Abstract
Therapeutic proteins contain a large number of post-translational modifications, some of which could potentially impact their safety or efficacy. In one of these changes, pyroglutamate can form on the N terminus of the polypeptide chain. Both glutamine and glutamate at the N termini of recombinant monoclonal antibodies can cyclize spontaneously to pyroglutamate (pE) in vitro. Glutamate conversion to pyroglutamate occurs more slowly than from glutamine but has been observed under near physiological conditions. Here we investigated to what extent human IgG2 N-terminal glutamate converts to pE in vivo. Pyroglutamate levels increased over time after injection into humans, with the rate of formation differing between polypeptide chains. These changes were replicated for the same antibodies in vitro under physiological pH and temperature conditions, indicating that the changes observed in vivo were due to chemical conversion not differential clearance. Differences in the conversion rates between the light chain and heavy chain on an antibody were eliminated by denaturing the protein, revealing that structural elements affect pE formation rates. By enzymatically releasing pE from endogenous antibodies isolated from human serum, we could estimate the naturally occurring levels of this post-translational modification. Together, these techniques and results can be used to predict the exposure of pE for therapeutic antibodies and to guide criticality assessments for this attribute.
Keywords: Antibodies, Drug Metabolism, Immunochemistry, Post-translational Modification, Protein Turnover, N-terminal Modification
Introduction
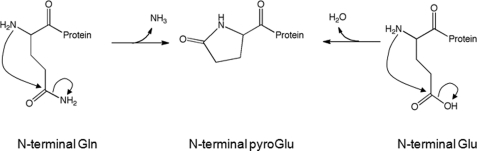
Pyroglutamate (pE),2 or pyrrolidone carboxylate, is a cyclic amino acid found at the N termini of some proteins and biological peptides (1). Formation occurs through the rearrangement of the originally synthesized glutamate or glutamine residues at this position (Fig. 1). Although this reaction proceeds spontaneously at reasonable rates, glutaminyl cyclase, an enzyme that catalyzes this reaction, is found in many plants and animals, including humans (2). Reaction rates for both the spontaneous reaction and the enzymatically catalyzed reaction appear to be much faster with glutamine than with glutamate residues (2).
FIGURE 1.
Pyroglutamate formation mechanism. The mechanism of pyroglutamate (pyroGlu) formation from Glu or Gln is shown.
Many antibodies contain pE on the light chain or heavy chain (3). Although pyroglutamate formation is more prevalent at N-terminal glutamine residues (4, 5), it can occur at glutamate residues as well (6–8). Antibody pE forms spontaneously in vitro (5), although it is not known whether glutaminyl cyclase accelerates this rate in blood.
For therapeutic monoclonal antibodies, pE can be one of many post-translational modifications observed during production and storage. Because of the loss of a primary amine in the glutamine to pE conversion, antibodies become more acidic. Incomplete conversion produces heterogeneity in the antibody that can be observed as multiple peaks using charge-based analytical methods, such as ion exchange chromatography or isoelectric focusing (4). Differences in pE formation have been considered a concern in drug production, because heterogeneity differences may indicate a lack of process control (5). Interestingly, heterogeneity as the result of glutamate to pE conversion would not be observable by these analytical methods, because antibodies with these N termini share the same charge state.
Recently, more effort has been placed on understanding how heterogeneity impacts product quality (9). Those attributes of particular concern are ones that affect the safety or efficacy of the drug. In contrast, attributes not impacting drug safety or efficacy would appear to be less of a concern. Focusing process control on critical quality attributes is the cornerstone of Quality by Design, an emerging paradigm in biotechnology development (10, 11). As part of an effort to ascertain the impact of N-terminal heterogeneity on the safety and efficacy of therapeutic antibodies, studies were conducted to monitor pE formation in vivo. Here we describe N-terminal glutamate to pE conversion, as opposed to the more prevalent glutamine to pE conversion, in vivo and how these studies are used, in part, to assess criticality of this attribute.
EXPERIMENTAL PROCEDURES
Materials
Three recombinant human IgG2 monoclonal antibodies (mAb1, mAb2, and mAb3) with Glu at the N termini of their heavy chains (HC) were studied for pE conversion. Two of these (mAb1 and mAb2) also express glutamate on the N termini of their light chains (LC). Both mAb1 and mAb2 bind to specific cell surface receptors in humans, whereas mAb3 binds to a circulating soluble ligand. mAb4, which is expressed with a glutamine at the N terminus of its heavy chain, was used as a control standard in the analysis of pE levels in endogenous proteins. All four mAbs were produced at Amgen Inc. (Thousand Oaks, CA) and expressed in Chinese hamster ovary cells. Soluble target ligands used for affinity purification were also expressed and purified at Amgen Inc. Actigel ALD Superflow and sodium cyanoborohydride were purchased from Sterogene Bioseparations. Lysyl endopeptidase (Lys-C) was ordered from Wako Chemicals USA. Pfu pyroglutamate aminopeptidease (PGAP) was obtained from Takara Biotechnology. DTT, sodium iodoacetate, ammonium bicarbonate, urea and l-pyroglutamic acid were purchased from Sigma-Aldrich. TFA and guanidine HCl were from Pierce. Formic acid (FA) was from ALFA AESAR.
