Abstract
CD40 is a tumor necrosis factor receptor (TNFR) family protein that plays an important role in B cell development. CD154/CD40L is the physiological ligand of CD40. We have determined the crystal structure of the CD40-CD154 complex at 3.5 Å resolution. The binding site of CD40 is located in a crevice formed between two CD154 subunits. Charge complementarity plays a critical role in the CD40-CD154 interaction. Some of the missense mutations found in hereditary hyper-IgM syndrome can be mapped to the CD40-CD154 interface. The CD40 interaction area of one of the CD154 subunits is twice as large as that of the other subunit forming the binding crevice. This is because cysteine-rich domain 3 (CRD3) of CD40 has a disulfide bridge in an unusual position that alters the direction of the ladder-like structure of CD40. The Ser132 loop of CD154 is not involved in CD40 binding but its substitution significantly reduces p38- and ERK-dependent signaling by CD40, whereas JNK-dependent signaling is not affected. These findings suggest that ligand-induced di- or trimerization is necessary but not sufficient for complete activation of CD40.
Keywords: Crystal Structure, Receptor Structure-Function, Signal Transduction, Site-directed Mutagenesis, Tumor Necrosis Factor (TNF), CD154, CD40, CD40L
Introduction
CD40 is a member of the TNF receptor superfamily and a type 1 transmembrane protein (1). It is widely expressed in diverse cell types including B lymphocytes, dendritic cells, monocytes, platelets, endothelial cells, and fibroblasts. CD40-deficient mice are defective in thymus-dependent immunoglobulin class switching and germinal center formation, demonstrating the indispensable role of CD40 in cellular and humoral immune responses (2). CD154, also called CD40L or gp39, is the physiological ligand of CD40 (3). It is mainly expressed on activated T lymphocytes and regulates B cell function by binding to CD40 on the B cell surface. Defects in CD154 cause the most common form of hyper-IgM syndrome, indicating that CD154 is important in normal lymphocyte development (4–6). Patients with hyper-IgM syndrome have high serum levels of IgM and low levels of IgG, IgA, and IgE, resulting in recurrent bacterial infections. The T lymphocytes of these patients express a defective CD154 protein that is either not stable or is unable to bind to CD40. The phenotypes of CD154-deficient mice are similar to those of CD40-deficient ones (7). CD40 and CD154 also play critical roles in cells other than B lymphocytes. For example, in monocytes CD154 induces expression of CD54, MHC class II, CD86, CD40, IL-1, IL-6, IL-8, and TNFα, leading to enhanced cell survival and inflammatory responses (8). Because of their crucial roles in immune regulation, CD40 and CD154 are being actively investigated as targets of novel immune regulatory drugs (9, 10).
Because CD154, like other TNF family proteins, exists as a stable trimer (11), it has been proposed that its binding leads to trimerization of CD40 and initiates intracellular signaling by recruiting TRAF4 adapter proteins to the cytoplasmic domain of CD40 (9). Mutagenesis studies have demonstrated that the intracellular region of CD40 has multiple TRAF binding sites. Membrane-distal sites are responsible for binding TRAF1, -2, and -3, and membrane-proximal sites for binding TRAF6 (9, 12). Because multiple TRAFs can bind directly or indirectly to the intracellular region of CD40, several signaling pathways, including NF-κB, MAPK, and PI3K pathways can be activated by CD40. It has also been reported that a TRAF-independent pathway involving Jak3 (Janus family kinase) is activated by CD40.
CD154 is a ∼30-kDa type II transmembrane protein containing a TNF homology domain responsible for receptor binding and activation (7). It has also been detected in supernatants of activated T cells as soluble forms with apparent molecular masses of 14, 18, and 29 kDa (13, 14). These bind CD40 and generate intracellular signals. Although sequence identity among TNF family proteins is relatively low, CD154 and other TNF family proteins are composed of sandwiched antiparallel β-sheets forming homologous trimeric complexes (11). The crystal structures of three TNF family proteins, TNFβ, TRAIL (TNF-related apoptosis-inducing ligand), and BAFF (B cell-activating factor), complexed with their cognate receptors have been reported (15–19). All these TNF receptor (TNFR) family proteins contains several cysteine-rich domains (CRDs) forming an elongated ladder-like structure. Sequence alignment with other TNFR family indicates that the extracellular region of CD40 has three CRDs. Each is composed of two structural modules stabilized by 1∼2 disulfide bridges (20). The canonical TNF-TNFR family complex consists of three TNFRs bound to one TNF trimer, with the binding sites located in the grooves between pairs of TNF monomers (19). Although BAFF complexed with BAFFR or BCMA (B cell maturation) forms a large virus-like cluster under certain conditions (16, 18), the basic unit of the cluster appears to be the homologous ligand trimer and receptor complex.
To study ligand specificity and activation mechanism, we undertook a crystallographic study of CD40-CD154. Here, we report the 3.5 Å resolution crystal structure of the extracellular domain of CD40 complexed with the TNF homology domain of CD154. Unexpectedly, CD40 and CD154 are found in a 2:3 ratio in the crystal. Charge interaction plays an important role in the CD40-CD154 binding, and the mutations found in hyper-IgM patients could be mapped on the interaction interface. Mutagenesis experiments of CD154 led us to identify a loop that is critical for receptor activation but not for receptor binding.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The signal sequence and extracellular domain of human CD40 (residues Met1∼Asp190) fused to the Fc domain of human immunoglobulin G1 was co-expressed with the TNF homology domain of human CD154 (residues Gly116∼Leu261) in Hi5 insect cells (Invitrogen) using pVL1393 and pAcGP67A baculovirus vectors (BD Biosciences). The secreted CD40-CD154 complex was purified by protein A Sepharose (GE Healthcare) affinity chromatography. The Fc-tag of the fusion protein was removed by overnight thrombin digestion at 4 °C. The cleaved CD40-CD154 complex was further purified by Hitrap Q anion exchange chromatography and Superdex 200 (GE Healthcare) gel filtration chromatography. Fractions from the gel filtration column containing the CD40-CD154 complex were pooled and concentrated to ∼20 mg/ml for crystallization and amino acid analysis. CD40 could be produced not only with the Fc-tag but also with a hexahistidine tag and purified using a nickel-nitrilotriacetic acid (Qiagen) affinity chromatography. After thrombin cleavage and further purification, the resulting CD40-CD154 could be crystallized in the same condition.
