Abstract
Hepatitis C virus (HCV) RNA replicates its genome on specialized endoplasmic reticulum modified membranes termed membranous web and utilizes lipid droplets for initiating the viral nucleocapsid assembly. HCV maturation and/or the egress pathway requires host sphingolipid synthesis, which occur in the Golgi. Ceramide transfer protein (CERT) and oxysterol-binding protein (OSBP) play a crucial role in sphingolipid biosynthesis. Protein kinase D (PKD), a serine/threonine kinase, is recruited to the trans-Golgi network where it influences vesicular trafficking to the plasma membrane by regulation of several important mediators via phosphorylation. PKD attenuates the function of both CERT and OSBP by phosphorylation at their respective Ser132 and Ser240 residues (phosphorylation inhibition). Here, we investigated the functional role of PKD in HCV secretion. Our studies show that HCV gene expression down-regulated PKD activation. PKD depletion by shRNA or inhibition by pharmacological inhibitor Gö6976 enhanced HCV secretion. Overexpression of a constitutively active form of PKD suppressed HCV secretion. The suppression by PKD was subverted by the ectopic expression of nonphosphorylatable serine mutant CERT S132A or OSBP S240A. These observations imply that PKD negatively regulates HCV secretion/release by attenuating OSBP and CERT functions by phosphorylation inhibition. This study identifies the key role of the Golgi components in the HCV maturation process.
Keywords: Golgi, Hepatitis Virus, Intracellular Trafficking, Lipid-binding Protein, Protein Kinases, Virus Assembly, CERT, HCV, OSBP, PKD
Introduction
HCV4 infects about 2–3% of the world population and is the leading causative agent of chronic liver disease (1, 2). HCV infection progresses to steatosis, cirrhosis, and hepatocellular carcinoma (1, 3). HCV is a positive strand RNA virus classified as Hepacivirus genus within the Flaviviridae family. HCV encodes a polyprotein of about 3000 amino acids that is proteolytically processed into three mature structural and seven nonstructural viral proteins (4, 5). The HCV RNA genome replicates within a ribonucleoprotein complex on the ER-derived modified membranous structures termed the “membranous web” (6–8). The viral replication complex is assembled in close proximity to cytosolic lipid droplets, and this arrangement promotes subsequent steps of viral assembly/morphogenesis. HCV alters host lipid metabolism and causes the redistribution and accumulation of lipid droplets around the perinuclear region (9, 10). The viral core protein closely associates with lipid droplets and recruits NS5A, and these interactions are critical for an efficient viral assembly process (11). Evidence suggests that HCV secretion is linked to cellular very low density lipoprotein (VLDL) secretion (12). HCV secretion is inhibited by silencing apolipoprotein B-100 (apoB), apoE, and apoC-I as well as inhibition of microsomal triglyceride transfer protein activity (13–15). These and other data strongly argue for the utilization of the VLDL secretory pathway by HCV for its maturation/secretion (12, 16). Although the VLDL secretion pathway is not completely characterized, it is believed to occur through the Golgi network (17, 18). The exact pathway that results in the association of HCV nucleocapsids (either enveloped or non-enveloped) with the VLDL particles en route to the Golgi compartment remains to be characterized. Similarly the role of lipid droplets in HCV morphogenesis remains to be clearly understood.
OSBP is a sterol sensor and facilitates trafficking of cholesterol or hydroxycholesterol from ER to Golgi (19, 20). OSBP binds to both vesicle-associated membrane protein-associated protein (VAP)-subtype A on the ER and phosphatidylinositol 4-phosphate (PI4P) on the Golgi to form a “membrane contact site” (MCS) to facilitate lipid transfer between opposing surfaces (21). CERT, which shares functional homology with OSBP, regulates the transport of ceramide from ER to the Golgi where the ceramide is converted to sphingolipids (22). OSBP modulates CERT activation and translocation to the Golgi and thereby integrates sterol homeostasis to sphingolipid biosynthesis (21, 23). We previously showed that OSBP mediates HCV secretion while binding to NS5A and vesicle-associated membrane protein-associated protein (VAP)-subtype A (24). Inhibition of CERT function effectively suppressed HCV release without affecting RNA replication (25). These studies indicate that these lipid transport proteins, CERT, and OSBP directly contribute to HCV morphogenesis/secretion.
PKD is a serine/threonine kinase and exists in three distinct isoforms (PKD1, PKD2, and PKD3). PKD regulates multiple cellular processes including cell survival, adhesion, motility, and differentiation (26–28). In addition, PKD promotes the fission of cargo vesicles from the TGN and thus regulates the secretion of these vesicles from the TGN to the plasma membrane (26, 28, 29). PKD is recruited to the Golgi through the interaction between diacylglycerol and its cysteine-rich C1a domain (27–29). The Golgi-associated PKD is activated by a novel PKC isoform, PKCη, by phosphorylation of serine residues in the “activation loop” of PKD (30). At the TGN, PKD activates PI4KIIIβ to generate PI4P, which mediates the Golgi localization of CERT and OSBP proteins via binding to their pleckstrin homology (PH) domains. PKD-mediated phosphorylation of CERT at Ser132 and OSBP at Ser240 impairs their Golgi localization and inhibits their functions in integrating the cholesterol and sphingomyelin (SM) metabolism (31, 32). Although active PKD is known to promote secretion of small cargo proteins (e.g. VSV-G), little is known about how PKD modulates the transport of large cargos like viral vesicles or encapsidated viral core particles in the TGN. In this study, we investigated the functional role of PKD in the HCV maturation and/or secretion process with an emphasis on its substrates, CERT and OSBP.
Our studies show that PKD negatively regulates HCV secretion via the attenuation of OSBP and CERT through phosphorylation of their specific serine residues. HCV infection mitigates PKD activation. RNA interference of PKD expression and inhibition of PKD activity led to an increase in HCV secretion. Overexpression of a constitutively active form of PKD caused suppression of HCV secretion. This suppression by PKD was subverted by the ectopic expression of CERT S132A mutant or OSBP S240A mutant. These studies identify the key role of the Golgi network in HCV secretion process.