Human Phamacokinetic mAb Study
Both mAbs (mAb1 and mAb2) used in the PK studies were administrated to healthy human subjects (male and female, ages 19–48 years) via either single intravenous or subcutaneous injections at different dosing levels: 1000 mg intravenously, 300 mg intravenously, 100 mg intravenously, or 300 mg subcutaneously. Blood samples were collected over several weeks at different time points. After allowing time to clot, the clot was separated from serum by centrifugation (2000 × g for 15 min). Serum was stored in cryotubes at −20 °C or colder until use. mAb concentrations in serum were determined with a sandwich ELISA assay. Human subjects were both female and male in the age range from 19 to 48 years old. mAbs were purified from the serum samples (0.5 ml) using the affinity purification procedures described elsewhere (12). The protease digestion procedure for low concentration PK mAb samples was described in a previously published paper (12).
In Vitro Incubations
To mimic physiological conditions in vitro, mAbs were incubated at 2 mg/ml in PBS (10 mm sodium phosphate, 150 mm sodium chloride) at 37 °C for up to 34 days. The pH of PBS was measured at 37 °C and adjusted to pH 7.4. A time point was collected to match those from the in vivo studies: 1 h and 2, 6, 10, 20, and 34 days. After incubation, the mAb samples were buffer-exchanged with 5 mm sodium acetate, pH 5, to prevent further change in pE conversion. A portion of the incubated mAb samples (100 μg of protein) was digested with Lys-C (12) and then analyzed by LC/MS.
The time course was repeated to test the effect of denaturant. mAb1 (2 mg/ml) was incubated in PBS, with and without 6 m guanidine HCl (final concentration), at 37 °C for 2 weeks. A small portion of the incubating sample was removed at 1, 4, 8, and 14 days. Following incubation, the samples were exchanged into 5 mm sodium acetate, pH 5, to quench the reaction. The samples were digested with Lys-C and analyzed by LC/MS as described.
LC/MS/MS Analysis with LTQ MS
The LC/MS/MS system consisted of an Agilent HP1100 HPLC system directly connected to a Thermo Scientific LTQ electrospray ion trap mass spectrometer.
The mAb digestion products were separated using a reversed phase HPLC column (Phenomenex Jupiter C5, 2 × 250 nm, 300 Å, 5 μ) with column temperature maintained at 50 °C. Mobile phase A was 0.1% FA in water, and mobile phase B was 0.1% FA and 90% acetonitrile in water. The gradient (hold at 2% B for 2 min, 2–22% B in 38 min, and then to 22–42% B in 82 min) was performed with a flow rate of 0.2 ml/min. Approximately 10–30 μg of each sample was injected. The chromatogram was monitored by both UV light absorbance (set at 214 nm wavelength) and mass spectrometry. Mass spectrometric detection included full scan in positive mode, as well as data-dependent ultra zoom scan and MS/MS of the most intense ion of the scan. Monoisotopic masses and charges were determined from ultra zoom scans. MassAnalyzer software developed in-house (13) was used for peptide identification by correlating the sequence of mAb to fragmentation mass spectra. UV chromatograms were used to quantify the pE formation peak versus main peptide peak.
LC/MS/MS Analysis with LTQ-Orbitrap
Some analyses were performed on an Agilent 1200SL HPLC system directly connected to a Thermo Scientific LTQ-Orbitrap for enhanced sensitivity and mass accuracy. Approximately 5 μg of each sample was injected into this system. The LC column (Waters 1.7 μ particle column (BEH300 C18, 2.1 × 100 mm)) was used at 50 °C with flow rate of 0.3 ml/min. Mobile phases were 0.04% TFA in water (A) and 0.04% TFA in acetonitrile (B). The gradient was 0.5–20% B in 40 min, 20–40% B in 80 min, and then 40–99% B in 6 min. For MS setup, a full MS scan was set on the Orbitrap with 60,000 resolution, and three CID MS/MS scans were used with dynamic exclusion. All of the data analyses were performed using a new prototype version of MassAnalyzer.