Quantitative Amino Acid Analysis
The purified CD40-CD154 complex was enzymatically deglycosylated to reduce the thickness of the SDS-PAGE bands. For analysis, 5 μg of 0.25 mg/ml of CD40-CD154 was incubated with 125 ng of peptide-N4-(N-acetyl-β-glucosaminyl)asparagine amidase at 37 °C for 4 h. The proteins were separated by 15% SDS-PAGE, excised, and analyzed for quantitation. The quantitative amino acid analysis was conducted at the amino acid analysis facility of Yale University.
Crystallization and Data Collection
Crystals of the CD40-CD154 complex were grown at 23 °C using the hanging-drop vapor diffusion method by mixing 1 μl of protein solution and 1 μl of crystallization buffer containing 12% PEG 6000, 0.1 m MOPS (pH 6.5), and 10% isopropyl alcohol. Crystals suitable for diffraction experiments grew within 1 week. All crystals were flash-frozen in a buffer containing 12% PEG 6000, 0.1 m MOPS (pH 6.5), and 30% isopropyl alcohol using boiling liquid nitrogen at −170 °C. Diffraction data were collected at the ID29 beam line of the European Synchrotron Radiation Facility (ESRF), and the program Mosflm/SCALA was used to index, integrate, and scale the diffraction data (supplemental Table S1). The S132W mutant form of CD40 in complex with CD154 was produced using the same method and crystallized in the identical condition. The mutant proteins gave smaller crystals that diffracted x-ray to 5 Å. Complete data collection and structure determination have not been attempted.
Structure Determination
The initial phases were calculated by molecular replacement using the program PHASER (21) and the receptor-free CD154 structure with Protein Data Bank code 1ALY as the search probe (11). The atomic models were built by iterative modeling and refinement using programs O and PHENIX (22, 23). The atomic models of the CD40 receptor were built into the strong and continuous electron density found in the 2Fo − Fc and Fo − Fc electron density maps. The structure of the TNFβ-TNFR1 complex (19) was used as a guide for model building. The resulting model was further refined using the program CNS (version 1.3) (24), and the deformable elastic network (DEN) method was employed to improve the refinement procedure. The previously reported structure of CD154 refined at 2.0 Å resolution was used to calculate the initial DEN table and the γ and ωDEN values were systematically optimized. The final refinement statistics are summarized in supplemental Table S1. Residues 126–131 and 146–190 of CD40 were not clearly visible in the electron density map and were excluded from the final model. No non-glycine residues were found in the disallowed region of the Ramachandran plot.
Protein Production Using HEK293E Cells
A sequence encoding the extracellular region of CD154 (residues Gly116–Leu261) was cloned between the BamHI and XbaI sites of a modified pcDNA3 vector (Invitrogen), which has an Epstein-Barr virus origin of replication (oriP) at the BglII site, the signal sequence of the immunoglobulin κ light chain between the HindIII and BamHI sites, and a FLAG sequence between the XbaI and ApaI sites. Site-directed mutagenesis of the extracellular domain of CD154 was performed by overlap PCR and confirmed by DNA sequencing. HEK293E (human embryonic kidney 293E) cells were maintained in high glucose DMEM (Welgene) supplemented with 5% FBS (Welgene). They were seeded 24 h before transfection in a 150-mm culture dish at a density of 1.0 × 107 cells/well and transiently transfected with 10 μg of the expression vector using TransfectinTM Lipid reagent (Bio-Rad). Cell culture supernatants were harvested every 4 days up to 24 days. The secreted proteins were purified using anti-FLAG M2 affinity resin (Sigma) and eluted with FLAG peptide (Sigma).
Activity Assay Using an NF-κB Reporter System
The gene encoding full-length human CD40 was inserted between the XhoI and XbaI sites of pcDNA3.1/V5-His vector. The CD40 plasmid was transfected into HEK293T cells using TransfectinTM Lipid reagent (Bio-Rad). Stably transfected clones expressing CD40 were screened with 200 μg/ml G418, and surface expression of CD40 was confirmed by flow cytometry using anti-CD40 antibody (Novus Biologicals). The stable cell lines were seeded 24 h before transfection in a 24-well plate at a density of 2.0 × 105 cells/well and transiently transfected with 190 ng of NF-κB-luciferase reporter plasmid and 10 ng of CMV-Renilla luciferase plasmid as an internal control using TransfectinTM Lipid reagent. After 3 h, cells were mixed with 5∼20 μg/ml of the wild type or mutant CD154 proteins for 24 h. Reporter activity was measured using the firefly and Renilla luciferase assay system (Promega).
B Cell Activation Assay
Human peripheral B lymphocytes were purified from peripheral blood of healthy donors with magnetic beads using a B cell isolation kit from Miltenyi Biotech. Purified human peripheral B lymphocytes were cultured in RPMI 1640 (Sigma Aldrich) supplemented with 10% FBS (Welgene), 2 mm l-glutamine (Invitrogen), and 1 mm sodium pyruvate (Invitrogen). Purified human B lymphocytes (5 × 105 cells/ml) were incubated in 96-well plates and stimulated with 1∼5 μg/ml of the wild type or mutant CD154 proteins and 20 ng/ml of recombinant human IL-4 (R&D Systems). The concentration of IL-6 in cell culture supernatants was determined at 7 days after stimulation using a human IL-6 ELISA kit (R&D Systems).
Flow Cytometric Analysis
One million HEK293T cells stably expressing CD40 were washed twice with FACS buffer containing phosphate-buffered saline and 3% bovine serum albumin. The washed cells were incubated with 10 μg/ml CD154 wild type or mutant proteins for 30 min at 4 °C. After another round of washing with FACS buffer, the cells were incubated with FITC-conjugated anti-FLAG monoclonal antibody (Sigma, catalog no. F4049) for 30 min at 4 °C. Cells were acquired using FACScan (BD Biosciences). Data were analyzed with ModFit LT software (version 3.0; BD Biosciences).
Western Blotting
BJAB (EBV-negative, Burkitt-like lymphoma) B lymphoblastoid cells were incubated with 5∼20 μg/ml wild type or mutant CD154 proteins for 15 min at 37 °C. Cultured cells were washed with phosphate-buffered saline and lysed in a lysis buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 1 mm Na3VO4, 1 mm PMSF, pH 7.5) with freshly added protease inhibitor mixture (Sigma). After centrifugation, the protein concentration in the supernatant was determined by BCA assay (Pierce). Samples containing 20 μg of protein were resolved by 12% SDS-PAGE and transferred to a PVDF membrane (Millipore) using a Mighty Small Transphor Unit (Amersham Biosciences). The primary antibodies used: anti-p38, anti-phosho-p38, anti-ERK1/2, anti-phospho-ERK1/2, anti-JNK/SAPK, and anti-phospho-JNK/SAPK were obtained from Cell Signaling (Danvers). Anti-CD154 was from R&D Systems. Rabbit or mouse secondary antibodies were obtained from Santa Cruz Biotechnology.