EXPERIMENTAL PROCEDURES
Plamsids
The plasmids pJC1 and p7-Rluc2A were kind gifts of Drs. F. Chisari (The Scripps Research Institute) and C. Rice (Rockefeller University), respectively. p7-Rluc2A is derived from JC1 (33). N-terminally HA-tagged human PKD1 expression vectors HA.PKD, HA.PKD.K/W, HA.PKD.S738E/S742E, and ΔPH were obtained from Dr. Alex Toker through Addgene (Cambridge, MA). pcDNA3-STrEP tagged (Strep)-PKD1 expression vectors were generated by PCR-mediated cloning using HA.PKD vector as template. The following pair of oligonucleotides was used for PCR: forward, 5′-cgggatccatggctagctggagccacccgcagttcgagaaaagcgcccctccggtcctg-3′; and reverse, 5′-caatctttgcactgcaagcc-3′. The PCR product was digested with BamHI and HindIII and then inserted into corresponding sites of pcDNA3-HA.PKD vectors. The Strep tag affinity procedure was performed according to the manufacturer's instructions (IBA, Gottingen, Germany). pSUPER-shRNA expression vector for targeting PKD1 expression was developed as described previously (34). The following pairs were inserted between BglII and HindIII sites of pSUPER vector: shPKD1-1, 5′-gatccccatgctgtgggggctggtacttcaagagagtaccagcccccacagcattttttggaaa-3′ and 5′-agcttttccaaaaaatgctgtgggggctggtactctcttgaagtaccagcccccacagcatggg-3′; and shPKD1-2, 5′-gatccccgttccctgaatgtggtttcttcaagagagaaaccacattcagggaactttttggaaa-3′ and 5′-agcttttccaaaaagttccctgaatgtggtttctctcttgaagaaaccacattcagggaacggg-3′. The designs of shPKD1-1 (35) and shPKD1-2 (36) were described previously. pcDNA3-FLAG-CERT and pcDNA3-FLAG-CERT S132A expression vectors were gifts from Dr. Juan Saus and Dr. Manilola Olayioye, respectively, and were described previously (31, 37). pFLAG-CMV-OSBP was described previously (24). pFLAG-CMV-OSBP S240A was constructed by PCR-mediated site-directed mutagenesis using the following primers: S240A, 5′-gctctgcagcgttctctcGCtgagctggagtccctgaa-3′ and 5′-ttcagggactccagctcaGCgagagaacgctgcagagc-3′. The PCR product was digested with AfeI and BclI and cloned into corresponding sites of the vector.
Lentiviral Packaging and Transfection
pCSII-EF1-MCS and a set of packaging plasmid vectors including pCMV-VSV-G-RSV-Rev and pCAG-HIVgp were kind gifts from Dr. Hiroyuki Miyoshi (Riken BioResource Center). cDNAs encoding Strep-tagged PKD1, FLAG-OSBP, and FLAG-CERT were recloned into a multicloning site of pCSII-EF1-MCS using oligolinkers and conventional molecular cloning techniques. HEK293FT cells were used as the packaging host to obtain lentiviral vectors. Packaging transfection was performed using TransIT-LT1 reagent (Mirus Bio) accordingly to the manufacturer's instructions.
Reagents and Antibodies
Anti-FLAG monoclonal antibody clone M2 was purchased from Sigma. Anti-PKD1 polyclonal antibody was obtained from Cell Signaling Technology. Anti-phospho-PKD1 (PKC sites, rabbit monoclonal, EP1493Y) and anti-phospho-PKD1 (autophosphorylation site, rabbit polyclonal) antibodies were purchased from Novus Biological and Cell Signaling Technology, respectively. The anti-PKD pMOTIF antibody was a generous gift from Dr. A Toker (Harvard Medical School) (38). Mouse monoclonal anti-NS5A antibody was a kind gift from Dr. C. Rice (Rockefeller University). Mouse monoclonal anti-Core antibody was purchased from Affinity Bioreagents. Rabbit polyclonal anti-human albumin and anti-calnexin antibody were from MP Biomedicals and Santa Cruz Biotechnology, respectively. PKC inhibitor Gö6983 and PKC/PKD inhibitor Gö6976 were purchased from Calbiochem. Inhibitors were dissolved in dimethyl sulfoxide (DMSO) (Hybri-Max, Sigma-Aldrich) and used at a final concentration of DMSO not exceeding 0.1% (v/v).
Immunoprecipitation
For immunoprecipitation of FLAG-CERT proteins, transfected cells in a 60-mm dish were lysed in 1 ml of lysis buffer (0.25% deoxycholic acid, 0.1% Triton X-100, 150 mm NaCl, 100 mm Tris/HCl, pH 8.0, plus phosphatase inhibitors). After brief sonification and centrifugation, 0.5 ml of cleared lysate was incubated with 2 μg of anti-FLAG monoclonal antibody M2 (Sigma-Aldrich) for 1 h at 4 °C. Immune complexes were captured with Protein G-Sepharose 4FF beads (GE Healthcare) by subsequent incubation at 4 °C for 1 h. Captured immune complexes were washed five times with 1 ml of lysis buffer and two times with Tris-buffered saline. Western blot analysis was performed as described previously (24).
Cell Culture
Huh7.5.1 cells were a kind gift from Dr. Frank Chisari (The Scripps Research Institute). Huh7 and Huh7.5.1 cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.1 mm minimum Eagle's medium nonessential amino acids (Invitrogen).
In Vitro RNA Transcription and RNA Transfection
Viral genomic RNAs were synthesized in vitro by T7 RNA polymerase using the Ribomax large scale RNA production system (Promega, Madison, WI). Transcribed RNAs were extracted by the acid guanidinium thiocyanate-phenol-chloroform method and quantified by spectrometry. Electroporation was performed using Cytomix buffer (39) as described previously (24).
HCV Infection, Focus-forming Unit Assay, and HCV Secretion Assay
Initial infection was carried out by transfecting Huh7.5.1 cells with in vitro synthesized JC1 RNA. Cultured supernatants were collected at 10 days after transfection and titrated by focus-forming unit assay as described previously (40). The intracellular infectivity assay was carried out as described previously (14). Briefly, the collected cells were resuspended in culture medium and lysed by four freeze-thaw cycles. The cell debris were pelleted by centrifugation at 4000 rpm for 5 min. The supernatant collected was used for the focus-forming unit assay. In the case of p7-Rluc2A, in vitro transcribed genomic RNA was electroporated into Huh7.5.1 cells. Cultured supernatants containing reporter virion particles were collected at the indicated time and centrifuged for clarification (900 × g for 3 min). 100 μl of cleared supernatant was overlaid onto naïve Huh 7.5.1 cells in a 96-well plate format (1 × 104 cells/well) and incubated for 48 h with a change of culture medium at 12 h postinoculation. The relative amounts of infectious reporter virions contained in the supernatant were assayed for Renilla luciferase activity (Promega) of infected naïve Huh-7 cells. Confocal immunofluorescence microscopy was performed as described previously (24).
ssHRP-FLAG Secretion Assay
The ssHRP-FLAG secretion assay was carried out as described previously (28). HCV-infected and uninfected Huh 7.5.1 cells were transfected with ssHRP-FLAG plasmid together with empty vector or Strep-tagged PKD1 expression vectors encoding wild type (WT), constitutively active (CA), PH domain deletion (ΔPH), or kinase-dead (KD) mutant forms of PKD1 at a ratio of 1:6.5. At 24 h post-transfection, culture medium was replaced with serum-deficient medium. Secreted ssHRP-FLAG in the 12-h culture supernatants was quantified by incubation of the clarified supernatants with HRP substrate, and absorbance was measured in an ELISA plate reader at 450 nm.
