Abstract
Hepatitis C Virus (HCV) nonstructural 5A (NS5A) is a pleiotropic protein involved in viral RNA replication and modulation of the cellular physiology in HCV-infected cells. To elucidate the mechanisms of the HCV life cycle, we identified cellular factors interacting with the NS5A protein in HCV-infected cells. Huh7.5 cells were electroporated with HCV Jc1 RNA. Cellular factors associated with HCV NS5A were identified by immunoprecipitation with Dynabead-conjugated NS5A antibody and LC-MS/MS. Phosphatidylinositol 4-kinase type IIIα (PI4KIIIα) was identified as a binding partner for the NS5A protein. NS5A derived from both genotypes 1b and 2a interacted with PI4KIIIα. NS5A interacted with PI4KIIIα through amino acids 401–600 of PI4KIIIα and domain I of NS5A. Interference of the protein interaction between NS5A and PI4KIIIα decreased HCV propagation. Knockdown of PI4KIIIα significantly reduced HCV replication in Huh7 cells harboring the subgenomic replicon and in Huh7.5 cells infected with cell culture grown virus (HCVcc). Silencing of PI4KIIIα further inhibited HCV release into the tissue culture medium. NS5A may recruit PI4KIIIα to the HCV RNA replication complex. These data suggest that PI4KIIIα is an essential host factor that supports HCV proliferation and therefore PI4KIIIα may be a legitimate target for anti-HCV therapy.
Keywords: Hepatitis Virus, Lipid, RNA Silencing, RNA Viruses, Viral Protein, Viral Replication, Virus
Introduction
Hepatitis C virus (HCV)2 is a single-stranded positive-sense RNA virus capable of establishing a chronic infection in 70–80% of HCV-infected patients. Approximately 3% of the population of the world are chronically infected with HCV. Over 30% of them will develop cirrhosis within 20 years, which subsequently may lead to hepatocellular carcinoma (1–3). A vaccine is not available and the optimal current therapy for patients infected with HCV is a combination of the pegylated interferon (IFN) and ribavirin. However, the therapy with these agents is prolonged, costly, and associated with a high rate of side effects. Furthermore, the sustained virologic response rate to the combination therapy varies with different genotypes of HCV. Although small molecule inhibitors of viral protease and RNA-dependent RNA polymerase are in clinical development, the error prone nature of the viral RNA polymerase leads to rapid emergence of viral-resistant mutations to these therapeutic candidates. Therefore, a new approach to control HCV proliferation could be identifying the host factors involved in the HCV life cycle. HCV relies heavily on host proteins for all steps of its life cycle, including viral entry, uncoating, replication, assembly, and virion release. Therefore, any step that can interrupt the HCV life cycle will be a putative target for HCV therapy.
Nonstructural 5A (NS5A) protein is a multifunctional phosphoprotein consisting of 447 amino acids residues. NS5A protein is localized in the cytoplasm and forms part of the HCV RNA replication complex (4). NS5A has been shown to interact with many host factors, including TRAF2, hVAP-33, PKR, and Grb2, to regulate viral replication and cellular signaling pathways (5–8). A growing body of evidence indicates that NS5A may play important roles in the HCV-induced liver pathogenesis. We have shown that NS5A modulated TNFα signaling of the host cells through the NS5A-TRAF2 interaction (5). In addition, we demonstrated that NS5A induced steatosis and hepatocellular carcinoma in transgenic mice (9). Recently, we also showed that NS5A modulated β-catenin signaling that might play an important role in HCV pathogenesis (10).