RT-PCR Analysis
Total RNA from cell lysates was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Promega), and semi-quantitative PCR was performed using PCR Master Mix with the following pairs of primers: human IL-1α, 5′-AAT GAC GCC CTC AAT CAA AG-3′ (forward) and 5′-TCC GTA TCT CAG GCA TCT CC-3′ (reverse); human IL-6, 5′-ATG AAC TCC TTC TCC ACA AGC GC-3′ (forward) and 5′-GAA GAG CCC TCA GGC TGG ACT G-3′ (reverse); human IL-8, 5′-ATG ACT TCC AAG CTG GCC GTG-3′ (forward) and 5′-TTA TGA ATT CTC AGC CCT CTT CAA AAA CTT CTC-3′ (reverse); human TNFα, 5′-ATG AGC ACT GAA AGC ATG ATC CG-3′ (forward) and 5′-TCA CAG GGC AAT GAT CCC AAA GT-3′ (reverse).
RESULTS
Structure Determination and Overall Structure
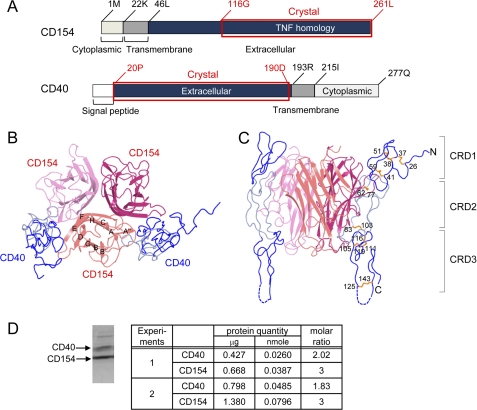
Using a recombinant baculovirus expression system, the TNF homology domain of the CD40 ligand was co-expressed with the extracellular domain of CD40 (Fig. 1A). After affinity purification, the fusion partner was removed by thrombin digestion and the resulting CD40-CD154 complex was purified and crystallized for structure determination. The crystals diffracted x-rays to 3.5 Å resolution, and the structure was determined by the molecular replacement technique using the previously reported structure of CD154 as the search probe (11). The binding sites for CD40 are located in the intersubunit grooves of the CD154 trimer. Unexpectedly, we found that one CD154 trimer interacted with only two CD40 in the crystal, and one of the three potential CD40 interaction sites in the CD154 trimer remained empty (Fig. 1, B and C). The 2:3 molar ratio of CD40 and CD154 was confirmed by quantitative amino acid analysis in the solution state. For this analysis, the purified complex was separated by SDS-PAGE after partial deglycosylation and the bands corresponding to CD40 and CD154 were quantitated by amino acid analysis (Fig. 1D).
FIGURE 1.
Overall structure of the CD40-CD154 complex. A, domain arrangements of human CD40 and CD154. The crystallized fragments are marked by red boxes and labeled. Top view (B) and side view (C) of the CD40-CD154 complex. The strands of CD154 are labeled. CRD1 and CRD3 of CD40 are colored dark blue, and CRD2 is colored light blue. D, molar ratio of CD40 and CD154 in solution. The purified complex of CD40 and CD154 were separated by SDS-PAGE after partial deglycosylation (see text). The protein bands were stained by Coomassie Brilliant Blue (left), excised, and quantitated by amino acid analysis (right). Samples from two independent preparations labeled as experiments 1 and 2 were analyzed. The minor band in the higher molecular weight region of the SDS-PAGE gel is that of peptide-N4-(N-acetyl-β-glucosaminyl)asparagine amidase added for deglycosylation. It was not subjected to the quantitation analysis. Significant figures of the analysis represent accuracy of the final quantitation step of HPLC chromatograms.
The CD154 monomer has a typical TNF family structure, containing a sandwich of two antiparallel β-sheets, with a Greek key fold (11, 25). The extensive trimerization interface between the subunits is formed mostly by hydrophobic residues. The overall structure of CD154 bound to CD40 does not differ from that of CD154 on its own, and the two CD154 structures can be superimposed with Cα root mean square difference of 0.717 Å. The greatest structural changes are located in the flexible loop regions and are not directly related to CD40 binding.
Structure of CD40 and Its Interaction with CD154
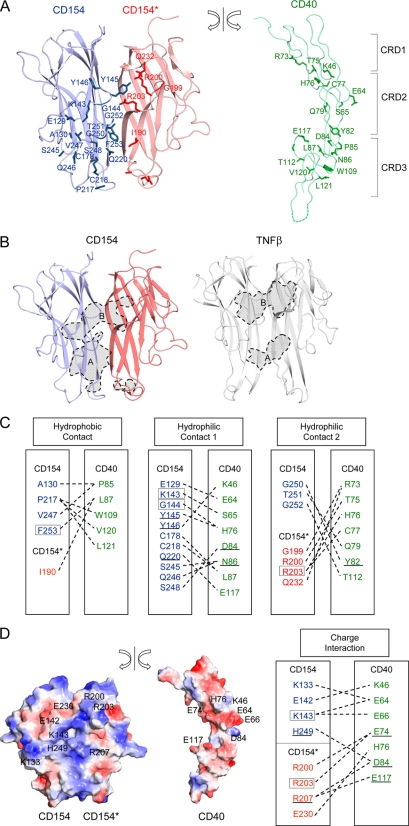
The crystallized extracellular domain of CD40 consists of 170 amino acids and contains three CRDs (supplemental Fig. S1). Each CRD has 2∼3 disulfide bridges that run roughly parallel to each other, forming a ladder-like elongated structure. These disulfide bridges play a critical role in stabilizing the structure of CD40. The extracellular domain of CD40 binds to a crevice area between pairs of CD154 subunits and interacts with both of them (Figs. 1B and 2A). In this article, the CD154 subunit on the left side of CD40 is labeled CD154 and that on the right side is labeled CD154*. The total interaction area buried by CD154 is 611 Å2, twice as large as that buried by CD154*, which is 378 Å2 (Fig. 2B). This asymmetry in interface size arises mainly because the interaction involving CRD3 and the second half of the CRD2 modules of CD40 is dominated by one of the CD154 subunits (Fig. 2B). The interaction involving CRD1 and the first half of CRD2 is approximately equally distributed between the two CD154 subunits.
FIGURE 2.