RESULTS
HCV Infection Impairs PKD1 Protein Activity
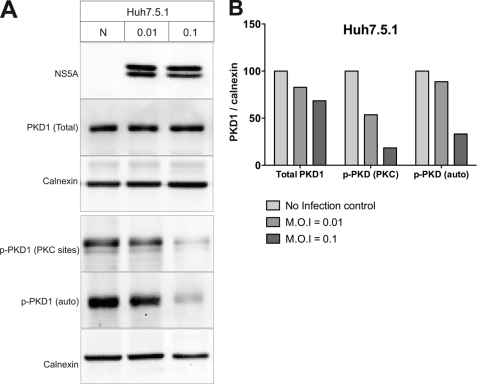
PKD regulates the trafficking of secretory vesicles by promoting the fission of these vesicles from trans-Golgi network (28). In this study, we investigated the functional role of PKD in the transport of HCV particles. First, we analyzed the expression levels and kinase activity of PKD1 in HCV-infected cells. PKCη is a novel PKC isoform known to activate PKD1 at the TGN by phosphorylating Ser738 and Ser742 residues in the PKD1 activation loop (30, 41, 43). The activated PKD1 then autophosphorylates the Ser910 residue in its C terminus, which correlates with the kinase-active form of PKD1 (42, 43). We infected Huh7.5.1 cells with culture-derived HCV virus particles at multiplicities of infection (m.o.i.) of 0.01 and 0.1. Six days postinfection, the whole cell lysates were analyzed by immunoblotting using phosphoserine antibodies that specifically recognize the phosphorylated Ser738 and Ser742 (PKC site) and Ser910 (PKD autophosphorylation site). The results show that the PKD1 kinase activation is impaired in HCV-infected cells, although total PKD1expression levels in uninfected and HCV-infected cells are the same (Fig. 1A). Consistent with the above data, the reduction of PKD1 phosphorylation is more pronounced in cells infected with the high titer of HCV virus relative to those infected with the lower titer (Fig. 1A, m.o.i. 0.1 versus m.o.i. 0.01). It has been shown that inhibition of PKD1 activity is associated with the decline in the general secretion capacity of the cell (28). In agreement with the above results, we observed a decline in secretion of secretory horseradish peroxidase (ssHRP-FLAG) from HCV-infected Huh7.5.1 cells compared with the uninfected cells (supplemental Fig. S1). This also holds true for infected and uninfected cells expressing PKD and its mutants. Together, these results suggest that HCV infection leads to impairment of PKD1 activity and in subsequent reduction in the secretory capacity of the host cell.
FIGURE 1.
HCV infection down-regulates PKD1 activation in Huh7.5.1 cells. A, analysis of PKD expression in HCV-infected cells. Huh7.5.1 cells were infected with culture-derived HCV particles by the indicated m.o.i. Six days after infection, whole cell lysates were analyzed by Western blot assay. Phosphorylated serine residues at 738 and 742 (PKC sites) or 910 (autophosphorylation site) were probed using phosphoserine substrate-specific antibody. Both phosphorylations represent the enzymatically active form of PKD1 (p-PKD1). B, quantitation of PKD1 expression and activation. Detected protein band intensities (shown in A) were calculated by 1D Image Analysis Software (Eastman Kodak Co.). PKD protein band intensity was normalized to calnexin. The values are represented as percentages relative to uninfected control (N).
PKD1 Inhibition Enhances HCV Virion Release
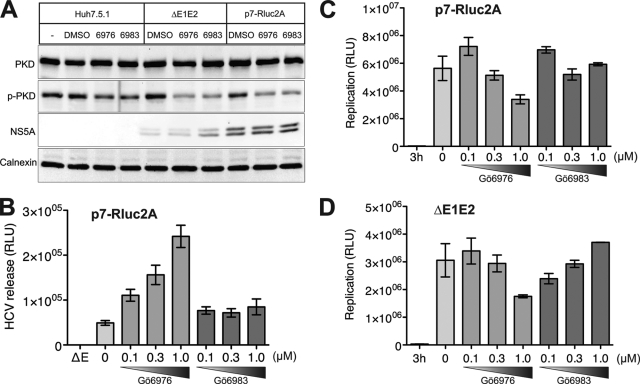
Next we evaluated the functional significance of PKD1 in HCV infection. We used two experimental approaches to analyze the effect of PKD inhibition on suppression of HCV replication and/or virion secretion. The first approach consisted of using a pharmacological inhibitor of PKD. Gö6976 is an ATP-competitive PKC inhibitor that suppresses both PKCα (IC50 = 2.3 nm) and PKCβI (IC50 = 6.2 nm) and also strongly inhibits PKD activity (IC50 = 20 nm) (15), whereas PKC inhibitor Gö6983 inhibits PKCα (IC50 = 7 nm), PKCβ (IC50 = 7 nm), PKCγ (IC50 = 6 nm), PKCδ (IC50 = 10 nm), and PKCζ (IC50 = 60 nm) but not PKD (IC50 = 20 m) (44). Thus, we used Gö6983 and Gö6976 to selectively elucidate the specific effect of PKD inhibition as opposed to the effect observed as a consequence of inhibition of other PKC isoforms. Huh7.5.1 cells were transfected with in vitro transcribed RNA of HCV chimera (p7-Rluc2A) containing Renilla luciferase (Rluc) reporter gene inserted between p7 and NS2 of the monocistronic JC1 RNA genome (33). The transfected cells were cultured in the presence of physiologically effective concentrations of these inhibitors to elucidate their effect on the HCV life cycle. Quantification of luciferase reporter activity in the lysates of the transfected cells was used to assess HCV RNA replication. Whereas the secretion of infectious HCV viral particle was assessed by infecting naïve Huh 7.5.1 cells with culture supernatants collected at 72 h post-transfection, the reporter activity in the infected cells was measured 48 h postinfection. Interestingly, the PKC/PKD inhibitor Gö6976 enhanced HCV secretion in a dose-dependent manner, whereas the PKC inhibitor Gö6983 had no notable effect on HCV secretion (Fig. 2B). The ΔE1E2 mutant, which contains a large in-frame deletion within the coding sequence of E1 and E2 envelope proteins in the p7-Rluc2A genome and is therefore incapable of producing infectious virus particles, was used as a negative control for the HCV secretion assay (Fig. 2B). In addition, we observed a gradual dose-dependent decline in replication of both p7-Rluc2A and its ΔE1E2 counterpart upon treatment with Gö6976 (Fig. 2, C and D). However, with the broad spectrum PKC inhibitor Gö6983, the results on replication of p7-Rluc2A and the ΔE1E2 deletion mutant were rather inconsistent. The intracellular infectivity of Gö6976-treated cells was also less than that of untreated control and Gö6983-treated cells (supplemental Fig. S2A), whereas the extracellular infectivity increased upon Gö7976 treatment (supplemental Fig. S2B). Together, these data suggest that the accelerated secretion of HCV observed upon inhibition of PKD is not a consequence of an enhanced rate of virus maturation/assembly. This implies that under normal physiological conditions PKD negatively regulates HCV secretion.
FIGURE 2.
