-helix B. Functional analysis revealed the importance of these conserved features for kinase activity and suggests that kinase activity is essential for the biological role of LYK3 in the establishment of the root nodule nitrogen-fixing symbiosis with rhizobia bacteria. The kinase domain of LYK3 has dual serine/threonine and tyrosine specificity, and mass spectrometry analysis identified seven serine, eight threonine, and one tyrosine residue as autophosphorylation sites in vitro. Three activation loop serine/threonine residues are required for biological activity, and molecular dynamics simulations suggest that Thr-475 is the prototypical phosphorylated residue that interacts with the conserved arginine in the catalytic loop, whereas Ser-471 and Thr-472 may be secondary sites. A threonine in the juxtamembrane region and two threonines in the C-terminal lobe of the kinase domain are important for biological but not kinase activity. We present evidence that the structure-function similarities that we have identified between LYK3 and IRAK-4 may be more widely applicable to plant RLKs in general.
Keywords: Molecular Dynamics, Plant, Protein Kinases, Protein Structure, Receptor Serine/Threonine Kinase, Legume, Receptor-like Kinase, Symbiosis
Introduction
Higher plants show a remarkable expansion of receptor-like kinases (RLKs)4; for example, there are over 400 RLK genes in Arabidopsis thaliana (1), which have diverged to fulfill different physiological roles. These genes all encode proteins with the same basic structure as follows: an extracellular region (containing various domains that presumably bind different types of ligands) followed by a single transmembrane-spanning segment and an intracellular region containing a Ser/Thr kinase-like domain. Phylogenetic analysis has shown that the kinase domain is monophyletic and shares a common evolutionary origin with the human IRAK and Drosophila Pelle group of soluble Ser/Thr kinases. These kinases are quite closely related to the animal receptor Tyr kinase and the Raf kinase families (1).
The structure of the human IRAK-4 kinase has been solved and has revealed several novel features, including a Tyr gatekeeper in the catalytic pocket, an N-terminal helix B, and regulation by activation loop phosphorylation that resembles Tyr kinases (2, 3). IRAK-4 contains an Arg before the catalytic Asp (an RD kinase), which interacts with a phosphorylated residue in the activation loop. It shares about 38% sequence identity with another family member, IRAK-1, which does not contain the Arg residue and thus is a non-RD kinase. Both IRAK-4 and IRAK-1 are involved in innate and adaptive immune responses and play central roles in signal transduction via the related Toll-like receptors and interleukin 1 receptors (TLR/IL-1R family).
In comparison with work on related animal kinases, the elucidation of the structure and activation of the kinase domains of plant RLKs is still in its infancy. The best studied examples are BRI1 (4, 5), BAK1 (5), SERK1 (6), and FLS2 (7) of A. thaliana, Xa21 of rice (8), SYMRK of Lotus japonicus (9), and NARK of soybean (10). Analysis of the kinase domains of these proteins in vitro and in planta have suggested that auto-phosphorylation is an important event for signal transduction and have defined certain phosphorylation sites required for kinase activity and for biological activity. For example, phosphorylation of the activation loop of BRI1 and BAK1 is important for transduction of the brassinosteroid signal (4, 5), whereas phosphorylation of the juxtamembrane region is important for Xa21-mediated immunity (8). By functional analysis and by reference to animal receptor Tyr kinases (11), it has been suggested that the mechanism of activation of plant non-RD RLKs, such as Xa21, may be different from RD kinases, such as BRI1 and BAK1, where activation loop phosphorylation is important (8). To date, no structural data are available for the kinase domain of plant RLKs, but structures of two bacterial effectors in complex with a related plant receptor-like cytoplasmic kinase (RLCK), called Pto, have been reported (12, 13).
We are studying the perception of bacterial lipochitooligosaccharidic signals (Nod factors), which are important for establishing the nitrogen-fixing symbiosis between rhizobia bacteria and legume plants. Perception of these factors by the plant leads to controlled infection and the development of nodules on the roots (nodulation), in which the bacteria fix atmospheric dinitrogen which supports the growth of the plant (14). Genetic studies have led to the identification of two lysin motif RLKs (LysM-RLKs) which are required for Nod factor responses and nodulation (15–20). In the model legume Medicago truncatula (barrel medic), one of these RLKs, NFP, has been shown to possess a dead kinase, whereas the other, LYK3, has an active kinase (16). In L. japonicus the two genes, NFR5/NFR1, potentially orthologous to NFP/LYK3 are both required for the earliest Nod factor signaling events, whereas in M. truncatula LYK3 appears to be essential only for later responses, leading to nodulation and infection (15, 17–20). In this study, we aimed to determine whether LYK3 kinase activity is essential for nodulation in M. truncatula and to identify regions of the kinase domain that are important for signal transduction. Our results suggest a remarkable structural conservation between the kinase domain of LYK3 and human IRAK-4, which may suggest that both kinases are activated via similar mechanisms. We further argue that this structural and functional homology extends to other members of the plant RLK/RLCK family.
EXPERIMENTAL PROCEDURES
Plant Constructs, Transformation, and Analysis
For plant expression, the coding sequence of LYK3 was cloned in-frame with a C-terminal 3×FLAG epitope tag and in-between the constitutive cauliflower mosaic virus (CaMV) 35S promoter and terminator. Point mutations were introduced using the QuikChangeTM site-directed mutagenesis kit (Stratagene); all constructs were sequenced to exclude PCR errors and to confirm the designed mutation(s). Agrobacterium rhizogenes ARqua1 strains, containing the constructs in the pBin+ vector, were used to produce M. truncatula lyk3-1 plants (19, 20) with transgenic roots that were selected on kanamycin (21). Complementation for root nodulation was analyzed following transfer into growth pouches and inoculation with Sinorhizobium meliloti strain 2011, carrying either a LacZ or GFP marker (22). Data were analyzed using the χ2. For constructs that did not complement (G334E, K349A, D441A, G504E, S471A/T472A/T475A, T475A, T512A, E362A, and T319A), at least eight plants were analyzed for expression of the fusion protein in roots by Western blotting with anti-FLAG antibodies; in all cases at least five of the tested plants expressed the fusion protein, like wild type. The experiments were repeated at least twice. Subcellular localization of mutant proteins was analyzed using confocal microscopy by comparison of the localization of the C-terminal fusions of these proteins with sYFP2 (23) with a plasma membrane and cytosolic marker proteins following their simultaneous production in Nicotiana benthamiana leaves, essentially as described (24) with the modifications described in supplemental Methods. All proteins tested localized to the plasma membrane, like wild type.