Structure of the CD40-CD154 interaction interface. A, the CD40 molecule is split from the bound CD154 and rotated to show the interaction interface. Residues directly contacting ligand or receptor are drawn. B, the CD40 contact area in CD154 and TNFR1 contact area in TNFβ are marked with broken lines and colored in gray. The contact areas can be divided into A and B patches (see text). C, residues making direct contacts are linked by broken lines, and residues found in hyper-IgM patients are boxed. The underlined residues were studied in previous mutagenesis research and proven to be critical for ligand-receptor interaction. D, surface representation of CD154 and CD40. The orientation is as in A. Positively and negatively charged surfaces are blue and red, respectively (left). Residues making critical charge interactions are linked by broken lines (right).
Hydrophilic and charge interactions play a major role in the CD40-CD154 interaction, and only one-third of the interaction interface is provided by nonpolar amino acids scattered throughout the interaction interface (Fig. 2, C and D). The interaction surfaces of CD40 and CD154 show clear charge complementarity; the interaction surface of CD154 is predominantly positively charged and that of CD40 negatively charged (Fig. 2D). Consistent with the structural observations, Singh et al. (26) reported that individual substitutions of the negatively charged residues Glu74, Asp84, and Glu117 in CD40, and Lys143, His249, Arg203, and Arg207 in CD154 prevented CD40-CD154 binding and receptor activation. The crystal structure and the mutagenesis data demonstrate the importance of the charge interaction in CD40-CD154 binding.
Mutations of CD154 can lead to hyper-IgM syndrome (4–6). Genetic analysis of such patients identified various mutations that block the normal function of CD154 (27). It has been reported that some of the altered residues are located in the protein core or trimerization interface, pointing to roles in structural stability or trimer formation (11). We found that the majority of the missense mutations exposed on the surface of the CD154 trimer in our crystal structure were either located in the direct CD40 contact area or involved in the critical charge interactions between CD40 and CD154. These residues are boxed in Fig. 2, C and D. This patient data demonstrates the importance for the immune response of the CD40-CD154 interaction revealed in our crystal structure. The physiological relevance of our structural observations is further supported by a series of site-directed mutagenesis experiments performed by Bajorath et al. (28–33). The locations of the residues involved are also given in Fig. 2, C and D.
Comparison with Structure of TNFβ-TNFR1 Complex
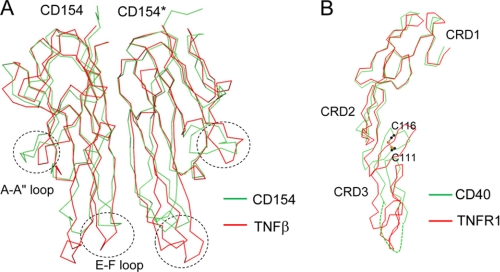
The structures of three TNF family proteins, TNFβ, TRAIL, and BAFF, together with their cognate receptors, have been reported (15–19). Of these, the CD40-CD154 structure most closely resembles that of TNFβ-TNFR1. Both CD154 and TNFβ have a common jelly roll type of fold, and the two structures can be superimposed with a Cα root mean square difference of 1.17 Å (Fig. 3A). The biggest structural changes are focused in the loops connecting the C and D strands and the E and F strands. CRD1 and CRD2 of the CD40 structure are superimposable with those of the TNFR1 structures with Cα root mean square difference of 1.05 Å (Fig. 3B). However, the structure of CRD3 deviates substantially from the canonical CRD structure due to a disulfide bridge formed between Cys111 and Cys116. This disulfide bridge is not found in the other TNFR family proteins with known structures, TNFR1 and DR5 (supplemental Fig. S1). These structural changes in CD154 and CD40 have an impact on the overall shape of the interaction interface (see below).
FIGURE 3.
Structural comparison of CD154 and CD40 with TNFβ and TNFR1. A, the structures of TNFβ and CD154 are aligned, and their Cα traces are superimposed. B, the structures of TNFR1 and CD40 are aligned. The side chains of Cys111 and Cys116 forming a disulfide bridge are shown as ball-and-stick models. Sulfurs and carbons are in orange and black, respectively.
The interaction interface between TNF and TNF receptor family proteins can be divided into two areas, patches A and B (Fig. 2B) (15, 16). The A patch interaction involves the second half of the CRD2 and CRD3 modules of CD40 and the D, E, and G strands and C–D, E–F, and G–H loops of CD154. The B patch interaction involves CRD1 and the first half of the CRD2 modules of CD40 and the A″, B′, E strands and A″–B′, D–E loops of CD154. The B patch interactions of TNFR1 and CD40 are highly homologous, and the two structures can easily be superimposed. However, the interactions of the A patches are substantially different in TNFR1 and CD40. In the TNFβ-TNFR1 structure, the A patch interaction area is equally divided between the two ligand subunits. In contrast, the A patch of CD40 interacts almost exclusively with one of the two CD154 subunits (Fig. 2B). This asymmetric distribution of the A patch interaction is due to structural changes in both ligands and receptors. The structural changes in CD154 involve mainly the loop connecting the C and D strands and the loop connecting the E and F strands. The C–D loop in CD154 is disordered, and the E–F loop undergoes 5∼7 Å structural shifts that alter the positions of the critical residues interacting with CD40. The structural changes in CD40 are focused in the CRD3 region. They appear to be triggered by the disulfide bridge between Cys111 and Cys116 of the receptor (Fig. 3B). This disulfide bridge distorts the direction of the ladder-like structure of CD40, pushing the CRD3 of CD40 away from the CD154* by ∼10 Å and increasing the interaction with CD154.