PKD inhibitor Gö6976 promotes HCV secretion in dose-dependent manner. A, effect of PKD/PKC (Gö6976) and PKC (Gö6983) inhibitors on protein expression as shown by Western blot assays. Huh7.5.1 cells were mock-transfected or transfected with in vitro transcribed p7-Rluc2A RNA as indicated. Transfected cells were treated with or without 1 μm Gö6976 or Gö6983 2 days after transfection and incubated for 3 additional days. Electroblotted membranes were probed with the following antibodies: anti-PKD, anti-phospho-PKD1 (p-PKD; PKC sites), anti-NS5A, and anti-calnexin. B, effect of PKC (Gö6983) and PKC/PKD (Gö6976) inhibitors on HCV virion release. Cultured supernatants were used to infect naïve Huh7.5.1 cells, and cellular lysates were evaluated for Renilla luciferase activity. This protocol detects the HCV virion release. The E1E2 mutant (ΔE), which contains an in-frame deletion within the E1 and E2 coding sequence and hence is incapable of producing infectious virus particles, was used as a negative control in the HCV virion release assay. C, effect of PKC (Gö6983) and PKC/PKD (Gö6976) inhibitors on HCV replication. Intracellular luciferase activities were measured for samples as described in B. This analysis represents intracellular viral RNA replication. D, effect of PKC (Gö6983) and PKC/PKD (Gö6976) inhibitors on HCV replication. p7-Rluc2A containing the E1E2 deletion (ΔE1E2) was used to monitor RNA replication in the presence of inhibitors. This mutant is defective in HCV particle formation. All experiments are carried out in triplicate and the data shown are mean ± S.E. RLU, relative luciferase units.
We also evaluated the effect of these inhibitors on VLDL secretion by Western analysis of apoB and apoE proteins in the culture supernatants. Our observations reveal that treatment with PKD inhibitor Gö6976 modestly decreased apoB and apoE secretion when compared with secretion by untreated control cells (supplemental Fig. S4). VLDL secretion occurs via the Golgi (17, 18). Inhibition of PKD1 activity is associated with a decline in the general secretion capacity of cells occurring via the Golgi compartment (supplemental Fig. S1B) (28). Similarly, VLDL secretion was also inhibited by PKD inhibitor Gö6976 (supplemental Fig. S4).
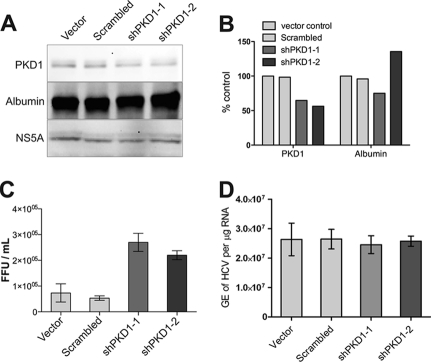
In the second approach, we used shRNA-mediated down-regulation of PKD1 using a DNA vector-based shRNA expression system to analyze its effect on the HCV life cycle. Using two different PKD-specific shRNAs, we could achieve a modest (∼50%) reduction of PKD1 protein expression (Fig. 3, A and B). The PKD1-shRNA vector-transfected Huh7.5.1 cells were electroporated with in vitro transcribed HCV JC1 genome, and HCV virus particle secretion in culture supernatants was assessed by performing the focus-forming unit assay (14, 24). Down-regulation of PKD expression was associated with a concomitant increase in secretion of infectious HCV virus particles (Fig. 3C), although the levels of HCV replication determined by quantitative RT-PCR analysis of intracellular RNA remained unchanged (Fig. 3D). Overall, these data, in concert with the data presented in Fig. 2, support the notion that PKD inhibition or down-regulation leads to an increase in the secretion of infectious HCV virus particles.
FIGURE 3.
PKD1 depletion promotes HCV secretion. Huh7.5.1 cells were transfected with plasmid vectors encoding shRNA as indicated. Four days after transfection, cells were electroporated with in vitro transcribed HCV JC1 RNA and incubated for 3 days. Whole cell lysates were subjected to Western blot assays (A). Panels represent expression of total PKD1, albumin, and HCV NS5A, respectively. B, the level of PKD1 depletion was quantified by 1D Image Analysis Software (Kodak). C, HCV secretion/release was measured by the focus-forming unit (FFU) assay as described under “Experimental Procedures.” D, HCV replication determined by quantitative RT-PCR analysis of HCV genome equivalents (GE)/μg of total intracellular RNA. The data shown are mean ± S.E. of three independent experiments.
Overexpression of Dominant Positive PKD1 Suppresses HCV Secretion
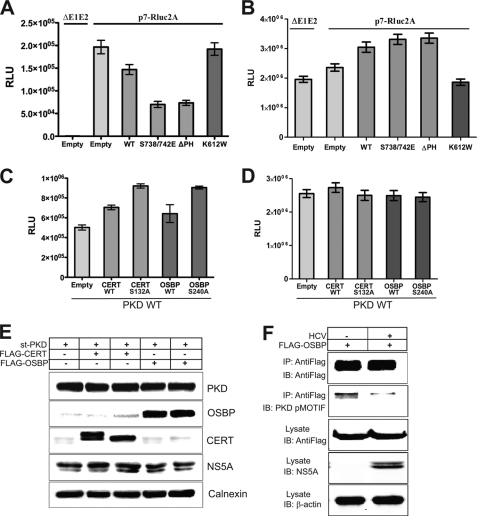
We next analyzed the effect of various functional domain mutants of PKD1 on HCV replication and secretion. We used the lentiviral expression system to overexpress various PKD1 mutants. Transduction of HCV-infected Huh7.5.1 cells with the dominant positive S738/742E or PH domain-lacking ΔPH PKD mutants hindered HCV secretion (Fig. 4A), although PKD wild type or the S738E/742E and ΔPH PKD mutants modestly increased the HCV replication (Fig. 4B). Both S738/742E and ΔPH PKD mutants are kinase-active forms of PKD (28). In contrast, the kinase-inactive/dead (K612W) PKD1 mutant did not enhance HCV secretion (Fig. 4A). The earlier results (Figs. 2B and 3C) suggested that inhibition or down-regulation of PKD1 is associated with enhanced rates of virus secretion, suggesting that PKD kinase activity exerts a negative effect on HCV secretion. The dominant positive and ΔPH mutants of PKD1 are constitutively active kinases, and hence their overexpression negatively impacted HCV secretion, whereas the overexpression of kinase-inactive PKD mutant had no obvious effect on the virus secretion.
FIGURE 4.