Expression in E. coli and Kinase Activity
For expression in E. coli, the predicted intracellular region (aa 257–620) was cloned behind the GST tag in the vector pGEX-6P-1 (16). Point mutations were introduced as above. Protein purification and kinase activity used the methods described (16) with minor modifications (supplemental Methods).
Western Blots
Proteins were separated on 10 or 12% SDS-polyacrylamide gels, transferred onto nitrocellulose or polyvinylidene fluoride (PVDF) membranes, and detected with antibodies and coupling to horseradish peroxidase (HRP) as indicated in each figure. The antibodies were used as follows: anti-FLAG-HRP (Sigma, 1:5000), anti-GST-HRP (GE Healthcare, 1:10,000), and rabbit anti-phospho-Thr (1:1000), and rabbit anti-phospho-Ser (1:200) (all from Invitrogen) in combination with goat anti-rabbit-coupled HRP (Millipore, 1:25,000) and mouse monoclonal anti-phospho-Tyr (1:666 Millipore clone 4G10) in combination with goat anti-mouse-coupled HRP (Millipore, 1:12,500). The specificity of the anti-Tyr(P) antibodies was shown by competition of reactivity in the presence of 20 mm Tyr(P) or 20 mm each of Thr(P) and Ser(P). HRP was detected by chemiluminescence using the Immobilon Western blotting detection system (Millipore) and a digital camera (G-Box, Syngene, UK).
Mass Spectrometry Analysis
About 50 μg of purified protein was run on a 10% SDS-polyacrylamide gel and in-gel digested with trypsin and/or V8 protease (25). The resulting peptides were analyzed by nano-liquid chromatography-tandem mass spectrometry (LC-MS/MS) using an Ultimate 3000 system (Dionex, Amsterdam, the Netherlands) coupled to an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Peptides were identified using Mascot software (version 2.2.03, Matrix Science, London, UK). For phosphopeptides, each MS/MS spectrum was manually inspected to ensure acceptable ion coverage and phosphorylation site identification (for details see supplemental Methods).
Homology Modeling and Molecular Dynamics (MDs) Simulations
The homology model of LYK3 kinase was built based on an alignment of the intracellular region of LYK3 (GenBankTM accession number CAM06621.1) with the IRAK-4 kinase sequence and on the protein crystal structure of IRAK-4 (Protein Data Bank code 2NRU) (2) using the Orchestrar modeling suite of SYBYL 7.3 (Tripos Associates, St. Louis). The parameters and verification of the model were carried out as described (supplemental Methods). For MD simulations, models of both phosphorylated and unphosphorylated kinase were soaked in a cubic box of water molecules and subjected to energy minimization and the equilibration step, followed by 7.3-ns long duration MD production using the AMBER8 program (University of California) with parameters for phosphorylated residues.
RESULTS
LYK3 Kinase Activity Is Required for its Biological Role
The predicted architecture of LYK3 (16, 17) is shown in Fig. 1A. The intracellular region (LYK3-IR, aa 257–620), containing a core protein kinase domain (aa 322–592), was used to search the Protein Data Bank using PSI_BLAST. Among all kinases for which the tridimensional structure is known, the human interleukin-1 receptor-associated kinase-4 (IRAK-4) showed the closest sequence identity as follows: 33% over the alignment shown in Fig. 1B, in comparison with 32% for the plant Pto kinase (12). Highly conserved residues/motifs characteristic of all active kinases are identical in both the human and plant sequences, including the nucleotide-binding Gly-rich loop, a catalytic Lys in the “VAIK” phosphotransfer motif, the catalytic Asp in the HRD motif of the catalytic loop, and the DFG at the start of the activation segment (26). The presence of the Arg in the HRD motif shows that both proteins are RD kinases (Fig. 1B).
FIGURE 1.
Domain architecture of LYK3 and its sequence homology with IRAK-4. A, domain architecture of the predicted coding region of LYK3, with numbering corresponding to the amino acid sequence. The architecture is based on GenBankTM CAM06621.1 and published information (16, 17). SP, signal peptide; LysM, lysin motif; TM, transmembrane segment; pKinase, protein kinase; ER, extracellular region; IR, intracellular region. B, alignment of the kinase domains of LYK3 and IRAK-4 with shading of conserved residues. The start and end of the core kinase domain are marked with arrows. The motifs and structural features of interest have been boxed; the gatekeeper Tyr is indicated by the letter Y, and the two residues determining an RD kinase are indicated by the letters RD. The position of the α-helices in IRAK-4 are underlined. In LYK3, residues that have been mutated in subsequent experiments are marked with *, and every 10th residue is marked by !.
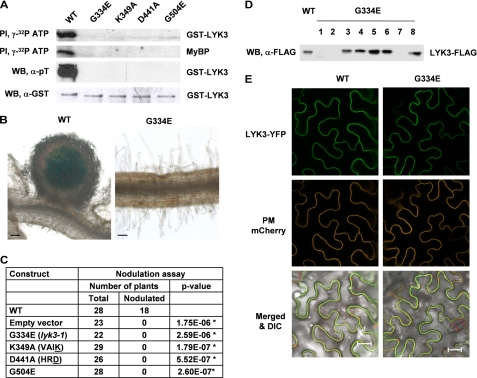
To determine whether kinase activity is required for the function of LYK3 in the development of root nodules in response to rhizobia bacteria (nodulation), site-directed mutagenesis was used to create specific mutants that were tested for both kinase activity and nodulation activity. The LYK3-IR was expressed in Escherichia coli, and the purified glutathione S-transferase (GST) fusion protein was shown to have both autophosphorylation and transphosphorylation activities (Fig. 2A). Proteins containing mutations in each of four highly conserved kinase domain residues (Gly-334, Lys-349, Asp-441, and Gly-504; Fig. 1B) all lost both in vitro kinase activities, as expected (Fig. 2A). These mutants included one in the Gly-rich loop, G334E, which occurs in the strong lyk3-1 mutant allele (20). Wild-type and four LYK3 constructs containing the above mutations were then analyzed for their ability to complement the lyk3-1 mutant for nodulation activity (Fig. 2B); constructs of all four kinase-dead mutants failed to complement (Fig. 2C). To verify that the proteins were expressed, Western blot analysis showed that roots of at least 60% of the M. truncatula transgenic plants tested expressed the LYK3 proteins (data for the G344E mutant protein is shown in Fig. 2D). To check for correct subcellular localization, yellow fluorescent protein (YFP) fusions of the wild-type and mutant proteins were expressed in the N. benthamiana leaf expression system; all the proteins localized uniformly to a thin layer at the cell boundary (data for the G344E mutant protein is shown in Fig. 2E), identical to the localization of a plasma membrane marker labeled with mCherry (PM mCherry). This localization is different from a fluorescent protein cytosolic marker where the fluorescence is nonuniform at the cell boundary and also includes cytoplasmic strands and nuclei (supplemental Fig. S1). Thus, both the wild-type LYK3 and the derived mutant proteins all localize to the plasma membrane. Together, the results suggest that kinase activity of LYK3 is required for its biological function.