Greatly Reduced Agonist Activity of S132W Mutant of CD154
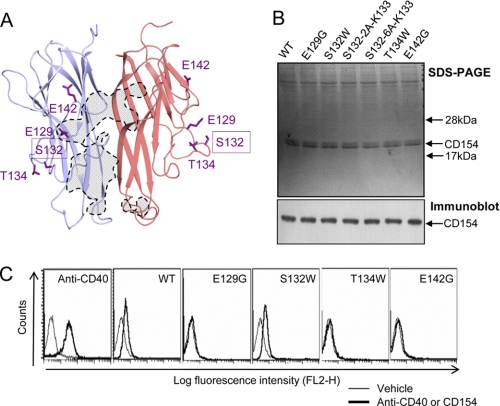
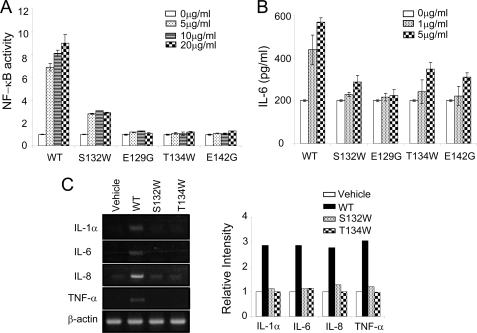
It has been proposed that the binding of trimeric CD154 induces multimerization of CD40, which in turn results in recruitment of TRAF to the intracellular domain. However, the multimeric structure of the receptor-ligand complex is poorly understood, particularly under membrane-attached conditions. Furthermore, several previous reports suggest that the higher order aggregation of CD40 may have a role in signal initiation (34–37). Therefore, we looked for CD154 mutants that were incapable of activating CD40 but still interacted with it. Several amino acid residues from 129 to 157 of CD154 were selected for site-directed mutagenesis because that region is exposed for interaction with other proteins or higher order multimerization of the receptors. Among them, soluble CD154 (residues Gly116–Leu261) mutants, E129G, S132W, T134W, and E142G were successfully expressed in HEK293E cells (Fig. 4, A and B). Using these mutants, we performed the following functional analyses. First, binding of the mutant ligands to CD40 was characterized. HEK293T cells stably transfected with CD40 were incubated with various CD154 wild or mutant types and analyzed by flow cytometry. The E129G, T134W, and E142G mutants failed to bind the receptor (Fig. 4C). The E129G and E142G mutants failed to bind because these residues either directly contact CD40 or make critical charge interactions with it (Fig. 2, C and D). In contrast, the S132W mutant retained a strong binding affinity for CD40, comparable indeed with that of wild type CD154. Ser132 is not located in the receptor binding region in our crystal structure. Second, to characterize downstream signaling, we measured CD40-dependent NF-κB activation in HEK293T cells stably transfected with CD40. Treatment with wild type CD154 resulted in a concentration-dependent increase of reporter activity, while the negative control mutants, E129G, T134W, and E142G, caused no induction (Fig. 5A). Surprisingly, the S132W mutant of CD154 had only ∼30% of the NF-κB activity of wild type CD154. We also analyzed IL-6 secretion in human primary B cells after treatment with CD154 wild type and mutants (Fig. 5B). IL-6 production in response to S132W was near the background level obtained with the negative controls, E129G, T134W, and E142G. We also studied the effect of mutant CD154 proteins on THP-1 monocytes, in which CD154 is known to play a proinflammatory role (8). As expected, wild type CD154 induced high levels of IL-1α, IL-6, IL-8, and TNFα, whereas the S132W mutant failed to induce any of these cytokines (Fig. 5C).
FIGURE 4.
CD40 binding activities of the CD154 mutants. A, positions of mutated residues in the crystal structure. The direct CD40 contact area in CD154 is marked with dashed lines. B, CD154 mutants used in the analysis. Equal amounts of affinity-purified wild type and mutant forms of soluble CD154 (residues Gly116–Leu261) were analyzed by SDS-PAGE and detected by Coomassie Blue staining and immunoblotting with anti-CD154 antibody. C, flow cytometric analysis. HEK293T cells stably transfected with a vector carrying full-length CD40 receptor were treated with FACS buffer alone (Vehicle, see “Experimental Procedures”) or FACS buffer containing 10 μg/ml WT or mutant forms of CD154 protein for 30 min at 4 °C. An anti-CD40 antibody was used to verify CD40 expression in the stably transfected HEK293T cells. Binding with FITC-conjugated anti-FLAG antibody was analyzed by flow cytometry. The results are representative of more than three independent experiments.
FIGURE 5.
Reduced NF-κB activation and IL-6 production in the S132W mutant of CD154. The S132W mutant has substantially reduced activity in NF-κB activation and IL-6 secretion assays. A, HEK293T cells stably transfected with CD40 were transiently transfected with NF-κB reporter and incubated with or without CD154 WT or mutant proteins (5∼20 μg/ml) for 24 h. The cells were then subjected to the luciferase assay. B, human primary B cells were prepared as described in “Experimental Procedures,” incubated with or without CD154 wild type or mutant proteins for 7 days. IL-6 protein secreted into supernatants was measured by ELISA. C, THP-1 cells were treated with 10 μg/ml wild type or the indicated mutant forms of CD154 for 12 h and then subjected to semiquantitative RT-PCR analysis (left). Relative intensities of the ethidium bromide stained bands were analyzed by TINA2.0 (Strauben, Hardt, Germany) (right). Data are the means ± S.E. of three independent experiments.
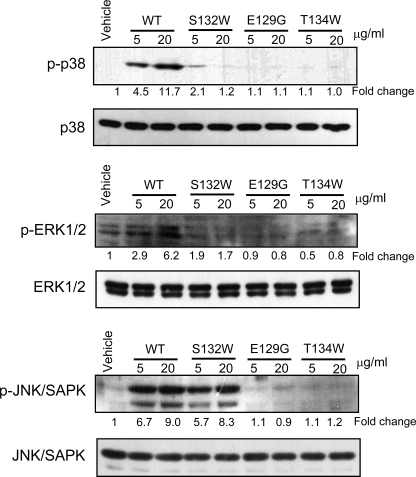
To further characterize the S132W mutant, we examined intracellular signaling by measuring the phosphorylation status of several MAPKs known to be activated by the CD40 receptor. After treatment of BJAB B lymphoblastoid cells with CD154 wild type and the mutants, we analyzed the phosphorylation of various kinases by immunoblotting. Wild type CD154 gave rise to high levels of phosphorylation of p38, ERK, and JNK, whereas the negative controls, E129G and T134W, which do not bind CD40, barely induced any phosphorylation of these kinases (Fig. 6). Interestingly the S132W mutant induced normal phosphorylation of JNK but virtually no phosphorylation of p38 and ERK. We conclude that the S132W mutant binds strongly to CD40 but induces significantly reduced activation of NF-κB and barely any phosphorylation of p38 and ERK. To assess the structural integrity of the mutant protein, we expressed the S132W mutant of CD154 together with the CD40 ectodomain in insect cells. The mutant protein was purified as a stable complex with CD40 and crystallized in the same conditions as the wild type protein. The crystals proved to belong to the same space group, with unit cell sizes practically identical to those of wild type crystals (supplemental Table S1). These data suggest that the mutation does not alter the overall structure of the receptor-ligand complex and together with our other observations demonstrate that the S132W mutant can bind tightly to the CD40 receptor but cannot activate it normally.
FIGURE 6.
Reduced activation of p38 and ERK1/2 by the S132W mutant. The S132W mutant cannot activate p38 and ERK1/2. BJAB cells were stimulated with CD154 WT or mutant forms for 15 min at 37 °C. Cell lysates were analyzed by immunoblotting with antibodies against phosphorylated p38, ERK1/2, and JNK. The blots were stripped and immunoblotted again with antibodies to total p38, ERK1/2, and JNK. Relative intensities of the phosphor forms in the autoradiograms were analyzed by TINA2.0. Data are representative of three independent experiments.