PKD regulates HCV secretion via phosphorylation of CERT and OSBP. A, effect of PKD overexpression on HCV secretion. Huh7.5.1 cells were infected with lentiviral vectors encoding PKD1 (WT), dominant active (S738/742E), PH domain deletion (ΔPH) mutant, and kinase-inactive mutant (K612W) as indicated. At 24 h postinfection, cells were electroporated with in vitro transcribed p7-Rluc2a RNA. At 72 h postelectroporation, the culture media were used for the HCV secretion assay. The graph bars represent rates of HCV secretion in a relative manner. B, effect of PKD overexpression on HCV replication. Cellular lysates used in A were used to determine replication by Renilla luciferase activity. C, effect of overexpression of CERT and OSBP wild type and mutants on HCV secretion. Huh7.5.1 cells were co-infected with a lentiviral vector encoding PKD1 wild type as well as those encoding WT or S132A CERT or WT or S240A OSBP, respectively. At 24 h postinfection, cells were electroporated with p7-Rluc2A RNA, and the HCV secretion assay was performed as described above. D, effect of overexpression of CERT and OSBP wild type and mutants on HCV replication. Samples used in C were used to determine replication by Renilla luciferase activity. E, Western blotting of cell lysates from samples in C. The Western analysis shows the expression of PKD1, OSBP, CERT, NS5A, and calnexin in the lysates. F, detection of PKD-specific phosphorylation of OSBP at Ser240. Huh7.5.1 cells were transduced with lentivirus encoding FLAG-tagged OSBP. At 24 h post-transduction, the cells were infected with HCV JC1 virus at an m.o.i. of 0.1., and 4 days later, the cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted (IB) with PKD pMOTIF antibody. The cell lysates were probed for the expression of FLAG-OSBP and HCV NS5A. β-Actin serves as a protein loading control. All the experiments are carried in triplicate and the data shown are mean ± S.E. RLU, relative luciferase units; st-PKD, strep-tagged PKD.
PKD1 Negatively Impacts HCV Secretion through CERT and OSBP Phosphorylation
Among the multiple substrates of PKD1, OSBP and CERT are key players in the Golgi lipid trafficking and biogenesis (21, 32). As we have discussed above, both CERT and OSBP positively contribute to HCV maturation/secretion (24, 25). Recent reports show that PKD phosphorylates OSBP at Ser240 and CERT at Ser132 and inhibits their activity (phosphorylation inhibition) (31, 32). We speculated that PKD1 negatively impacts HCV secretion through phosphorylation inactivation of CERT and OSBP. To evaluate this point, we analyzed the status of PKD-specific OSBP phosphorylation at the Ser240 residue by using PKD pMOTIF antibody. Huh 7.5.1 cells transduced with lentivirus encoding FLAG-tagged OSBP were infected with JC1 virus at an m.o.i. of 0.1. Four days postinfection, cellular lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted using pMOTIF antibody. The results show that PKD-mediated phosphorylation of OSBP in HCV-infected cells was significantly less than that of uninfected control cells (Fig. 4F), thus confirming that HCV infection impairs PKD1 activity. Hence, we assume that the serine mutants of CERT and OSBP that do not serve as PKD substrate would therefore alleviate or subvert the negative regulation of PKD1 on HCV virus particle secretion. To test this hypothesis, we analyzed the effect of coexpressing PKD1 with the respective S240A and S132A serine mutants of OSBP and CERT on HCV secretion. Huh7.5.1 cells were infected with lentivirus encoding wild type PKD1 followed by superinfection with lentivirus encoding the respective serine mutants of OSBP or CERT. The transduced cells were then subjected to electroporation with in vitro transcribed RNA of p7-Rluc2A at 24 h, and infectivity of culture supernatants collected at 72 h postelectroporation was evaluated. Coexpression of the CERT or OSBP serine mutant, respectively, with PKD1 led to a 2-fold increase in the infectious HCV virus particles released in the culture supernatants compared with that of PKD1 expression alone (Fig. 4C) in contrast to the HCV replication levels that remained fairly constant in all cases (Fig. 4D). This modest 2-fold increase is significant considering the presence of endogenous wild type forms of OSBP and CERT. In addition, we observed that the expression of wild type CERT and OSBP also partially increased HCV secretion although not as significantly as the expression of mutant forms (Fig. 4C). Similar results were obtained when wild type or S132A CERT mutant was expressed in the absence of ectopic PKD expression (supplemental Fig. S5). Together, these data support our hypothesis that PKD1 negatively influences HCV secretion through the phosphorylation inhibition of OSBP and CERT proteins that otherwise serve as positive regulators of HCV virus secretion.
HCV Infection Induces Morphological Changes of TGN
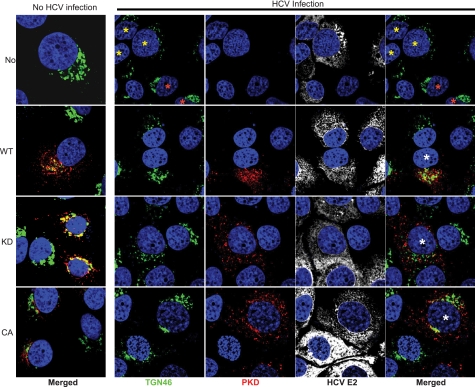
Previous studies showed the functional reliance of HCV on TGN-localizing PI4P-binding proteins such as CERT, OSBP, and PI4KIIIβ (24, 25). These proteins serve as PKD substrates and contribute to the integrity and functionality of TGN in sorting and shipping vesicular cargo to their intended destination. Little is known about the functional role of TGN in HCV morphogenesis. In attempts to gain insight into this aspect, we investigated the morphological changes of TGN in HCV-infected cells and those overexpressing the various mutants of PKD1. The HCV-infected Huh7 cells were transduced with lentiviruses encoding wild type, kinase-inactive (KD), or dominant active (CA) forms of PKD1. At 48 h post-transduction, the cells were processed for immunofluorescence staining of TGN using anti-TGN46 antibody and counterstained with anti-Strep tag (PKD1) and anti-HCV E2 antibody. In HCV-infected Huh7 cells the TGN46 staining pattern was dispersed and displayed a distorted/fragmented pattern of Golgi staining in contrast to the characteristic perinuclear staining pattern observed in uninfected Huh7 cells (Fig. 5, top row, cells are labeled with yellow versus red asterisks). Quantitative analysis revealed that nearly 90% of HCV-infected cells displayed a dispersed or fragmented pattern of the Golgi in contrast to 10% of uninfected cells (supplemental Fig. S3). As reported previously (28), the ectopically expressed WT or dominant active (CA) PKD showed partial localization at the TGN in uninfected Huh7 cells, whereas the kinase-dead/inactive PKD mutant strongly localized to the TGN, which displayed a condensed staining pattern because of a block in vesicle fission (Fig. 5, first column). The ectopic expression of WT or dominant active (CA) PKD in HCV-infected Huh7 cells resulted in partial restoration of the dispersed TGN staining pattern typically observed in HCV-infected cells (Fig. 5, second and fourth rows, see cells marked with white asterisks). In contrast, the expression of the kinase-inactive PKD1 mutant (KD) in HCV-infected cells did not lead to any restoration of TGN organization (Fig. 5, third row, see cells marked with white asterisks). These observations highlight the potential of HCV gene expression in causing the Golgi fragmentation to positively promote secretion reminiscent of the Golgi fragmentation observed during herpesvirus or chlamydia infections (45–47). As described above, the overexpression of dominant active PKD hindered HCV virus secretion, and in agreement with this result, we observed a partial restoration of the Golgi fragmentation in HCV-infected cells expressing dominant active or wild type PKD. Together, these observations suggest that Golgi distortion/fragmentation may aid HCV maturation/secretion.