Isolation of Endogenous Antibodies
Endogenous IgGs were isolated from healthy human donors by protein A affinity chromatography as previously described (14). Briefly, repeated injections of serum of 60 μl, isolated as described above, were passed over a Poros A/20 protein A column (Applied Biosystems, part number: 1-5022-24) equilibrated in 20 mm Tris, 150 mm NaCl, pH 7. After washing the column with the same solution until no protein elution was detected (280 nm), the bound IgG was eluted with 20 mm NaOAc, 150 mm NaCl, pH 3. The collected IgG fraction from the protein A column was concentrated by ultrafiltration and exchanged into a stable buffer (5 mm sodium acetate, pH 5) prior to Lys-C digestion.
Analysis of pE Levels from Endogenous Antibodies
A protocol using PGAP to remove pE from N termini of IgGs (15) was applied to endogenous human IgGs with some modifications. A volatile buffer (0.1 m NH4HCO3, pH 7.9) was substituted for the PGAP phosphate digestion buffer, and glycerol was omitted from the procedure. Briefly, a 0.5-mg sample of endogenous IgG or mAb4 control was denatured and reduced in 1 ml of 8 m guanidine, 0.35 m Tris-HCl, 55 mm DTT, pH 8.5, at 60 °C for 90 min. The protein was then alkylated with sodium iodoacetate. Following a DTT quench step, the mixture was buffer-exchanged into the volatile digestion buffer using Sephadex G-25 (NAP-5 column). The concentrations of eluted proteins were measured by absorbance at 280 nm, using 1.4 cm−1 ml mg−1 as an extinction coefficient. Eluted samples were then digested by PGAP in the presence of 12 mm DTT at 37 °C for 3 days. A portion of PGAP-digested mAb4 was digested with trypsin and analyzed by RP-HPLC-MS (7) to assess the efficiency of PGAP digestion. Pyroglutamate removal was judged complete based on 100% loss of the pE from the mAb4 heavy chain N-terminal peptide using mass and MS/MS analysis.
The pE released from the PGAP digestion was detected using a LC/MS system by a protocol similar to one previously described (16). Major differences included using a volatile buffer, modifications to the separation gradient and solvents, and use of an ion trap MS (Thermo Scientific LTQ XL electrospray ion trap MS) instead of a triple quad MS. Mass detection of the protonated molecular pE (130.1 Da) ion was sufficient for quantification, so monitoring of the daughter ion was deemed unnecessary. The pyroglutamate was separated using a reversed phase HPLC column (Agilent Zorbax 300SB-C18, 4.6 × 150 nm, 5 μm) with the column temperature maintained at 40 °C. The mobile phase A was 0.1% FA in water, and the mobile phase B was 0.1% FA and 90% acetonitrile in water. The gradient (hold at 0% B for 5 min, 0–5% B in 4 min and 5–45% B in 3 min, and then hold at 45% B for 5 min) was performed with a flow rate of 0.2 ml/min. Approximately 60 μl of each sample was injected. The chromatogram was monitored by mass spectrometry full scan with mass range from 100 to 400 Da. Xcalibur software (Thermo Scientific) was used to extract the ion mass. The peak area of pE was integrated to calculate the level. A standard curve from 10 to 500 pmol (R2 = 0.9997) was generated using commercial l-pyroglutamic acid (Sigma-Aldrich).
RESULTS
Identification and Quantification of pE by LC/MS/MS
Three IgG2 therapeutic monoclonal antibodies were analyzed for the presence of N-terminal pyroglutamate. Two of these mAbs (mAb1 and mAb2) express Glu at the N termini of their HC and LC, and the third (mAb3) expresses glutamate at the N terminus of the HC but aspartate at the N terminus of the LC. To generate N-terminal pE for identification and quantification purposes, the purified mAbs were incubated in PBS, pH 7.4, at 37 °C for up to 1 month. These conditions previously have been used to form pyroglutamate from N-terminal glutamate on selected antibodies (6). Pyroglutamate was identified by peptide mapping with protease Lys-C. The identification of the modification of the N-terminal peptide was based on the loss of 18 mass units that was specific for the N-terminal residue (MS/MS analysis; not shown) (6, 7).