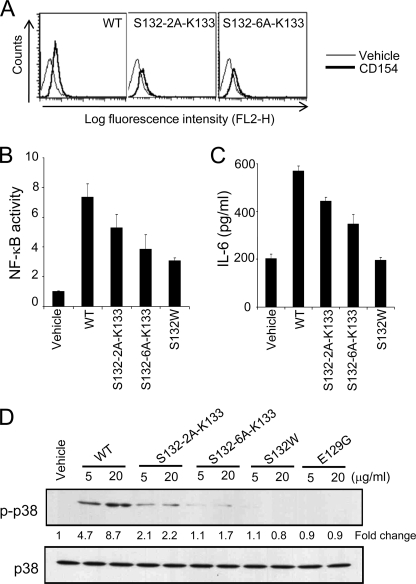
Because a small serine residue is substituted by a bulky tryptophan in the S132W mutant, we postulated that spatial conflict might lead to its reduced agonist activity. To test this hypothesis, we generated spatial conflict in the Ser132 loop by inserting alanines between Ser132 and Lys133. First, we verified expression of the two and six alanine insertion mutants, S132–2A-K133 and S132–6A-K133 (Fig. 4B). Both mutants bound normally to membrane-bound CD40 (Fig. 7A), and their activities were reduced to similar extents: S132–2A-K133 and S132–6A-K133 gave ∼30 and ∼50% reductions of NF-κB activity and IL-6 secretion, respectively (Fig. 7, B and C). Phosphorylation of p38 was also reduced as a function of the number of alanines inserted (Fig. 7D). These mutational data suggest that spatial conflict in the Ser132 loop inhibits complete activation of CD40.
FIGURE 7.
Effects of alanine insertions in the Ser132 loop of CD154. The loop-insertion mutant, S132–6A-K133, displays low NF-κB stimulating activity similar to the S132W mutant. A, HEK293T cells stably transfected with CD40 were incubated with CD154 wild type or mutant forms and analyzed by flow cytometry as in Fig. 4C. B, HEK293T cells stably transfected with CD40 were transiently transfected with an NF-κB reporter and incubated with or without CD154 wild type or mutant proteins for 24 h. Cells were then subjected to the luciferase assay. C, human primary B cells were prepared as described under “Experimental Procedures” and incubated with or without CD154 wild type or mutant proteins for 7 days. IL-6 secretion was measured by ELISA. D, BJAB cells were incubated with CD154 wild type or mutant proteins for 15 min at 37 °C. Cell lysates were analyzed by immunoblotting with antibodies to phosphorylated or total p38 as in Fig. 6. Data are the means ± S.E. of three independent experiments.
DISCUSSION
We have determined the first crystal structure of the CD40-CD154 complex. The overall interaction between CD40 and CD154 closely resembles the canonical pattern of the TNF-TNFR family interaction. However, substantial differences were found in several aspects of the interaction that controls its ligand binding and activation specificity. Charge complementarity has a more prominent role in the CD40-CD154 interaction. Some of the previously reported mutations disturb this charge interaction and thus disrupt the CD40-CD154 complex. The CRD3 of CD40 has a disulfide bridge in an unusual position that leads to an unequal distribution of the interaction area between the two CD154 subunits. Mutations of the Ser132 loop of CD154 did not block receptor binding but interfered with p38 and ERK signaling, suggesting that ligand binding and the resulting receptor di- or trimerization is necessary but not sufficient for the initiation of complete CD40-dependent signaling.
The most unexpected finding from our CD40-CD154 structure concerns the molar ratio of receptors and ligands. In our CD40-CD154 complex, only two of the three potential receptor binding sites are engaged with the receptor and the remaining site is unoccupied (Fig. 1B). This is not a crystallization artifact because CD40 and CD154 form a complex with a 2:3 molar ratio in solution as well as in crystals (Fig. 1D). All TNF-TNFR family structures known so far have a 3:3 molar ratio. This has led to the prevailing hypothesis of receptor activation according to which the stable trimeric structure of the TNF ligand family induces trimerization of TNF receptor family proteins, which in turn induces recruitment of TRAF adaptor proteins and initiates intracellular signaling. The fact that TRAFs form stable trimeric complexes further supports this hypothesis (37, 38).
In a survey of the literature, we found that the CD40-CD154 complex is not the first example with a 2:3 binding ratio; Eldredge et al. (39) reported that the purified extracellular domain of LTβR binds to its ligand LIGHT (LT-related inducible ligand that competes for glycoprotein D binding to herpesvirus entry mediator on T cells) with a 2:3 binding ratio. LIGHT and LTβR are also TNF ligand and receptor family proteins, respectively. The authors estimated the molar ratio of receptor to ligand by several biophysical methods including analytical ultracentrifugation, gel filtration chromatography, isothermal calorimetry, and static light scattering. The structure and molecular mechanisms controlling this unusual molar ratio have not been discussed in the case of the LTβR-LIGHT complex. Recently, two independent structural studies of the Fas signaling complex have questioned the current hypothesis (40, 41). Fas is a TNF receptor family member containing an intracellular death domain. The Fas death domains were found to form tetrameric or pentameric multimers when bound to the signaling adaptor FADD (Fas-associated protein with death domain). These Fas-FADD complex structures lack 3-fold symmetries, suggesting that receptor trimerization is not a prerequisite for activation of at least some TNF receptor family members.
It is not clear why the third receptor site in the CD154 trimer remains empty because all three potential receptor binding sites have identical structures, and there is no obvious steric repulsion between the receptors when the third receptor molecule is modeled into the empty binding site. There are many possible explanations for this observation. The preferred one is that charge repulsion may have a role in determining the molar ratio. As described in “Results,” charge interactions play an important role in the CD40-CD154 interaction; the binding interfaces of the CD40 receptors are predominantly negatively charged and those of the CD154 proteins positively charged (Fig. 2). Mutations substantially disturbing this charge complementarity disrupt stable receptor binding. We think that binding of the third CD40 molecule may be hindered by electrostatic repulsion from the two receptors already occupying the ligand trimer. Future mutagenesis studies are required to clarify the role of the charge distribution in the stoichiometry of the complex. It is important to note that our structural observations are based on truncated receptors without transmembrane and intracellular domains. We believe that the binding stoichiometry can be altered if there is favorable interaction in the transmembrane or intracellular domains of CD40. In physiological membrane-anchored conditions, full-length CD40 and other TNF family receptors form “preligand aggregates” that may affect binding stoichiometry as well (42, 43). Further research is required to analyze the binding kinetics and stoichiometry of CD154 under more physiologically relevant conditions.