FIGURE 5.
HCV infection induces morphological changes of TGN. A diffused pattern of the TGN (green) is seen associated with HCV infection. HCV-infected and uninfected Huh7 cells were transduced with respective lentiviral vectors encoding empty ORF (No), PKD WT, PKD K612W (KD), or PKD S738/742E (CA) as indicated on the left. Left column, merged images of the TGN (green) and Strep-tagged PKD (red) in uninfected Huh7 cells. The 4 × 4 panels on the right show images of TGN46 (green), Strep-tagged PKD (red), and HCV envelope (E2) (white) staining as indicated in HCV-infected Huh7 cells. The extreme right column shows the merged view of TGN and PKD. All panels were counterstained for nuclei with DAPI (blue). In the upper row, cells highlighted with yellow and red asterisks represent HCV-infected and uninfected cells, respectively. The cells highlighted with white asterisks in the extreme right column represent HCV-infected cells transduced with respective lentiviral vectors, as indicated.
DISCUSSION
The establishment of productive HCV infection culture system has enabled investigators to pursue inquiries relating to the HCV entry, replication, maturation, and secretion processes of viral life cycle (40, 48, 49). Several studies attempted to delineate cellular pathways leading to release of viral particles. These studies correlated viral export with lipoprotein trafficking and revealed the reliance of HCV viral particle trafficking on the VLDL secretory pathway (13–16, 50). It was shown that silencing apoB and apoE or inhibiting microsomal triglyceride transfer protein activity abrogated HCV secretion. In earlier studies, apoE and apoB were found on the surface of infectious viral particles, suggesting that lipoproteins associate with viral particles to produce lipoviroprotein particles (51–54). These findings raise the possibilities of viral entry via LDL or SR-B1 receptors, and indeed there have been studies that support this view (51, 52, 55, 56). These studies further link the functional significance of the viral maturation/secretion pathway in contributing to subsequent binding/entry events of the viral life cycle (55, 57).
An enveloped viral particle is topologically identical to a cargo vesicle while they are en route to cell surface for egress. Large cargos such as chylomicrons, some viruses, and procollagen reach the TGN by an atypical route. The egress pathway of these cargos from the TGN to the cell surface has not been characterized. PKD isoforms differentially regulate the basolateral transport of proteins from the TGN toward the plasma membrane and also modulate the transport of cargo in polarized cells (26, 47, 58). PKD regulates Golgi lipid homeostasis through the phosphorylation of PI4KIIIβ, CERT, and OSBP substrates. PKD1 phosphorylates PI4KIIIβ to stimulate lipid kinase activity, leading to the production of PI4P at the Golgi. PI4P in turn contributes to the formation of PI4P-rich vesicles to carry cargo proteins. Thus, PI4KIIIβ, which is localized in the Golgi, regulates anterograde transport of cargo proteins destined for the plasma membrane (15, 36). Recent work shows that PKD-mediated phosphorylation of CERT attenuates its function in SM synthesis, and phosphorylation of OSBP causes its dislocalization from the Golgi (32). These activities of PKD comprise a regulatory role in the maintenance of Golgi homeostasis. Both CERT and OSBP play crucial roles in HCV secretion at different levels. A functional role of SM synthesis and Golgi-localized OSBP in HCV secretion has been reported previously (24, 25). Mutations in the OSBP PH domain including its deletion abrogated HCV secretion (24). PKD phosphorylation of CERT and OSBP at respective serine residues collectively cripples their functions. Our studies show decreased HCV secretion upon ectopic overexpression of dominant active PKD mutants (CA and ΔPH), consistent with the role of PKD in negatively regulating OSBP and CERT functions. Hence, mutating the serine residues of CERT and OSBP that serve as PKD substrates should counter this negative influence of PKD. We observed that overexpression of the serine mutants of CERT (S132A) and OSBP (S240A) subverted the negative effect of PKD1 on HCV secretion (Fig. 4C). We further observed that overexpression of both wild type CERT and OSBP also marginally increased HCV secretion (Fig. 4C). Because CERT and OSBP both serve as PKD substrates, transient overexpression of these proteins may overwhelm the basal capacity of PKD-mediated phosphorylation, resulting in the presence of some unphosphorylated active CERT and OSBP proteins that can eventually enhance HCV secretion. In addition, inhibition of PKD activity by the PKD-specific inhibitor Gö6976 enhanced the rate of viral release without any effect on intracellular RNA replication, thus indicating that this inhibition is exerted at post-translation/replication steps of the viral life cycle (Fig. 2, B and C). Based on a body of literature on PKD, OSBP, and CERT in Golgi trafficking, a recent model was proposed by Toker and co-workers (32) that integrates the functions of these key players in Golgi trafficking. We extrapolate these observations in the context of HCV infection and accordingly propose the following model (Fig. 6). In this model, HCV gene expression causes down-regulation of PKD activation (autophosphorylation). Mechanisms of this down-regulation are not known. PKD phosphorylation of OSBP disables its association with TGN. OSBP phosphorylation may lead to an inactive state in which the interaction between its PH domain and PI4P is inhibited that will abrogate further functions including its interactions with CERT. Similarly, PKD phosphorylation of CERT decreases the interaction of its PH domain with PI4P, which will lead to decreased transport of ceramide and hence attenuation of SM synthesis. In either case, the phosphorylation of OSBP and CERT leads to inactivation of these proteins, leading to abrogation of their principal functions that ultimately manifest in the overall inhibition of the HCV secretion process in the TGN. A recent report demonstrates that HCV particles are relatively enriched in SM, cholesterol, and cholesterol ester (59).
FIGURE 6.
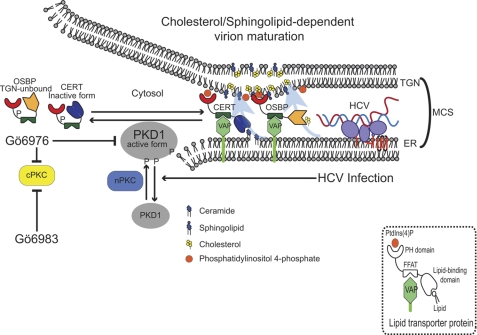
Model depicting role of protein kinase D in HCV secretion. PKD1 regulates HCV secretion by regulating enzymatic activities of CERT and OSBP thorough phosphorylation inhibition. OSBP and CERT form a membrane contact site between the ER and the TGN by binding to both VAP-A and PI4P. CERT protein transports ceramide from the ER to the TGN where transported ceramides are converted to sphingolipids. OSBP transports cholesterols and oxysterols to the TGN. These transported lipids contribute to the formation of microdomains enriched with cholesterols and sphingolipids. HCV particles have been shown to be sensitive to sphingomyelinase and methyl-β-cyclodextrin treatments (cholesterol depletion) (25). CERT inhibitor (HPA-12) attenuates HCV secretion. OSBP facilitates HCV secretion (24). PKD1 phosphorylates OSBP to sequester it from the TGN. PKD1 phosphorylates CERT to inactivate its enzyme activity. In this study, PKD1-specific siRNA enhanced HCV secretion. PKD1 inhibition by the PKD-specific inhibitor Gö6976 also promoted HCV secretion. Overexpression of a dominant active form of PKD1 suppressed HCV secretion. Coexpression of OSBP S240A or CERT S132A mutant subverted this suppression. PKD1 negatively regulates the HCV maturation/secretion pathway through the phosphorylation of CERT and OSBP. To circumvent these effects, HCV down-regulates PKD activation (Fig. 1). nPKC, novel PKC; cPKC, conventional PKC; PtdIns(4)P, phosphatidylinositol 4-phosphate; VAP-A, vesicle-associated membrane protein-associated protein (VAP) subtype A; FFAT, diphenylalanine (FF) in an acidic tract.