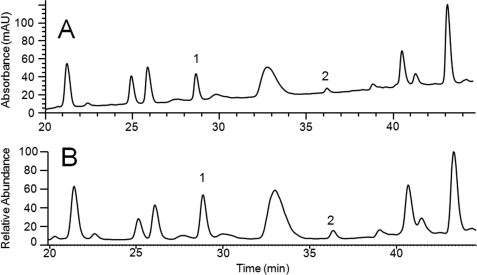
Fig. 2 shows the region of the Lys-C peptide map of mAb1 containing the heavy chain N-terminal peptides. On the UV (214 nm) chromatogram (Fig. 2A), the modified (pE-containing) and the unmodified (Glu-containing) peptides are well resolved from each other and other interfering peptides, allowing quantification based on integration of these peaks. A comparison with the total ion current chromatogram of the same chromatographic region is shown in Fig. 2B. The extracted ion current indicates that no interfering signals were found in either peak. Pyroglutamate quantification is relative, based on the total signal for HC or LC N-terminal peptides. Comparison of this approach using the UV or the total ion current peak integration indicated that MS-based quantification underestimated the pE levels for the mAb1 HC by ∼5% with the specific MS parameters used in this study. These results indicate that the pE modification did not significantly alter the ionization efficiency of this peptide. Similar results were obtained when comparing the 214-nm UV and the MS-based quantification of the mAb3 HC N-terminal peptide. However, the length (12 amino acids) and the charge states (two acidic residues and one basic residue) are similar between these peptides. The UV- and MS-based quantitation was also compared using the much larger (40 amino acids; two acidic residues and three basic residues) Lys-C generated N-terminal peptide of mAb2 LC. The presence of Trp and Tyr in this peptide allowed 280 nm-based UV absorbance to be used instead of 214 nm-based UV absorbance, allowing quantification in the presence of peptides lacking nonaromatic residues. Again, MS-based quantitation matched UV-based quantitation fairly well. Moreover, truncation of this peptide to 18 amino acids (two acidic residues and one basic residue) by trypsin digestion did not affect MS-based quantitation. Because the N-terminal modification was shown to not significantly impact the MS signal on multiple peptides, we based pE quantification on MS analysis, even for those peptides in which resolution did not allow confirmation of this approach by UV-based analysis.
FIGURE 2.
mAb1 peptide map. A portion of mAb1 peptide map chromatogram at (A) 214 nm (UV) and (B) mass spectrometry total ion current is shown. Peak 1, HC N-terminal peptide; peak 2, pE modified HC N-terminal peptide.
Measuring Glu and pE Levels in Vivo
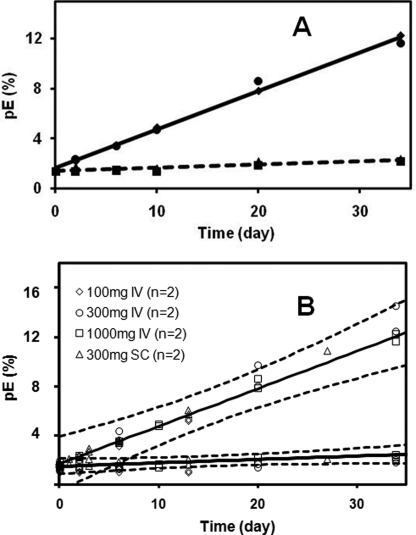
Relative levels of pE in circulating mAbs were monitored in serum taken from pharmacokinetic studies. The therapeutic antibodies were administered to healthy human subjects in single dose intravenous or subcutaneous injections. Affinity resin was then used to purify the antibody from the serum samples collected at specific times after dosing (12). Peptide mapping, described above, was used to analyze the pE and Glu on the N-terminal peptides in the isolated mAbs. Control experiments showed that the affinity-isolated mAb1 or mAb2 spiked into serum did not impact the pE present in the original sample (not shown). Fig. 3A shows the changes in the relative pE levels over time in vivo on both the mAb1 HC and LC for one dosing scheme (1000 mg intravenously). Increases in the HC pE levels were readily observed, linearly increasing from less than 2% to ∼12% over the 34 days tested (∼0.3%/day). Although the mAb1 LC also had an N-terminal Glu residue, little change was detected over the same period. Assuming that these changes follow first order kinetics, a rate constant of 0.00332 day−1 could be calculated for the mAb1 HC, for a predicted half-life of 209 days (Table 1). Estimates for the mAb1 LC rate constants were less accurate because of the small changes detected but nonetheless indicate rates more than 10 times slower than with the mAb1 HC. Similar HC pE changes were observed for different intravenous dosing levels (100, 300, and 1000 mg) and for subcutaneous administration (Table 1). Confidence intervals (95%) placed around the 1000-mg intravenous dose data (Fig. 3B) encompass all the data from the other dosings. Confidence intervals on the calculated rate constants for each dosing are presented in Table 1.
FIGURE 3.
mAb1 N-terminal pE levels in vivo for the 1000-mg intravenous dosing. A, circles, HC pE levels in subject 1; diamonds, HC pE levels in subject 2; squares, LC pE levels in subject 1; triangles, LC pE levels in subject 2. The trend lines represent the averages from the two subjects. B, comparisons of all the in vivo dosings of mAb1. Two subjects for each dosing are shown. Upper curve, relative HC pE levels. Lower curve, relative LC pE levels in vivo.
TABLE 1.