FIGURE 2.
LYK3 has autophosphorylation and transphosphorylation activity, and kinase activity is required for its biological role. A, protein phosphorylation activity of LYK3-IR. The purified GST-LYK3-IR (GST-LYK3) protein and four mutant proteins with point mutations in conserved kinase residues were analyzed in vitro for autophosphorylation activity and transphosphorylation activity (using myelin basic protein, MyBP) using radiolabeled ATP ([γ-32P]ATP). The purified proteins were also subjected to Western blot analysis using anti-phospho-Thr antibodies (α-pT). Anti-GST antibodies (α-GST) were used to check the protein loadings on the gel. Only the wild-type protein showed auto- and transphosphorylation activity in vitro and Thr phosphorylation in E. coli. B, nodulation assay for biological activity. Full-length wild-type LYK3-FLAG protein and the corresponding G334E mutant were analyzed for complementation of the M. truncatula lyk3-1 mutant for nodulation. Transgenic roots with the wild-type construct but not the G334E mutant form nodules following inoculation with the bacterial symbiont, S. meliloti. LacZ staining shows the presence of the bacteria in the nodule. C, complementation for nodulation. Similar testing of all four mutants shows that none of the kinase-dead mutants complement lyk3-1 for nodulation; * represents a highly significantly difference (p value <0.01) to the wild-type construct. D, expression of fusion proteins in roots. To check for expression in M. truncatula, roots of eight individual plants transformed with the G334E construct were analyzed by Western blot using anti-FLAG antibodies; 60% of the plants express easily detectable levels of the LYK3-FLAG protein. E, fusion protein subcellular localization. To check for correct subcellular localization, full-length wild type (WT) or the G334E mutant (G334E) of LYK3 were fused to sYFP2 (LYK3-YFP) and were each co-expressed in N. benthamiana leaves with a plasma membrane marker, HVR-ROP7 labeled with mCherry (PM-mCherry), and the leaves were analyzed by confocal microscopy. Superposition of the fluorescence images shows clear co-localization of the LYK3 WT and the G334E mutant YFP proteins with the plasma membrane mCherry marker at the cell boundary (merged and differential interference contrast image, DIC). Bars, 100 μm in B and 20 μm in E. PI, Phosphorimage; WB, Western blot.
LYK3 Is a Dual Specificity Kinase and Autophosphorylates on Ser/Thr and Tyr Residues
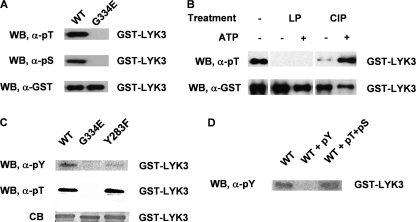
There is substantial evidence showing that IRAK/Pelle kinases and their plant RLK homologues are Ser/Thr kinases. Using antibodies to specific phosphorylated amino acids, we showed that the wild-type LYK3-IR protein, expressed in E. coli, is phosphorylated on both Ser and Thr residues (Fig. 3A). As the G334E mutant did not react with either of the antibodies, we can conclude that the phosphorylation of the wild-type LYK3 protein is due to autophosphorylation. To identify the phosphorylation sites, the wild-type protein was dephosphorylated by treatment with either λ-phosphatase or calf intestinal phosphatase and then rephosphorylated in vitro. The λ-phosphatase was more efficient in dephosphorylation, but the protein was not able to rephosphorylate during the time of the assay, whereas the calf intestinal phosphatase-treated protein successfully rephosphorylated (Fig. 3B). Both the untreated and the calf intestinal phosphatase-treated/rephosphorylated proteins were used for identification of the phosphorylated residues by LC-MS/MS analysis (supplemental Table S1). By using both trypsin and V8 protease digestion, almost 95% of the LYK3 sequence was covered in the MS spectra (Fig. 4). In total, the position of seven phosphorylated serines and eight phosphorylated threonines were identified (Fig. 4 and supplemental Fig. S2). Seven of them were found in the juxtamembrane (JM) region, two in the C-terminal tail (C-tail), and five in the core kinase domain, including two threonines (Thr-472 and Thr-475) in the activation loop. In addition, two peptides suggested that the activation loop Ser-471 is also phosphorylated, although the phosphorylation position remains ambiguous (supplemental Fig. S2). It is noteworthy that phosphorylated Thr-433 was detected only after de- and rephosphorylation of the LYK3 kinase. This treatment of the protein also revealed that Tyr-283 in the JM region is a likely site of Tyr phosphorylation, although the phosphorylation position could not be confirmed with certainty by the LC-MS/MS analysis (supplemental Fig. S2). To further investigate this observation, the Tyr phosphorylation of a Y283F mutant protein was compared with the wild-type and to the kinase-dead G334E proteins using anti-phospho-Tyr antibodies. The antibodies did not react to the G334E protein, suggesting that the Tyr phosphorylation of the wild-type protein is due to autophosphorylation (Fig. 3C). The poor reactivity of the Y283F protein to the antibody suggests that Tyr-283 is a major site of Tyr phosphorylation but may not be the only site as weak reactivity was still observed (Fig. 3C). The antibodies used were highly specific as reactivity of the anti-phospho-Tyr antibody to the wild-type protein could be competed with Tyr(P) but not by Thr(P) and Ser(P) (Fig. 3D), and also the Y283F mutant still reacted like wild-type to the anti-phospho-Thr antibodies (Fig. 3C). Together, these results suggest that LYK3 has mixed Ser/Thr and Tyr kinase activity and autophosphorylates multiple Ser/Thr residues and probably at least two Tyr residues.
FIGURE 3.