Inhibition of PKD has a negative effect on the general secretion of protein occurring through the Golgi compartment (supplemental Fig. S1). PKD inhibition led to a modest decrease in the VLDL secretion (supplemental Fig. S4). This observation suggests that PKD affects VLDL secretion like other secretory proteins trafficking through the Golgi network. This study, however, revealed that PKD inhibition has a positive effect on HCV secretion via CERT and OSBP phosphorylation. Several studies suggest that HCV secretion parallels VLDL secretion (13–16, 50). At this point, it is not known what fraction of VLDL is recruited for HCV secretion and at what stage of its transport does HCV piggybacks or associate with lipoprotein particles.
HCV gene expression appears to cause a general distortion of the Golgi compartment (Fig. 5, first row, compare cells with yellow versus red asterisks). Based on this observation, we conclude that this altered TGN pattern imparts a positive influence on the HCV maturation process reminiscent of Golgi fragmentation aiding herpesvirus morphogenesis or chlamydia infection (45–47). These changes modestly affected general secretion of proteins in HCV-infected cells (supplemental Fig. S1).
Several recent reports have highlighted the negative regulation of PKD in cell migration and motility. PKD is considered a central regulator of the cofilin signaling network via direct phosphorylation of slingshot I-like protein (60). PKD also phosphorylates cortactin at Ser298, which leads to its reduced F-actin binding (61). So active PKD inhibits the functions of slingshot I-like and cortactin, and kinase-inactive forms strongly enhance motility and invasiveness mediated by these proteins. The unrecognized function of PKD as a regulator of polarized cell motility and F-actin polymerization reinforces its mode of action via phosphorylation inhibition of these functions. Therefore, in keeping with this mode of action, PKD negatively regulates HCV secretion and release. Understanding the various steps of HCV morphogenesis will open newer avenues for the design of antiviral strategies.
Supplementary Material
Acknowledgments
We are grateful to Dr. T. Wakita (National Institute of Infectious Diseases, Tokyo, Japan) for kindly providing the infectious JFH1 molecular clone, Charles Rice (Rockefeller University, New York, NY) for p7-Rluc2A and monoclonal anti-NS5a antibody, Frank Chisari (The Scripps Research Institute, La Jolla, CA) for Huh7.5.1 cells and JC1, Dennis Burton (The Scripps Research Institute) for recombinant human monoclonal anti-E2 antibody, and Vivek Malhotra (Center for Genomic Regulation, Barcelona, Spain) for SS HRP-FLAG plasmid, and Alex Toker (Harvard Medical School) for pMOTIF antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants DK077704 and AI085087 (to A. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- HCV
- hepatitis C virus
- PKD
- protein kinase D
- CERT
- ceramide transfer protein
- OSBP
- oxysterol-binding protein
- TGN
- trans-Golgi network
- ER
- endoplasmic reticulum
- PI4P
- phosphatidylinositol 4-phosphate
- MCS
- membrane contact site
- PI4KIIIβ
- phosphatidylinositol 4-kinase IIIβ
- PH
- pleckstrin homology
- SM
- sphingomyelin
- VSV-G
- vesicular stomatitis virus G
- ssHRP
- signal sequence-tagged HRP
- CA
- constitutively active
- KD
- kinase-dead
- m.o.i.
- multiplicity of infection
- Rluc
- Renilla luciferase.
REFERENCES
- 1. Di Bisceglie A. M. (2002) J. Vasc. Interv. Radiol. 13, S169–171 [DOI] [PubMed] [Google Scholar]
- 2. Pawlotsky J. M. (2004) Trends Microbiol. 12, 96–102 [DOI] [PubMed] [Google Scholar]
- 3. Negro F., Sanyal A. J. (2009) Liver Int. 29, Suppl. 2, 26–37 [DOI] [PubMed] [Google Scholar]
- 4. Tellinghuisen T. L., Evans M. J., von Hahn T., You S., Rice C. M. (2007) J. Virol. 81, 8853–8867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Appel N., Schaller T., Penin F., Bartenschlager R. (2006) J. Biol. Chem. 281, 9833–9836 [DOI] [PubMed] [Google Scholar]
- 6. Penin F., Dubuisson J., Rey F. A., Moradpour D., Pawlotsky J. M. (2004) Hepatology 39, 5–19 [DOI] [PubMed] [Google Scholar]
- 7. Gosert R., Egger D., Lohmann V., Bartenschlager R., Blum H. E., Bienz K., Moradpour D. (2003) J. Virol. 77, 5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dubuisson J., Penin F., Moradpour D. (2002) Trends Cell Biol. 12, 517–523 [DOI] [PubMed] [Google Scholar]
- 9. Syed G. H., Amako Y., Siddiqui A. (2010) Trends Endocrinol. Metab. 21, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boulant S., Douglas M. W., Moody L., Budkowska A., Targett-Adams P., McLauchlan J. (2008) Traffic 9, 1268–1282 [DOI] [PubMed] [Google Scholar]
- 11. Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. (2007) Nat. Cell Biol. 9, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 12. Jones D. M., McLauchlan J. (2010) J. Biol. Chem. 285, 22733–22739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang K. S., Jiang J., Cai Z., Luo G. (2007) J. Virol. 81, 13783–13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gastaminza P., Cheng G., Wieland S., Zhong J., Liao W., Chisari F. V. (2008) J. Virol. 82, 2120–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang J., Luo G. (2009) J. Virol. 83, 12680–12691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ye J. (2007) PLoS Pathog. 3, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gusarova V., Seo J., Sullivan M. L., Watkins S. C., Brodsky J. L., Fisher E. A. (2007) J. Biol. Chem. 282, 19453–19462 [DOI] [PubMed] [Google Scholar]
- 18. Siddiqi S. A. (2008) Biochem. J. 413, 333–342 [DOI] [PubMed] [Google Scholar]
- 19. Ridgway N. D., Dawson P. A., Ho Y. K., Brown M. S., Goldstein J. L. (1992) J. Cell Biol. 116, 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang P. Y., Weng J., Anderson R. G. (2005) Science 307, 1472–1476 [DOI] [PubMed] [Google Scholar]
- 21. Peretti D., Dahan N., Shimoni E., Hirschberg K., Lev S. (2008) Mol. Biol. Cell 19, 3871–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumagai K., Yasuda S., Okemoto K., Nishijima M., Kobayashi S., Hanada K. (2005) J. Biol. Chem. 280, 6488–6495 [DOI] [PubMed] [Google Scholar]