Changes in the relative mAb1 pE levels in vivo
k is the first order rate constant calculated by averaging the data for two patients at each dosing. IV, intravenously; SC, subcutaneously.
| Dosing | Heavy chain |
Light chain |
||
|---|---|---|---|---|
| k | 95% confidence interval | k | 95% confidence interval | |
| day−1 | day−1 | |||
| 100 mg IV | 0.00304 | 0.002734–0.003347 | −0.000007 | −0.000201–0.000186 |
| 300 mg IV | 0.00374 | 0.003133–0.004353 | 0.000215 | 0.000135–0.000296 |
| 1000 mg IV | 0.00332 | 0.003152–0.003493 | 0.000261 | 0.000162–0.000360 |
| 300 mg SC | 0.00353 | 0.003324–0.003733 | 0.000155 | −0.000033–0.000342 |
| Average | 0.00336 | 0.00014 | ||
Similar experiments were performed in vivo with a second IgG2 antibody, mAb2. mAb2, like mAb1, contained glutamate on both its LC and HC N termini. A single 1000-mg intravenous dose was administered as part of a human PK study, with serum samples collected at specified time points, as described previously for mAb1. Using the soluble mAb2 protein ligand, mAb2 was affinity-purified and analyzed by similar methodology. The results showed linear increases in both mAb2 LC and HC pE levels with time. In this case, however, the changes were similar for both termini on the antibody. A summary of the in vivo results, reduced to first order rate constants, is shown in Table 2. Comparisons of the two antibodies suggest that variations can be found within chains on the same antibody and between the same chains on different antibodies.
TABLE 2.
N-terminal Glu to pE conversion rates in vivo and in vitro
The values (k) represent the first order rate constants averaged from two patients (1000 mg intravenously) for each of the two mAbs listed. Other values were obtained from the MS signals.
| Heavy chain |
Light chain |
|||
|---|---|---|---|---|
| k | 95% confidence interval | k | 95% confidence interval | |
| day−1 | day−1 | |||
| mAb1 in vivo | 0.00309a | 0.002730–0.003447 | 0.000105 | −0.000006–0.000217 |
| mAb1 in vitro | 0.00324a | 0.002601–0.003877 | 0.000158 | −0.000075–0.000241 |
| mAb2 in vivo | 0.00149 | 0.001018–0.001968 | 0.001304 | 0.000640–0.001967 |
| mAb2 in vitro | 0.00180 | 0.001756–0.001838 | 0.000715 | 0.000631–0.000798 |
a The UV absorbances of the peptides were used for quantitation purposes.
Glu to pE Conversion in Vitro
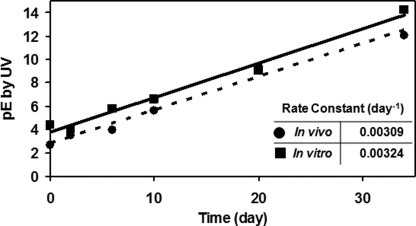
Increases to the pE level over time in vivo could be explained by either Glu to pE conversion or more rapid clearance of the Glu form. These two mechanisms can be distinguished by comparisons with in vitro changes under physiological conditions. To mimic physiological conditions, mAb1 and mAb2 samples were incubated in PBS at the blood pH of 7.4 and at 37 °C. Time course samples were then analyzed by the Lys-C peptide mapping method. A comparison of in vivo time course plot for the mA1 HC alongside the in vitro time course is provided in Fig. 4. Both time courses are linear with similar slopes. In all of the cases studied, the in vivo pE level time courses could be replicated with simple PBS incubations in vitro (Table 2). Together, these results support Glu to pE conversion as the mechanism for the changes observed in vivo and that the presence of pE does not impact clearance. Moreover, they suggest that the Glu to pE cyclization mechanism in vivo is also pH-controlled, so that simple in vitro incubations can accurately mimic Glu to pE conversion in vivo.
FIGURE 4.
Comparison of in vivo and in vitro pE formation rates. N-terminal Glu to pE conversion of mAb1 HC in vivo (circles) and in vitro in PBS at 37 °C at pH 7.4 (squares). The data represent the averages of two data sets for each experiment.
Differences in the Glu to pE conversion rates in vivo and in vitro could possibly be due to primary sequence or structural differences in the N-terminal regions of the antibodies studied. The mAb1 HC Framework 1 sequence is from the VH1 germ line, initiating with the sequence EVQL. mAb2 HC, from the VH3 germ line, begins with the same four amino acids on the N terminus but diverges shortly afterward. N-terminal sequences on the light chain are even more similar between the two antibodies, diverging only after the eighth residue from the N terminus. The mAb1 LC, from the VKVI germ line, begins with the sequence EIVLTQSPDFQ, whereas mAb2, from the VKIII germ line, begins with EIVLTQSPGTL.