The S132W mutant of CD154 had differential effects on the various kinds of CD40-dependent signaling. The NF-κB, ERK, and p38 pathways were severely affected in HEK293T and B lymphoblastoid cells, and IL-1α, IL-6, IL-8, and TNFα could not be induced in monocytes (Figs. 5 and 6). In contrast, JNK was almost optimally phosphorylated. This result is consistent with the report of Pearson et al. (44) that blocking of ERK using the specific inhibitor, PD98059, suppresses secretion of IL-6, IL-8, and TNFα. Several findings suggest that the effect of the S132W mutation is not due to disturbance of the structure of the CD154-CD40 complex. First of all, the Ser132 loop is separated from the ligand trimerization and ligand-receptor interaction sites in the crystal structure (Fig. 4A). The S132W protein is expressed at a similar level to the wild type protein and is stable (Fig. 4B). The mutant form of CD154 binds tightly to CD40, and the resulting complex is not dissociated by the various column purification steps used or by flow cytometric analysis (Fig. 4C). Moreover, the complexes of CD154 wild type and the S132W mutant with CD40 crystallize under the same conditions and as identical unit cells, demonstrating that the mutation does not change the global arrangement of the complex (supplemental Table S1). Finally, the effect of the insertion of six alanines into the Ser132 loop was similar to that of the S132W change (Fig. 7), indicating that the Ser132 loop can tolerate alterations without leading to global structural collapse and that the effect of the substitution is highly specific. From all these results we conclude that the Ser132 site is specifically and directly involved in activation of CD40 but not in receptor binding or ligand trimerization.
Experiments with mutants of the TRAF binding sites in the cytoplasmic domain of CD40 have indicated that CD154 activates the p38 and ERK pathways of CD40 in a TRAF6-dependent fashion (12, 45). Because the S132W mutation specifically blocked p38- and ERK-dependent signaling, it may primarily interfere with recruitment of TRAF6. Several mechanisms can be suggested to explain this effect. The first possibility is that this site interacts with other regulatory molecules or co-ligands specific to TRAF6 downstream signaling. For example, it has recently been shown that CD154 can bind to integrins on many cell types playing a role for inflammation and vascular disease (46–48). The second possibility, and the one we favor, is that this site is involved in higher order clustering of CD40. Many workers have reported evidence that higher order clustering of CD40 in the cell membrane plays a role in receptor activation (34–36). Furthermore, Pullen et al. (37) have reported that higher order clustering of CD40 is necessary for optimal signaling through TRAF6 binding. They argued that the TRAF6 binding site has 25- and 125-fold lower affinity than the TRAF3 and TRAF2 sites, respectively, and therefore requires the clustering of receptors to increase the avidity for TRAF6. However, we believe that conclusions should be drawn with caution and call for further biophysical characterization of CD40 clustering. Nonetheless, our S132W mutant promises to be a valuable tool in this work, and these results suggest that ligand-induced di- or trimerization is not sufficient for complete activation of CD40.
Supplementary Material
Acknowledgments
We thank the staff of the beam lines ID29 at the ESRF and BL41XU at SPring-8 for help with data collection and Dr. Julian Gross for critical reading of the manuscript.
This work was supported by the National Research Foundation (2010-0000101) and the Japanese-Korean Cooperative Program (2008-00539) of the Ministry of Education, Science, and Technology of Korea.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
The atomic coordinates and structure factors (code 3QD6) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TRAF
- TNF receptor-associated factor
- CRD
- cysteine-rich domain.
REFERENCES
- 1. van Kooten C., Banchereau J. (2000) J. Leukoc. Biol. 67, 2–17 [DOI] [PubMed] [Google Scholar]
- 2. Kawabe T., Naka T., Yoshida K., Tanaka T., Fujiwara H., Suematsu S., Yoshida N., Kishimoto T., Kikutani H. (1994) Immunity. 1, 167–178 [DOI] [PubMed] [Google Scholar]
- 3. Armitage R. J., Fanslow W. C., Strockbine L., Sato T. A., Clifford K. N., Macduff B. M., Anderson D. M., Gimpel S. D., Davis-Smith T., Maliszewski C. R., et al. (1992) Nature. 357, 80–82 [DOI] [PubMed] [Google Scholar]
- 4. Aruffo A., Farrington M., Hollenbaugh D., Li X., Milatovich A., Nonoyama S., Bajorath J., Grosmaire L. S., Stenkamp R., Neubauer M., et al. (1993) Cell. 72, 291–300 [DOI] [PubMed] [Google Scholar]
- 5. Korthäuer U., Graf D., Mages H. W., Brière F., Padayachee M., Malcolm S., Ugazio A. G., Notarangelo L. D., Levinsky R. J., Kroczek R. A. (1993) Nature. 361, 539–541 [DOI] [PubMed] [Google Scholar]
- 6. DiSanto J. P., Bonnefoy J. Y., Gauchat J. F., Fischer A., de Saint Basile G. (1993) Nature. 361, 541–543 [DOI] [PubMed] [Google Scholar]
- 7. Xu J., Foy T. M., Laman J. D., Elliott E. A., Dunn J. J., Waldschmidt T. J., Elsemore J., Noelle R. J., Flavell R. A. (1994) Immunity 1, 423–431 [DOI] [PubMed] [Google Scholar]
- 8. Kiener P. A., Moran-Davis P., Rankin B. M., Wahl A. F., Aruffo A., Hollenbaugh D. (1995) J. Immunol. 155, 4917–4925 [PubMed] [Google Scholar]
- 9. Elgueta R., Benson M. J., de Vries V. C., Wasiuk A., Guo Y., Noelle R. J. (2009) Immunol. Rev. 229, 152–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vonderheide R. H. (2007) Clin. Cancer Res. 13, 1083–1088 [DOI] [PubMed] [Google Scholar]