- 23. Perry R. J., Ridgway N. D. (2005) Biochim. Biophys. Acta 1734, 220–234 [DOI] [PubMed] [Google Scholar]
- 24. Amako Y., Sarkeshik A., Hotta H., Yates J., 3rd, Siddiqui A. (2009) J. Virol. 83, 9237–9246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aizaki H., Morikawa K., Fukasawa M., Hara H., Inoue Y., Tani H., Saito K., Nishijima M., Hanada K., Matsuura Y., Lai M. M., Miyamura T., Wakita T., Suzuki T. (2008) J. Virol. 82, 5715–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Van Lint J., Rykx A., Maeda Y., Vantus T., Sturany S., Malhotra V., Vandenheede J. R., Seufferlein T. (2002) Trends Cell Biol. 12, 193–200 [DOI] [PubMed] [Google Scholar]
- 27. Baron C. L., Malhotra V. (2002) Science 295, 325–328 [DOI] [PubMed] [Google Scholar]
- 28. Bossard C., Bresson D., Polishchuk R. S., Malhotra V. (2007) J. Cell Biol. 179, 1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maeda Y., Beznoussenko G. V., Van Lint J., Mironov A. A., Malhotra V. (2001) EMBO J. 20, 5982–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Díaz Añel A. M., Malhotra V. (2005) J. Cell Biol. 169, 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fugmann T., Hausser A., Schöffler P., Schmid S., Pfizenmaier K., Olayioye M. A. (2007) J. Cell Biol. 178, 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nhek S., Ngo M., Yang X., Ng M. M., Field S. J., Asara J. M., Ridgway N. D., Toker A. (2010) Mol. Biol. Cell 21, 2327–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jones C. T., Murray C. L., Eastman D. K., Tassello J., Rice C. M. (2007) J. Virol. 81, 8374–8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brummelkamp T. R., Bernards R., Agami R. (2002) Science 296, 550–553 [DOI] [PubMed] [Google Scholar]
- 35. Storz P., Toker A. (2003) EMBO J. 22, 109–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hausser A., Storz P., Märtens S., Link G., Toker A., Pfizenmaier K. (2005) Nat. Cell Biol. 7, 880–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raya A., Revert F., Navarro S., Saus J. (1999) J. Biol. Chem. 274, 12642–12649 [DOI] [PubMed] [Google Scholar]
- 38. Döppler H., Storz P., Li J., Comb M. J., Toker A. (2005) J. Biol. Chem. 280, 15013–15019 [DOI] [PubMed] [Google Scholar]
- 39. van den Hoff M. J., Moorman A. F., Lamers W. H. (1992) Nucleic Acids Res. 20, 2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D. R., Wieland S. F., Uprichard S. L., Wakita T., Chisari F. V. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Waldron R. T., Rozengurt E. (2003) J. Biol. Chem. 278, 154–163 [DOI] [PubMed] [Google Scholar]
- 42. Matthews S. A., Rozengurt E., Cantrell D. (1999) J. Biol. Chem. 274, 26543–26549 [DOI] [PubMed] [Google Scholar]
- 43. Rybin V. O., Guo J., Steinberg S. F. (2009) J. Biol. Chem. 284, 2332–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gschwendt M., Dieterich S., Rennecke J., Kittstein W., Mueller H. J., Johannes F. J. (1996) FEBS Lett. 392, 77–80 [DOI] [PubMed] [Google Scholar]
- 45. Heuer D., Rejman Lipinski A., Machuy N., Karlas A., Wehrens A., Siedler F., Brinkmann V., Meyer T. F. (2009) Nature 457, 731–735 [DOI] [PubMed] [Google Scholar]
- 46. Avitabile E., Ward P. L., Di Lazzaro C., Torrisi M. R., Roizman B., Campadelli-Fiume G. (1994) J. Virol. 68, 7397–7405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rémillard-Labrosse G., Mihai C., Duron J., Guay G., Lippé R. (2009) Traffic 10, 1074–1083 [DOI] [PubMed] [Google Scholar]
- 48. Lindenbach B. D., Evans M. J., Syder A. J., Wölk B., Tellinghuisen T. L., Liu C. C., Maruyama T., Hynes R. O., Burton D. R., McKeating J. A., Rice C. M. (2005) Science 309, 623–626 [DOI] [PubMed] [Google Scholar]
- 49. Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Kräusslich H. G., Mizokami M., Bartenschlager R., Liang T. J. (2005) Nat. Med. 11, 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang H., Sun F., Owen D. M., Li W., Chen Y., Gale M., Jr., Ye J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5848–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Agnello V., Abel G., Elfahal M., Knight G. B., Zhang Q. X. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12766–12771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. André P., Komurian-Pradel F., Deforges S., Perret M., Berland J. L., Sodoyer M., Pol S., Bréchot C., Paranhos-Baccalà G., Lotteau V. (2002) J. Virol. 76, 6919–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nielsen S. U., Bassendine M. F., Burt A. D., Martin C., Pumeechockchai W., Toms G. L. (2006) J. Virol. 80, 2418–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nielsen S. U., Bassendine M. F., Martin C., Lowther D., Purcell P. J., King B. J., Neely D., Toms G. L. (2008) J. Gen. Virol. 89, 2507–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Burlone M. E., Budkowska A. (2009) J. Gen. Virol. 90, 1055–1070 [DOI] [PubMed] [Google Scholar]
- 56. Molina S., Castet V., Fournier-Wirth C., Pichard-Garcia L., Avner R., Harats D., Roitelman J., Barbaras R., Graber P., Ghersa P., Smolarsky M., Funaro A., Malavasi F., Larrey D., Coste J., Fabre J. M., Sa-Cunha A., Maurel P. (2007) J. Hepatol. 46, 411–419 [DOI] [PubMed] [Google Scholar]
- 57. Owen D. M., Huang H., Ye J., Gale M., Jr. (2009) Virology 394, 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rémillard-Labrosse G., Lippé R. (2009) Commun. Integr. Biol. 2, 434–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Merz A., Long G., Hiet M. S., Brügger B., Chlanda P., Andre P., Wieland F., Krijnse-Locker J., Bartenschlager R. (2011) J. Biol. Chem. 286, 3018–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Eiseler T., Döppler H., Yan I. K., Kitatani K., Mizuno K., Storz P. (2009) Nat. Cell Biol. 11, 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Eiseler T., Hausser A., De Kimpe L., Van Lint J., Pfizenmaier K. (2010) J. Biol. Chem. 285, 18672–18683 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.