To determine whether structural elements play a role in the Glu to pE conversion rates, the impact of denaturation on the reaction kinetics in vitro was measured. mAb1 in vitro time course studies, similar to those outlined above, were repeated, but in the presence or absence of 6 m guanidine HCl. Under native, physiological conditions, the N-terminal glutamate on the mAb1 HC converted much faster than the LC, both in vivo and in vitro. The addition of 6 m guanidine HCl in vitro increased the rate constant of the LC pE formation ∼10-fold (from 0.00019 to 0.0022 day−1) but had little impact on HC pE formation rates (k: 0.0025 day−1 for native and 0.0027 day−1 in 6 m guanidine HCl). After denaturation, the pE formation rate constant on the mAb1 LC was similar to the HC (Table 3). Taken together, the results indicate that structural considerations, and not the primary sequence, are the main factors that generate Glu to pE conversion rate differences between antibody polypeptide chains. In contrast to the mAb1 LC N-terminal Glu, there appears to be no structural constraints on its HC. Increasing the LC flexibility through denaturation greatly accelerates the LC pE conversion rate but had little impact on HC conversion rates. The similar Glu to pE conversion rates observed between the mAb1 LC and HC in denaturant provide additional evidence that the MS-based analysis accurately quantifies the pE levels.
TABLE 3.
The effect of denaturant on mAb1 Glu to pE conversion rates in vitro
The values (k) are first order rate constants. +, with 6 m guanidine HCl; −, without 6 m guanidine HCl.
|
k, +/− guanidine HCl |
||
|---|---|---|
| − | + | |
| day−1 | ||
| Light chain | 0.00019 | 0.0022 |
| Heavy chain | 0.0025 | 0.0027 |
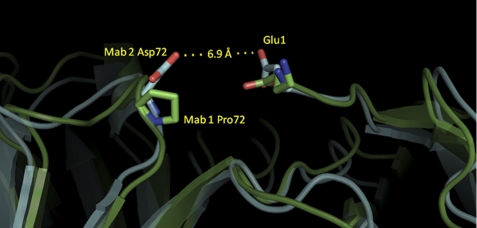
Modeling of the mAb1 and mAb2 using highly homologous Fab structures reveals features that could account for the pE formation rate differences between these two antibodies. mAb2, with the relatively fast LC pE formation rate, contains an Asp at position 72 on the HC (Kabat numbering), which models to within 7 Å of the LC N-terminal Glu (Fig. 5). In contrast, mAb1 has a Pro at the corresponding position. The Asp-72 may provide a repulsive force either directly with the Glu1 or simply increases the overall negative charge in that environment. Either mechanism would likely promote intramolecular cyclization because the side chain of Glu1 would not be expected to be tightly bound to the surface of mAb2. Without this repulsion, the Glu1 of mAb1 may bind to the surface and limit the N-terminal dynamics, resulting in lower pE formation rates.
FIGURE 5.
LC N-terminal region homology models of mAb1 and mAb2. Ribbons in green and light blue represent the model structures for mAb1 and mAb2, respectively. Side chain residues for light chain Glu-1, Pro-72 (mAb1), and Asp-72 (mAb2) are depicted as sticks (Kabat numbering). The distance from the Asp-61 delta oxygen and Glu-1 epsilon oxygen on the mAb2 model is given.
Patient Exposure to pE Modified mAb1 at Different Dosing
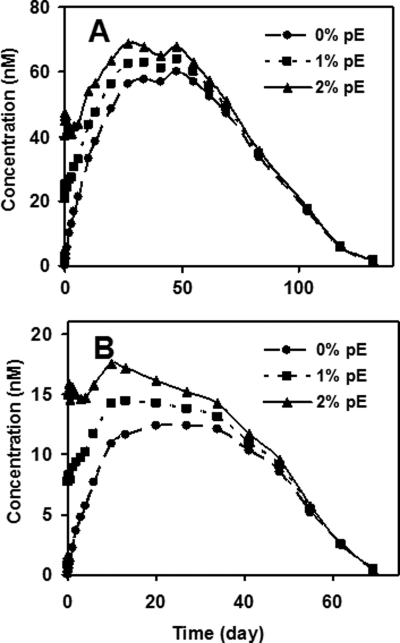
A patient exposure profile for an attribute formed or converted in vivo can be generated by the product of the overall drug elimination time course and the attribute conversion rate (17). The human pE exposure profiles for the mAb1 HC are shown in Fig. 6. Panel A illustrates the impact of drug lots differing in the initial levels of pE on the attribute concentration over time in vivo with a single 1000-mg intravenous dose, where the half-life removal from serum is ∼14 days (17). Lots with initial pE levels of 0, 1, and 2% are shown in this example. An increase of the initial pE level from 1 to 2%, which represents a 100% increase in relative pE level, generates an approximate 7% increase in total exposure, measured as the area under the curve, yet with the lower mAb1 dosing (300 mg intravenously) shown in Fig. 6B, the impact is greater. A similar increase of 1 to 2% in the initial relative pE level results in a 15% area under the curve increase. Because the relative differences in pE levels diminish with time in vivo, faster clearance of the antibody increases the exposure differences created by initial attribute level differences. In the case of mAb1, lower dosing leads to faster elimination of the antibody and thereby leads to greater exposure differences of the attribute. The impact of lot to lot variability would be expected to be even greater on mAb2, which has a short half-life (∼3 days) at the highest intravenous dosing and slower conversion. Thus, the impact of dosing on clearance, as well as initial levels and the rate of conversions, are all needed to obtain patient exposure profiles of pE in mAb1. Such modeling may be particularly useful in cases where the attribute is a safety concern, because the impact of lot to lot variation on patient exposure can be predicted.