LYK3 autophosphorylates on Ser, Thr, and Tyr residues and can be rephosphorylated in vitro, following partial dephosphorylation. A, analysis of the phosphorylation status of the GST-LYK3-IR wild-type (WT) and mutant G334E fusion proteins, purified from E. coli, using anti phospho-Thr (α-pT) and anti-phospho-Ser (α-pS) antibodies. Anti-GST (α-GST) antibodies show the protein loading. The wild-type protein but not the mutant is phosphorylated on both amino acids. B, dephosphorylation and rephosphorylation of LYK3-IR. The wild-type protein is totally dephosphorylated with λ-phosphatase (LP) and did not rephosphorylate with ATP in the presence of phosphatase inhibitors during the assay (+ATP), whereas the protein only partially dephosphorylated with calf intestinal phosphatase rephosphorylated in the same conditions. C, analysis of the tyrosine phosphorylation status of GST-LYK3-IR fusion proteins. Using anti-phospho-Tyr (α-pY) antibodies, the Y283F mutant protein shows very low tyrosine phosphorylation in comparison with the wild-type protein, whereas the kinase-dead G334E protein shows no Tyr phosphorylation. Both wild-type and Y283F proteins show similar Thr phosphorylation. The Coomassie Blue staining (CB) shows the protein loading. D, specificity of the anti-Tyr(P) (pY) antibodies. Reactivity to the wild-type protein (WT) was competed by Tyr(P) and not by Thr(P) + Ser(P) (pT+pS). WB, Western blot.
FIGURE 4.
Autophosphorylation sites in the intracellular region of LYK3. The GST-LYK3-IR autophosphorylated protein was purified from E. coli, and the phosphoresidues were identified by LC-MS/MS. Identified phosphorylated residues of LYK3 are shaded and underlined; those that are ambiguous are not underlined. Regions not covered by the MS analysis are in gray. Regions N- and C-terminal of the core kinase domain, corresponding, respectively, to the juxtamembrane and C-tail regions are shown in bold italics.
LYK3 Kinase Domain Shows Structural Features Common to the Human IRAK-4 Kinase Domain
We used a homology modeling approach using the IRAK-4 crystal structure as template (2) to provide new insights into the architecture of the LYK3 kinase. In this structure (Protein Data Bank code 2NRU) two activation loop residues, Thr-345 and Ser-346, are phosphorylated (2) and both are required for full kinase activity (27). To mimic the active state, the three identified phosphorylated residues in the activation loop of LYK3 (Ser-471, Thr-472, and Thr-475) were included in their phosphorylated forms.
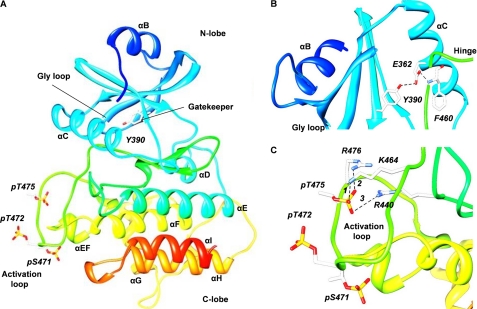
The model (Fig. 5A) shows the typical two-lobe structure of a kinase. The N-terminal lobe consists of the five-stranded antiparallel β-sheet and the prominent α-helix, termed helix C (aa 355–369). The LYK3 model, similarly to the IRAK-4 fold, contains an N-terminal extension with an α-helix, (helix B, aa 311–319; Fig. 1B), which packs against the β-sheet. The characteristic Gly-rich loop (aa 329–334), which anchors the nontransferable phosphates of ATP, follows this region and is located between the first two β-strands. The N-terminal lobe is connected to the C-terminal one through a hinge loop that is immediately preceded by a tyrosine (Tyr-390), which is correctly located to play the role of the “gatekeeper,” controlling access to the back of the pocket. In the LYK3 model, Tyr-390 establishes a hydrogen bond with the highly conserved glutamate, Glu-362 (Fig. 5B). These two residues are part of a stable hydrogen bond network because Glu-362 also acts as a hydrogen bond acceptor for the nitrogen of the phenylalanine (Phe-460) backbone in the DFG motif (Fig. 5B). This network is identical to the one involving Tyr-262, Glu-233, and Phe-350 in the IRAK-4 crystal structure. The C-terminal lobe is formed by several α-helices, small flexible loops, and the larger activation segment (aa 459–487) containing the activation loop (aa 466–477).
FIGURE 5.
Homology model of the LYK3 kinase domain based on the crystal structure of IRAK-4. A, homology model of the whole LYK3 kinase domain, showing the two-lobe structure and the position of key features. B, detail of the model surrounding the gatekeeper residue, Tyr-390, in the N-terminal lobe and connections via Glu-362 to Phe-460 in the C-terminal lobe. C, detail of the triply phosphorylated activation loop in the C-terminal lobe showing the hydrogen bond network around phospho-Thr-475.
Activation Loop Phosphorylation of LYK3 Resembles That of IRAK-4
The structure of IRAK-4 in its active state revealed that Thr-345 is the prototypical phosphorylated residue of the activation loop that interacts with the arginine in the HRD motif of the catalytic loop (2). This type of interaction is important for the activation of many RD kinases (11, 28). Phosphorylation may also occur on other activation loop residues (secondary sites), such as Thr-342 and Ser-346 in IRAK-4 (27), and can lead to changes in the conformation of the activation segment, thus activating the kinase and allowing substrate access (28).
To explain how phosphorylation might influence the structural dynamics of the activation loop of LYK3, the model of LYK3, triply phosphorylated in the activation loop, was compared with a model of the unphosphorylated protein using molecular dynamics (MD) simulations. The analysis of the energies for both models along the 7.3-ns simulations in explicit water revealed that the lowest energies were reached in the phosphorylated protein (supplemental Fig. S3A). The overall geometries of the models remained stable along the simulations as deduced by the calculation of the root-mean-square deviations of the α-carbons with respect to their initial states (supplemental Fig. S3B). Superimposition of the unphosphorylated LYK3 structure extracted at the end of the MD simulation on the phosphorylated LYK3 confirmed that the overall domain structure is stable, with the two structures sharing the same three-dimensional features, with the exception of surface loops (supplemental Fig. S4).
Attention was then focused on the activation segment (aa 459–487). Detailed analysis of the activation loop indicates that the phosphorylated Thr-475 is part of a polar network that remains stable for more than 60% of the simulation. The phosphate group is involved in salt bridges with Arg-476 located in the activation loop, and with Arg-440 and Lys-464 that are located, respectively, in the conserved HRD motif and in the activation segment (Fig. 5C). During the simulation, the phosphorylated Thr-472 loses an initial salt bridge with Lys-497 and points out of the protein-like residue Ser(P)-471; both residues expose the phosphate groups to the solvent environment (Fig. 5C and supplemental Fig. S4B). In the case of the unphosphorylated model, the activation loop does not contain this network of salt bridges and displays a more flexible behavior and a higher energy level. The absence of phosphorylated residues also influences the electrostatic properties of the activation loop, as the three positively charged amino acids are not neutralized by the phosphate groups (supplemental Fig. S4C).