- 11. Karpusas M., Hsu Y. M., Wang J. H., Thompson J., Lederman S., Chess L., Thomas D. (1995) Structure. 3, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 12. Pullen S. S., Dang T. T., Crute J. J., Kehry M. R. (1999) J. Biol. Chem. 274, 14246–14254 [DOI] [PubMed] [Google Scholar]
- 13. Graf D., Müller S., Korthäuer U., van Kooten C., Weise C., Kroczek R. A. (1995) Eur. J. Immunol. 25, 1749–1754 [DOI] [PubMed] [Google Scholar]
- 14. Mazzei G. J., Edgerton M. D., Losberger C., Lecoanet-Henchoz S., Graber P., Durandy A., Gauchat J. F., Bernard A., Allet B., Bonnefoy J. Y. (1995) J. Biol. Chem. 270, 7025–7028 [DOI] [PubMed] [Google Scholar]
- 15. Hymowitz S. G., Christinger H. W., Fuh G., Ultsch M., O'Connell M., Kelley R. F., Ashkenazi A., de Vos A. M. (1999) Mol. Cell. 4, 563–571 [DOI] [PubMed] [Google Scholar]
- 16. Kim H. M., Yu K. S., Lee M. E., Shin D. R., Kim Y. S., Paik S. G., Yoo O. J., Lee H., Lee J. O. (2003) Nat. Struct. Biol. 10, 342–348 [DOI] [PubMed] [Google Scholar]
- 17. Mongkolsapaya J., Grimes J. M., Chen N., Xu X. N., Stuart D. I., Jones E. Y., Screaton G. R. (1999) Nat. Struct. Biol. 6, 1048–1053 [DOI] [PubMed] [Google Scholar]
- 18. Liu Y., Hong X., Kappler J., Jiang L., Zhang R., Xu L., Pan C. H., Martin W. E., Murphy R. C., Shu H. B., Dai S., Zhang G. (2003) Nature. 423, 49–56 [DOI] [PubMed] [Google Scholar]
- 19. Banner D. W., D'Arcy A., Janes W., Gentz R., Schoenfeld H. J., Broger C., Loetscher H., Lesslauer W. (1993) Cell 73, 431–445 [DOI] [PubMed] [Google Scholar]
- 20. Naismith J. H., Sprang S. R. (1998) Trends Biochem. Sci. 23, 74–79 [DOI] [PubMed] [Google Scholar]
- 21. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 22. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 23. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 24. Schröder G. F., Levitt M., Brunger A. T. (2010) Nature. 464, 1218–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karpusas M., Lucci J., Ferrant J., Benjamin C., Taylor F. R., Strauch K., Garber E., Hsu Y. M. (2001) Structure. 9, 321–329 [DOI] [PubMed] [Google Scholar]
- 26. Singh J., Garber E., Van Vlijmen H., Karpusas M., Hsu Y. M., Zheng Z., Naismith J. H., Thomas D. (1998) Protein Sci. 7, 1124–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piirilä H., Väliaho J., Vihinen M. (2006) Hum. Mutat. 27, 1200–1208 [DOI] [PubMed] [Google Scholar]
- 28. Bajorath J., Stenkamp R., Aruffo A. (1993) Protein Sci. 2, 1798–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bajorath J., Chalupny N. J., Marken J. S., Siadak A. W., Skonier J., Gordon M., Hollenbaugh D., Noelle R. J., Ochs H. D., Aruffo A. (1995) Biochemistry 34, 1833–1844 [DOI] [PubMed] [Google Scholar]
- 30. Bajorath J., Marken J. S., Chalupny N. J., Spoon T. L., Siadak A. W., Gordon M., Noelle R. J., Hollenbaugh D., Aruffo A. (1995) Biochemistry 34, 9884–9892 [DOI] [PubMed] [Google Scholar]
- 31. Bajorath J., Seyama K., Nonoyama S., Ochs H. D., Aruffo A. (1996) Protein Sci. 5, 531–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bajorath J., Aruffo A. (1997) Proteins 27, 59–70 [PubMed] [Google Scholar]
- 33. Bajorath J. (1998) J. Biol. Chem. 273, 24603–24609 [DOI] [PubMed] [Google Scholar]
- 34. Haswell L. E., Glennie M. J., Al-Shamkhani A. (2001) Eur. J. Immunol. 31, 3094–3100 [DOI] [PubMed] [Google Scholar]
- 35. Kilinc M. O., Mukundan L., Yolcu E. S., Singh N. P., Suttles J., Shirwan H. (2006) Exp. Mol. Pathol. 80, 252–261 [DOI] [PubMed] [Google Scholar]
- 36. Miconnet I., Pantaleo G. (2008) Vaccine 26, 4006–4014 [DOI] [PubMed] [Google Scholar]
- 37. Pullen S. S., Labadia M. E., Ingraham R. H., McWhirter S. M., Everdeen D. S., Alber T., Crute J. J., Kehry M. R. (1999) Biochemistry 38, 10168–10177 [DOI] [PubMed] [Google Scholar]
- 38. Park Y. C., Burkitt V., Villa A. R., Tong L., Wu H. (1999) Nature 398, 533–538 [DOI] [PubMed] [Google Scholar]
- 39. Eldredge J., Berkowitz S., Corin A. F., Day E. S., Hayes D., Meier W., Strauch K., Zafari M., Tadi M., Farrington G. K. (2006) Biochemistry 45, 10117–10128 [DOI] [PubMed] [Google Scholar]
- 40. Wang L., Yang J. K., Kabaleeswaran V., Rice A. J., Cruz A. C., Park A. Y., Yin Q., Damko E., Jang S. B., Raunser S., Robinson C. V., Siegel R. M., Walz T., Wu H. (2010) Nat. Struct. Mol. Biol. 17, 1324–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scott F. L., Stec B., Pop C., Dobaczewska M. K., Lee J. J., Monosov E., Robinson H., Salvesen G. S., Schwarzenbacher R., Riedl S. J. (2009) Nature 457, 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chan F. K., Chun H. J., Zheng L., Siegel R. M., Bui K. L., Lenardo M. J. (2000) Science 288, 2351–2354 [DOI] [PubMed] [Google Scholar]
- 43. Chan F. K. (2007) Cytokine 37, 101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pearson L. L., Castle B. E., Kehry M. R. (2001) Int. Immunol. 13, 273–283 [DOI] [PubMed] [Google Scholar]
- 45. Kashiwada M., Shirakata Y., Inoue J. I., Nakano H., Okazaki K., Okumura K., Yamamoto T., Nagaoka H., Takemori T. (1998) J. Exp. Med. 187, 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Léveillé C., Bouillon M., Guo W., Bolduc J., Sharif-Askari E., El-Fakhry Y., Reyes-Moreno C., Lapointe R., Merhi Y., Wilkins J. A., Mourad W. (2007) J. Biol. Chem. 282, 5143–5151 [DOI] [PubMed] [Google Scholar]
- 47. Hassan G. S., Merhi Y., Mourad W. M. (2009) Trends Immunol. 30, 165–172 [DOI] [PubMed] [Google Scholar]
- 48. Prasad K. S., Andre P., He M., Bao M., Manganello J., Phillips D. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12367–12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.