FIGURE 6.
Patient exposure profiles based on dosing. The time course profile is based on mAb1 pharmacokinetic profile and the Glu to pE conversion rate for the mAb1 HC. A, 1000-mg intravenous dosing; B, 300-mg intravenous dosing.
Determination of pE Levels for Endogenous Antibodies
Many of the monoclonal antibodies and myeloma antibodies described in published studies contain pE on the N terminus of their heavy chains, which is mainly the result of glutamine conversion. Glutamines at the HC N termini of antibodies rapidly convert to pE and are found in the pE form by the time the protein has been purified from cell culture (5).3 All of the human heavy chain genes express either glutamate or glutamine as the N-terminal residue on the processed HC protein. Examination of the V base directory database reveals that 29 of the 49 VH germ line sequences contain glutamine as the N-terminal residue. Because of the rapid glutamine to pE conversion, one might suspect that a large fraction of endogenous antibodies could contain pE on their HC N termini.
Direct determination of the pE levels in endogenous antibodies was performed with IgG isolated from healthy human subjects. Analysis of the N-terminal residue by the peptide mapping approach, used for the mAbs, was not possible here because of the heterogeneity of the Framework 1 region in this polyclonal mixture. Instead, the level of pE was quantified by enzymatically releasing it from the protein followed by reversed phase HPLC/mass spectrometry. A polyclonal mixture of endogenous antibodies, containing IgG1, IgG2, and IgG4, was purified from human serum by protein A affinity and then treated with the pE cleaving enzyme pyroglutamate aminopeptidase. The treatment conditions used were shown to completely remove pE on mAb4 by Lys-C peptide mapping with mass spectrometric detection (not shown). Pure pE standards were used to generate a linear (R2 = 0.9997) calibration curve from 10 to 500 pmol. Released pE levels were then compared with the known concentration of antibody to determine the number of pE residues per antibody. Endogenous IgG from two healthy human subjects each contained ∼1.8 mol of pE/mol of antibody. A mAb4 antibody control generated 1.5 to 1.6 mol of pE/mol of antibody. Because the mAb4 was known to contain 2 mol of pE/mole of antibody, some protein loss may have occurred in sample handling or analysis that led to underestimation of the relative pE level. If similar losses occurred with the endogenous IgG sample, the generated values may represent a lower estimate of the pE levels. Although this approach does not determine the origin of the released pE, it does indicate that N-terminal pE is naturally occurring on endogenous antibodies at relatively high levels. Modifications considered unnatural may raise greater safety concerns on mAb drugs than ones found to be naturally occurring.
DISCUSSION
In previous studies, glutamate on the N termini of antibodies was shown to spontaneously convert to pyroglutamate under fairly mild conditions in vitro. Here we observed that similar conversion occurs on antibodies in vivo. For two antibodies containing N-terminal Glu on both heavy and light chains, conversion could be observed in circulation, although the rates differed between antibodies and between the polypeptide chains on the same antibody. A simple PBS model under physiological pH and temperature replicated the in vivo conversion rates reasonably well. These results strongly suggest that Glu to pE formation in vivo is nonenzymatic. Differences in the conversion rates in vitro could be attributed to structural differences between the proteins. The addition of denaturant increased the conversion rates and eliminated differences between polypeptides.
Pyroglutamate formation has been proposed as a stabilization mechanism for proteins and peptides in vivo. Because this cyclized residue is resistant to amino peptidases (18), it is reasonable to propose that its presence could slow physiological turnover. In this study, the turnover of antibodies appeared unaffected by the presence of pE. This conclusion is based on the observation that pE formation rates in vivo matched those measured in vitro. If pE-containing antibodies were cleared at a different rate than the original form, differences in the apparent conversion rates would have been expected. Thus, pyroglutamate appears to be a spontaneous and naturally occurring post-translational modification of antibodies in vivo that does not impact protein turnover.