Functional Analysis Identifies Key Residues for LYK3 Activity in the Kinase Domain
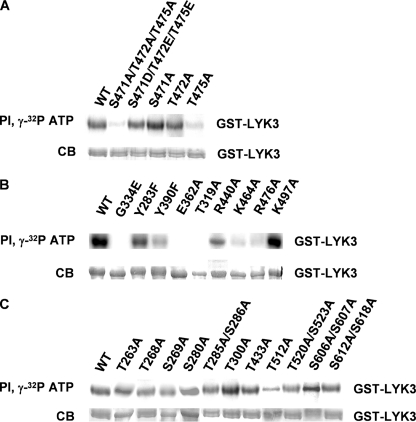
Site-directed mutagenesis was used to analyze the importance of key features of the model for kinase activity. Simultaneous substitution of the three activation loop phosphorylation sites by Ala (S471A/T472A/T475A) nearly abolished LYK3 kinase activity in vitro (Fig. 6A). Mimicking phosphorylated residues by replacing the Ser with Asp and the two Thr residues with Glu (mutant S471D/T472E/T475E) resulted in kinase activity similar to the wild-type protein (Fig. 6A). Mutation of each residue individually showed that Thr-475 is essential for kinase activity, whereas mutations in Ser-471 or Thr-472 had relatively little effect. Mutations in Arg-440 and in the two activation segment residues that interact with phosphorylated Thr-475 (Lys-464 and Arg-476) also led to substantial reduction in kinase activity, whereas a mutation in Lys-497, which is proposed to lose a salt bridge, following phosphorylation of Thr-472, did not alter kinase activity (Fig. 6B). Next, key residues in other parts of the model of LYK3 were analyzed. Ala substitutions showed that Thr-319 at the end of helix B is essential for kinase activity, as is the highly conserved Glu-362. A phenylalanine substitution in the gatekeeper, Tyr-390, also led to substantially reduced kinase activity and had a greater effect than a similar mutation in the potentially autophosphorylated residue, Tyr-283.
FIGURE 6.
Functional analysis of the LYK3 intracellular region identifies key residues for catalytic activity. A, analysis of mutants in the phosphorylated residues of the activation loop. Analysis of kinase activity using the radiolabeled ATP autophosphorylation assay ([γ-32P]ATP) of GST-LYK3-IR mutant proteins reveals the importance of Thr-475 for kinase activity. B, analysis of kinase activity of mutants in conserved residues implicated in kinase activity regulation as revealed by homology modeling with IRAK-4. The autophosphorylation assay ([γ-32P]ATP) reveals the importance of the following six residues for kinase activity (Thr-319, Glu-362, Tyr-390, Arg-440, Lys-464, and Arg-476). C, analysis of mutants in (potentially) phosphorylated serine and threonine residues. The autophosphorylation assay ([γ-32P]ATP) of mutants in experimentally determined or NetPhos predicted phosphorylated residues reveals that all proteins retain kinase activity. PI, Phosphorimage; CB, Coomassie Blue protein staining.
To examine the effects of the mutations on biological activity, the mutations were analyzed for their effect on the ability of the LYK3 constructs to rescue the lyk3-1 mutant for nodulation. The two kinase-dead mutants (T319A and E362A) were unable to complement, whereas the kinase-partially active mutant in the gatekeeper residue, Tyr-390, was still biologically active (Table 1). In the activation loop, the construct with the three Ala substitutions (S471A/T472A/T475A) did not complement, whereas transformation with the “phospho-mimicking” construct (S471D/T472E/T475E) resulted in nodulation, similar to wild type (Table 1). The T475A mutant protein was completely unable to complement lyk3-1 for nodulation as expected from its loss of kinase activity. However, the S471A and T472A mutations also both led to reduced nodulation activity (Table 1), despite the proteins retaining strong kinase activity (Fig. 6A).
TABLE 1.
Biological activity of LYK3 intracellular region mutants
FLAG-tagged constructs containing LYK3 mutations were transformed into the M. truncatula lyk3-1 mutant, and transgenic plants were scored for complementation for nodulation. Phosphorylation site mutants included sites that were identified to be phosphorylated by expression in E. coli (Fig. 4) and some that have a high prediction to be phosphorylated using the NetPhos programme.
| Construct | Nodulation assay |
||
|---|---|---|---|
| No. of plants |
p value | ||
| Total | Nodulated | ||
| WT | 28 | 18 | |
| Vector | 23 | 0 | 1.75E-06a |
| Kinase domain | |||
| T319A | 20 | 0 | 5.74E-06a |
| E362A | 16 | 0 | 3.02E-05a |
| Y390F | 23 | 15 | 9.45E-1 |
| S471A/T472A/T475A | 24 | 0 | 1.19E-06a |
| S471D/T472E/T475E | 28 | 16 | 5.84E-01 |
| S471A | 32 | 7 | 8.86E-04a |
| T472A | 26 | 3 | 7.10E-05a |
| T475A | 46 | 0 | 4.08E-10a |
| Phosphorylation sites | |||
| T263A | 15 | 11 | 5.46E-1 |
| T268A | 10 | 8 | 3.59E-1 |
| S269A | 12 | 8 | 8.85E-1 |
| S273A/T274A (NetPhos) | 14 | 6 | 1.86E-1 |
| S280A | 10 | 6 | 8.09E-1 |
| Y283F | 17 | 12 | 6.64E-01 |
| T285A/S286A | 30 | 21 | 6.43E-01 |
| T300A | 33 | 4 | 2.36E-05a |
| T433A | 26 | 4 | 2.58E-04a |
| T512A (NetPhos) | 27 | 0 | 3.78E-07a |
| T520A/S523A | 50 | 22 | 8.55E-02 |
| S606A/S607A (NetPhos) | 15 | 9 | 7.82E-01 |
| S612A/S618A | 21 | 13 | 8.64E-01 |
a Values indicate mutants that are totally or partially defective in nodulation activity (p value < 0.01) compared with the wild-type (WT) construct.
A Threonine Residue in the Juxtamembrane Region and Two in the Kinase Domain Are Important for Biological Activity but Not for Catalytic Activity
In addition to the activation loop, Ser/Thr phosphorylation of other parts of the protein can affect both catalytic and/or biological activity of a kinase. We thus tested Ala substitutions of Ser/Thr residues that had either been shown to be autophosphorylated in vitro (Fig. 4) or predicted by the NetPhos program to have a high probability to be phosphorylated. In some cases, closely neighboring residues were mutated together. In addition, the potentially phosphorylated Tyr-283 was mutated to phenylalanine. All of the tested mutant proteins retained autophosphorylation activity, following expression in E. coli, although T512A showed reduced activity (Fig. 6, B and C). In plants, the complementation test identified two Thr residues in the C-terminal lobe of the kinase domain with importance for biological activity; plants with the T512A mutation expressed the protein but were unable to nodulate, whereas the T433A mutation led to greatly reduced nodulation (Table 1). Residues in the C-tail and JM position (Fig. 4) were similarly tested, showing that the four Ser residues near the C terminus of the protein are not important for biological activity, whereas Ala substitution of Thr-300 in the JM region led to a protein with reduced biological activity. Mutants in the other JM phosphorylation sites, including Y283F, all retained biological activity suggesting that potential in vivo phosphorylation of these residues is not essential for signal transduction.