The analysis and monitoring of pE during the manufacturing and storage of therapeutic antibodies can be achieved with charged based methods, such as cation exchange (4), but only if it originates from glutamine, not glutamate. Glutamate conversion to pE does not result in a charge state difference, so this change might be missed using simple chromatography-based release assays. To monitor glutamate cyclization, more complex assays, such as peptide mapping, would be required (5, 6). Whether pE is monitored and controlled for a given therapeutic antibody should be based on its impact to safety and efficacy. Because the N-terminal residues of both the light chains and the heavy chains are in the variable region and near the CDR, pE formation potentially could impact target binding, but this may vary for individual antibodies. An impact on efficacy would be expected to increase the criticality of this attribute. Other results, described in this study, could also bear on the criticality assessment. In particular, pE exposure in patients, through its natural occurrence in endogenous antibodies and through pE formation on the therapeutic antibody in vivo, may lessen the safety concerns for this attribute. In cases where pE impacts efficacy, patient exposure profiles can be used to estimate how initial levels of pE could affect activity over time in vivo and help guide rational manufacturing limits on such labile attributes.
Acknowledgments
We thank Zhongqi Zhang and Bhavana Shah for sample analysis; Yvonne Pan, Lilly Xu, Cynthia Li, Eugene Babcock, and Fuat Doymaz for assistance; and Szilan Fodor, Pavel Bondarenko, and Drew Kelner for providing suggestions during the course of these studies.
Y. D. Liu, A. M. Goetze, R. B. Bass, and G. C. Flynn, unpublished data.
- pE
- pyroglutamate
- HC
- heavy chains
- PGAP
- pyroglutamate amino peptidase
- FA
- formic acid.
REFERENCES
- 1. Blomback B. (1967) in Methods in Enzymology (Hirs C. ed) Vol. 11, pp. 389–411, Academic Press, New York [Google Scholar]
- 2. Schilling S., Wasternack C., Demuth H. U. (2008) Biol. Chem. 389, 983–991 [DOI] [PubMed] [Google Scholar]
- 3. Dayhoff M. O. (ed) (1972) Atlas of Protein Sequence and Structure, National Biomedical Research Foundation, Silver Spring, MD [Google Scholar]
- 4. Ouellette D., Alessandri L., Chin A., Grinnell C., Tarcsa E., Radziejewski C., Correia I. (2010) Anal. Biochem. 397, 37–47 [DOI] [PubMed] [Google Scholar]
- 5. Dick L. W., Jr., Kim C., Qiu D., Cheng K. C. (2007) Biotechnol. Bioeng. 97, 544–553 [DOI] [PubMed] [Google Scholar]
- 6. Chelius D., Jing K., Lueras A., Rehder D. S., Dillon T. M., Vizel A., Rajan R. S., Li T., Treuheit M. J., Bondarenko P. V. (2006) Anal. Chem. 78, 2370–2376 [DOI] [PubMed] [Google Scholar]
- 7. Yu L., Vizel A., Huff M. B., Young M., Remmele R. L., Jr., He B. (2006) J. Pharm. Biomed. Anal. 42, 455–463 [DOI] [PubMed] [Google Scholar]
- 8. Liu H., Gaza-Bulseco G., Sun J. (2006) J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 837, 35–43 [DOI] [PubMed] [Google Scholar]
- 9. Chirino A. J., Mire-Sluis A. (2004) Nat. Biotechnol. 22, 1383–1391 [DOI] [PubMed] [Google Scholar]
- 10. Rathore A. S. (2009) Trends Biotechnol. 27, 546–553 [DOI] [PubMed] [Google Scholar]
- 11. Rathore A. S., Winkle H. (2009) Nat. Biotechnol. 27, 26–34 [DOI] [PubMed] [Google Scholar]
- 12. Liu Y. D., Chen X., Enk J. Z., Plant M., Dillon T. M., Flynn G. C. (2008) J. Biol. Chem. 283, 29266–29272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Z. (2004) Anal. Chem. 76, 6374–6383 [DOI] [PubMed] [Google Scholar]
- 14. Flynn G. C., Chen X., Liu Y. D., Shah B., Zhang Z. (2010) Mol. Immunol. 47, 2074–2082 [DOI] [PubMed] [Google Scholar]
- 15. Mozdzanowski J., Bongers J., Anumula K. (1998) Anal. Biochem. 260, 183–187 [DOI] [PubMed] [Google Scholar]
- 16. Qu J., Chen W., Luo G., Wang Y., Xiao S., Ling Z., Chen G. (2002) Analyst 127, 66–69 [DOI] [PubMed] [Google Scholar]
- 17. Liu Y. D., van Enk J. Z., Flynn G. C. (2009) Biologicals 37, 313–322 [DOI] [PubMed] [Google Scholar]
- 18. Rink R., Arkema-Meter A., Baudoin I., Post E., Kuipers A., Nelemans S. A., Akanbi M. H., Moll G. N. (2010) J. Pharmacol Toxicol Methods 61, 210–218 [DOI] [PubMed] [Google Scholar]