DISCUSSION
Our structure-function analysis of M. truncatula LYK3 has revealed a remarkable similarity in the kinase domains of this plant RLK and the human IRAK-4 kinase. Because of the common origins of the animal IRAK/Pelle kinases and plant RLKs (1, the similarities that we have reported may be widely applicable to the large plant RLK family. There are four features of the IRAK-4 kinase that are particularly relevant as follows: (i) the Tyr gatekeeper residue; (ii) the N-terminal extension (helix B); (iii) the activation loop; and (iv) the similarity to Tyr kinases.
First, the gatekeeper is a pivotal residue that controls access to the back of the ATP binding pocket. In the human kinome, 20% of the proteins possess a Thr, 40% possess a Met, others have a Phe and only the IRAK kinases possess a Tyr in this position (2). In IRAK-4, the gatekeeper Tyr-262 interacts with the conserved Glu-233 in helix C and effectively closes off the back pocket. Homology modeling of LYK3 suggests that this may also be the case in the plant RLK (Tyr-390 interacting with Glu-362). Examination of the alignment of 610 proteins of the Arabidopsis RLK/RLCK family (1) reveals that 507 of them (83%) contain Tyr in the gatekeeper position, and of these proteins, the predicted interacting Glu residue is conserved in 89%. There are some notable exceptions; for example, the well studied FLS2 receptor contains a Leu in the gatekeeper position and the tomato LePRK1 and -2 contain a Ser and Phe, respectively (29). In BRI1, this residue appears to be a site of phosphorylation, and mutation to Phe completely abolished its kinase activity (30), thus showing its importance in catalysis. In LYK3, a gatekeeper mutant (Y390F) retained partial kinase activity, and the protein was still biologically active. Clearly, the implications of the Tyr gatekeeper on the structure and regulation of kinase activity in plant RLKs needs to be further evaluated. Moreover, development of inhibitors for the IRAK kinases, which takes into account the unique structural features of the ATP-binding pocket, may be beneficial to the plant RLK community in providing effective and specific inhibitors.
Second, the model suggests that the plant RLK contains an N-terminal helix (helix B) that packs back on the β-sheet of the N-terminal lobe (Fig. 5). This helix occurs before the minimum homology sequence of a kinase domain, which in LYK3 starts at Phe-322 (Fig. 1). Although a helix B occurs in the structure of many animal kinases, the sequence is not conserved. However, it is clear from the alignment in Fig. 1 that the helix B region shows strong homology between the plant RLK and IRAK-4, including in the conservation of the residues (Leu-315 and Thr-319 of LYK3) that interact with the β-sheet. This region and these residues are conserved in many plant RLKs (29). Although its function in IRAK-4 is unknown, the equivalent region of IRAK-1 is required for enzymatic activity. Moreover, the conserved Thr at the C-terminal end of the helix (corresponding to LYK3 Thr-319) is autophosphorylated in IRAK-1 and controls a conformational change in the kinase, allowing subsequent phosphorylation of the activation loop and full kinase activity (31). In LYK3 (Fig. 6 and Table 1) and Xa21 (8), Ala substitution mutants in this threonine completely lose kinase activity, and the proteins are biologically inactive. This threonine has been shown to be phosphorylated in several plant RLKs, notably BRI1, SYMRK, and Xa21 (4, 8–9) and more recently NFR1 (32). In BRI1, the corresponding Thr is not essential for catalytic activity, but at the N-terminal end of the predicted helix B, Thr-872 (corresponding to Ser-169 in IRAK-4 and Thr-311 in LYK3) has been shown to be phosphorylated, and mutations in this residue lead to a hyperphosphorylated kinase (4). Interestingly, this latter residue appears only to be conserved as Ser/Thr in RD and not non-RD kinases of the plant leucine-rich repeat-RLK superfamily (4). Together, these results suggest that an N-terminal extension, including helix B, may be a common element of the animal, soluble IRAK/Pelle kinases and the plant membrane RLK kinases, and should perhaps be considered as an integral part of the N-terminal lobe regulating kinase activity rather than part of an RLK juxtamembrane region.
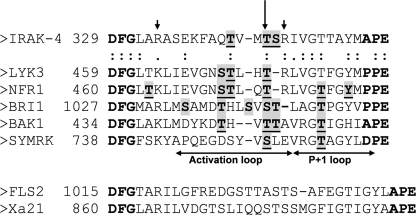
Third, our plant RLK shows remarkable similarity to IRAK-4 in the structure and potential regulation via activation loop phosphorylation. In IRAK-4, Thr-345 is the prototypical phosphorylated residue that forms a water-mediated hydrogen bond with the Arg residue (Arg-310) in the catalytic loop (HRD motif) (2). Homology modeling and MD analysis of LYK3 suggests that Thr-475 plays a similar role, and indeed mutagenesis revealed that it is essential for catalytic and biological activity. Examination of an alignment of the activation segment of well studied plant RLKs (Fig. 7) pinpoints a candidate for this prototypical residue in RD kinases, which in NFR1 (Thr-476), BAK1 (Thr-449), SYMRK (Ser-754), and possibly BRI1 (Ser-1044) are autophosphorylated (5, 9, 32). In the RLCK Pto, structural analysis has confirmed the role of an equivalent residue (12, 13). This residue is conserved as a Ser/Thr in 62% of the plant RLK/RCLK sequences (4), but the value is much lower for just the non-RD kinases, reinforcing the observation that activation loop phosphorylation of non-RD RLKs is generally less important than in RD kinases (11, 33). In IRAK-4, the phosphate on Thr-345 compensates for the positively charged pocket formed by Arg-310 and two Arg residues in the activation segment (2). In LYK3, the corresponding residues are Arg-440, Lys-464, and Arg-476 (Fig. 7), and each of them is required for full catalytic activity (Fig. 6B). In addition, LYK3 has two secondary phosphorylation sites, Thr-472 (which corresponds to Thr-342 in IRAK-4) and Ser-471. Phosphorylation of Thr-472 appears to change the orientation of this residue so that, like Ser(P)-471, it points out from the protein. As these two sites are required for biological but not catalytic activity, they may be involved in substrate binding in vivo. Other plant RLKs show potential secondary activation loop phosphorylation sites (Fig. 7). In addition, phosphorylation of the highly conserved Thr in the P + 1 loop, which follows the activation loop in the activation segment, may also be important for activity (5, 9–10, 32).
FIGURE 7.
Alignment of the activation segments of IRAK-4, LYK3, and other plant RLKs. In the alignment of IRAK-4 and LYK3 conserved residues are marked with a colon. The activation segment of the two plant non-RD RLKs (FLS2 and Xa21) are only aligned together and not with the other proteins. Phosphorylation sites are shaded in gray and underlined; ambiguous ones are not underlined. The prototypical phosphorylated residue in the activation loop of IRAK-4 is arrowed, and its two interacting, activation segment residues are marked with arrowheads.
Fourth, as pointed out by Wang et al. (2), both sequence and structural analyses suggest that IRAK-4 is more closely related to receptor Tyr kinases than Ser/Thr kinases suggesting that it could be a dual specificity kinase, although this has yet to be demonstrated biochemically. This observation is not surprising due to the common phylogenetic origins of these kinases (1) and the phylogenetic position of the IRAK kinases in the human kinome (34). We have shown that LYK3 autophosphorylates Tyr residues in addition to Ser/Thr residues, thus supporting the structural similarity to Tyr kinases and a functional dual specificity (Fig. 3). Although Tyr phosphorylation in plants has had a controversial history, it is now clear that it is widespread (35) and that certain RLKs exhibit dual Tyr and Ser/Thr specificity, including BRI1 (30) and NFR1 (32). For BRI1, Tyr-831 in the JM region is autophosphorylated and is important for biological activity (30), whereas none of the three in vitro Tyr phosphorylation sites in NFR1 are required for nodulation (32). We identified Tyr-283 as a major in vitro phosphorylated tyrosine residue in LYK3; this residue is also not essential for nodulation activity, although our assay does not exclude that it may play a more subtle role during infection. As pointed out by Oh et al. (30), it will be important to identify the residues that control dual specificity of catalytic activity in plant RLKs and indeed to determine whether IRAK-4 also exhibits Tyr kinase activity.
Our work has also shed light on the functioning of LYK3 in planta. We have shown that the lyk3-1 mutant contains an allele with a nonfunctional kinase, and we have shown that other catalytically inactive mutants fail to complement, suggesting that LYK3 requires kinase activity for its role in nodulation, although we cannot rule out that the indicated amino acid mutations have unexpected effects unrelated to phosphorylation that render LYK3 biologically inactive in vivo. The lyk3-1 allele may represent a nonfunctional null allele, and the phenotype of this mutant (19, 20) would suggest that LYK3 is not essential for initial responses to rhizobial Nod factors, which in M. truncatula requires NFP, a LysM-RLK with a dead kinase (16). Thus, the role of LYK3 appears to be different from its potential orthologue in L. japonicus, NFR1, which is required for early responses (15). Recently, studies on NFR1 have shown that it requires kinase activity for its role in nodulation and have shown that its kinase domain can phosphorylate the intracellular region of the putative NFP orthologue, NFR5, in vitro (32). In our experiments, no evidence was found for a specific transphosphorylation of NFP-IR by LYK3-IR (data not shown). The difference between the two legumes may be due to different requirements for early Nod factor signaling in the two legumes or to partial functional redundancy between the LYK genes in M. truncatula (16, 17).
In common with other plant RLKs (36), including NFR1 (32), we have found that LYK3 autophosphorylates on many Ser/Thr residues outside of the core kinase domain, including seven in the juxtamembrane region and two in the C-tail. Unlike BRI1, where phosphorylation of the C-tail is required for releasing autoinhibition (37), we have not found any evidence for a function of the two autophosphorylated C-tail Ser residues identified in LYK3. However, Thr-300 in the juxtamembrane region, which is phosphorylated in vitro, is important for biological activity. Three additional sites in the kinase domain (outside of the activation loop) are also autophosphorylated (Fig. 4). The autophosphorylated Thr-433 and the NetPhos-predicted Thr-512 have not previously been identified as phosphorylation sites in plant RLKs but are required for biological activity of LYK3. As Ala substitutions in Thr-300, Thr-433, and Thr-512 still lead to catalytically active proteins, these sites may be involved in signal transduction in vivo.
In conclusion, this work has shown the importance of kinase activity and key kinase domain residues in the biological function of the LysM-RLK LYK3 in the establishment of the important legume-rhizobia symbiosis and has shown how a homology model based on the human IRAK-4 kinase has related functional data to the predicted structure of this plant RLK. Because of the common evolutionary origins of the plant RLKs and the IRAK/Pelle kinases, sequence alignment to IRAK-4 and subsequent homology modeling provides a rapid and convenient method to construct a structural framework to interpret and compare the accumulating data on the large family of plant RLKs. Furthermore the special features of plants (ease of mutation, transformation, and phenotyping) will provide a wealth of structure-function data applicable to studies on animal IRAK/Pelle kinases.
Supplementary Material
Acknowledgments
We thank students Amandine Guillonneau and Vincent Duplan for help with parts of the project and Christine Hervé, Judith Fliegmann, and Clare Gough for critically reading the manuscript.
This work was supported in part by the European Community Marie Curie Research Training Network Programme Contract MRTN-CT-2006-035546 “NODPERCEPTION,” French National Research Agency Contracts “NodBindsLysM” and “Sympasignal,” Fondation pour la Recherche Médicale Contract “Grands Equipements,” the Toulouse Midi-Pyrénées Génopole, and the Midi-Pyrénées Regional Council Grant CR07003760.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1, supplemental Figs. S1–S4, supplemental Methods, and additional references.
- RLK
- receptor-like-kinase
- aa
- amino acid
- IR
- intracellular region
- IRAK
- interleukin-1 receptor-associated kinase
- LysM
- lysin motif
- MD
- molecular dynamics
- RLCK
- receptor-like cytoplasmic kinase
- JM
- juxtamembrane.
REFERENCES
- 1. Shiu S. H., Bleecker A. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang Z., Liu J., Sudom A., Ayres M., Li S., Wesche H., Powers J. P., Walker N. P. (2006) Structure 14, 1835–1844 [DOI] [PubMed] [Google Scholar]
- 3. Kuglstatter A., Villaseñor A. G., Shaw D., Lee S. W., Tsing S., Niu L., Song K. W., Barnett J. W., Browner M. F. (2007) J. Immunol. 178, 2641–2645 [DOI] [PubMed] [Google Scholar]
- 4. Wang X., Goshe M. B., Soderblom E. J., Phinney B. S., Kuchar J. A., Li J., Asami T., Yoshida S., Huber S. C., Clouse S. D. (2005) Plant Cell 17, 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang X., Kota U., He K., Blackburn K., Li J., Goshe M. B., Huber S. C., Clouse S. D. (2008) Dev. Cell 15, 220–235 [DOI] [PubMed] [Google Scholar]
- 6. Karlova R., Boeren S., van Dongen W., Kwaaitaal M., Aker J., Vervoort J., de Vries S. (2009) Proteomics 9, 368–379 [DOI] [PubMed] [Google Scholar]
- 7. Gómez-Gómez L., Boller T. (2002) Trends Plant Sci. 7, 251–256 [DOI] [PubMed] [Google Scholar]
- 8. Chen X., Chern M., Canlas P. E., Jiang C., Ruan D., Cao P., Ronald P. C. (2010) J. Biol. Chem. 285, 10454–10463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoshida S., Parniske M. (2005) J. Biol. Chem. 280, 9203–9209 [DOI] [PubMed] [Google Scholar]
- 10. Miyahara A., Hirani T. A., Oakes M., Kereszt A., Kobe B., Djordjevic M. A., Gresshoff P. M. (2008) J. Biol. Chem. 283, 25381–25391 [DOI] [PubMed] [Google Scholar]
- 11. Johnson L. N., Noble M. E., Owen D. J. (1996) Cell 85, 149–158 [DOI] [PubMed] [Google Scholar]
- 12. Xing W., Zou Y., Liu Q., Liu J., Luo X., Huang Q., Chen S., Zhu L., Bi R., Hao Q., Wu J. W., Zhou J. M., Chai J. (2007) Nature 449, 243–247 [DOI] [PubMed] [Google Scholar]
- 13. Dong J., Xiao F., Fan F., Gu L., Cang H., Martin G. B., Chai J. (2009) Plant Cell 21, 1846–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oldroyd G. E., Downie J. A. (2008) Annu. Rev. Plant Biol. 59, 519–546 [DOI] [PubMed] [Google Scholar]
- 15. Radutoiu S., Madsen L. H., Madsen E. B., Felle H. H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003) Nature 425, 585–592 [DOI] [PubMed] [Google Scholar]
- 16. Arrighi J. F., Barre A., Ben Amor B., Bersoult A., Soriano L. C., Mirabella R., de Carvalho-Niebel F., Journet E. P., Ghérardi M., Huguet T., Geurts R., Dénarié J., Rougé P., Gough C. (2006) Plant Physiol. 142, 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. (2003) Science 302, 630–633 [DOI] [PubMed] [Google Scholar]
- 18. Madsen E. B., Madsen L. H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003) Nature 425, 637–640 [DOI] [PubMed] [Google Scholar]
- 19. Catoira R., Timmers A. C., Maillet F., Galera C., Penmetsa R. V., Cook D., Dénarié J., Gough C. (2001) Development 128, 1507–1518 [DOI] [PubMed] [Google Scholar]
- 20. Smit P., Limpens E., Geurts R., Fedorova E., Dolgikh E., Gough C., Bisseling T. (2007) Plant Physiol. 145, 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boisson-Dernier A., Chabaud M., Garcia F., Bécard G., Rosenberg C., Barker D. G. (2001) Mol. Plant-Microbe Interact. 14, 695–700 [DOI] [PubMed] [Google Scholar]
- 22. Lévy J., Bres C., Geurts R., Chalhoub B., Kulikova O., Duc G., Journet E. P., Ané J. M., Lauber E., Bisseling T., Dénarié J., Rosenberg C., Debellé F. (2004) Science 303, 1361–1364 [DOI] [PubMed] [Google Scholar]
- 23. Kremers G. J., Goedhart J., van Munster E. B., Gadella T. W., Jr. (2006) Biochemistry 45, 6570–6580 [DOI] [PubMed] [Google Scholar]
- 24. van Ooijen G., Mayr G., Kasiem M. M., Albrecht M., Cornelissen B. J., Takken F. L. W. (2008) J. Exp. Bot. 59, 1383–1397 [DOI] [PubMed] [Google Scholar]
- 25. Borderies G., Jamet E., Lafitte C., Rossignol M., Jauneau A., Boudart G., Monsarrat B., Esquerré-Tugayé M. T., Boudet A., Pont-Lezica R. (2003) Electrophoresis 24, 3421–3432 [DOI] [PubMed] [Google Scholar]
- 26. Hanks S. K., Quinn A. M., Hunter T. (1988) Science 241, 42–52 [DOI] [PubMed] [Google Scholar]
- 27. Cheng H., Addona T., Keshishian H., Dahlstrand E., Lu C., Dorsch M., Li Z., Wang A., Ocain T. D., Li P., Parsons T. F., Jaffee B., Xu Y. (2007) Biochem. Biophys. Res. Commun. 352, 609–616 [DOI] [PubMed] [Google Scholar]
- 28. Nolen B., Taylor S., Ghosh G. (2004) Mol. Cell 15, 661–675 [DOI] [PubMed] [Google Scholar]
- 29. Geldner N., Robatzek S. (2008) Plant Physiol. 147, 1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oh M. H., Wang X., Kota U., Goshe M. B., Clouse S. D., Huber S. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kollewe C., Mackensen A. C., Neumann D., Knop J., Cao P., Li S., Wesche H., Martin M. U. (2004) J. Biol. Chem. 279, 5227–5236 [DOI] [PubMed] [Google Scholar]
- 32. Madsen E. B., Antolin-Llovera M., Grossmann C., Ye J., Vieweg S., Broghammer A., Krusell L., Radutoiu S., Jensen O. N., Stougaard J., Parniske M. (2010) Plant J. 65, 404–417 [DOI] [PubMed] [Google Scholar]
- 33. Liu G. Z., Pi L. Y., Walker J. C., Ronald P. C., Song W. Y. (2002) J. Biol. Chem. 277, 20264–20269 [DOI] [PubMed] [Google Scholar]
- 34. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 35. de la Fuente van Bentem S., Hirt H. (2009) Trends Plant Sci. 14, 71–76 [DOI] [PubMed] [Google Scholar]
- 36. Nühse T. S., Stensballe A., Jensen O. N., Peck S. C. (2004) Plant Cell 16, 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang X., Li X., Meisenhelder J., Hunter T., Yoshida S., Asami T., Chory J. (2005) Dev. Cell 8, 855–865 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.