Phosphatidylinositol 4-kinase type IIIα (PI4KIIIα) is a lipid kinase that is encoded by the PI4KCA gene in human (11). PI4KIIIα phosphorylates phosphatidylinositol (PtdIns) to phosphatidylinositol 4-P, which can be further phosphorylated by PIP5 kinases to phosphatidylinositol (4,5)-P2. PI4KIIIα is localized primarily to the endoplasmic reticulum and regulates endoplasmic reticulum exit sites (12, 13). Recently, PI4KIIIα has been identified as a cellular factor involved in the HCV life cycle using siRNA library screening (14–19). However, how PI4KIIIα regulates HCV proliferation is not clearly understood. In this study, we identified PI4KIIIα as a binding partner for the NS5A protein. Silencing of PI4KIIIα significantly reduced HCV replication and virion release in HCV-infected cells. These data suggest that HCV may modulate cellular PI4KIIIα for its own RNA replication and virion production in the HCV life cycle.
EXPERIMENTAL PROCEDURES
Plasmid Construction
cDNA corresponding to the NS5A coding sequence of HCV was amplified by PCR using the Korean isolate of HCV (genotype 1b) and subcloned into the pEF6 vector (Invitrogen). Full-length PI4KIIIα was amplified by PCR using cDNAs prepared from Huh7.5 cells and cloned into pFLAG-CMV2 or p3XFLAG-CMV (Sigma). PI4KIIIα mutants were also constructed using full-length PI4KIIIα as a template. Plasmid pFK-Jc1 was kindly provided by Dr. Ralf Bartenschlager (University of Heidelberg).
Cell Culture and DNA Transfection
All cell lines were grown in Dulbecco's modified Eagles' medium (DMEM) supplemented with 10% fetal calf serum, and 1% penicillin/streptomycin in 5% CO2 at 37 °C. Huh7 cells harboring subgenomic replicon and IFN-cured cells were described previously (20). For the transfection experiment, ∼5 × 105 cells plated on 60-mm dishes were transfected with plasmid DNA by using polyethyleneimine (Sigma) as described previously (10).
Preparation of Infectious Virus
The infectious cell culture grown virus (HCVcc) was generated as described previously (21). Briefly, the monolayer of Huh7.5 cells was washed twice in phosphate-buffered saline (PBS), trypsinized, and resuspended at a concentration of 6 × 106 cells/ml in Opti-MEM (Invitrogen). After centrifugation at 1,000 × g for 5 min, the cells were resuspended in 400 μl of cytomix solution containing 2 mm ATP and 5 mm glutathione, then mixed with 10 μg of Jc1 viral RNA, and electroporated using a Gene Pulser Xcell (Bio-Rad) in a 4-mm gap cuvette. Cell culture supernatants were collected 4 days after electroporation.
Identification of Host Proteins
Cross-linking of NS5A antibodies with Protein A Dynabeads (Dynal) was performed according to the manufacturer's instructions using dimethyl pimelimidate dihydrochloride (Pierce). Whole cell extracts were incubated with NS5A antibodies cross-linked to Dynabead for 2 h at 4 °C on a rotator. After being washed three times with a lysis buffer (50 mm HEPES, pH 7.6, 150 mm NaCl, 5 mm EDTA, 0.2% Nonidet P-40, and 1 mm pheylmethylsulfonyl fluoride), the beads were resuspended in a sample buffer and heated for 5 min. The proteins were separated on 8% SDS-PAGE and visualized by silver staining. The interested protein bands were excised and analyzed by LC/MS/MS. The individual spectra from MS/MS were processed using TurboSEQUEST software (Thermo Quest, San Jose, CA). The generated peak list files were used to query either the MSDB data base or NCBI using the MASCOT program.
Immunoblot Analysis
Cells were lysed in 400 μl of cell lysis buffer A containing 50 mm HEPES, pH 7.6, 150 mm NaCl, 5 mm EDTA, 0.2% Nonidet P-40, and 1 mm pheylmethylsulfonyl fluoride. The cell lysates were further centrifuged at 15,000 × g for 10 min at 4 °C and the pellets were discarded. The protein concentration was determined by the Bradford assay (Bio-Rad). Equal amounts of proteins were immunoblotted with the indicated antibodies. Proteins were detected using an ECL kit (Amersham Biosciences). Rabbit anti-PI4KIIIα antibody was purchased from Cell Signaling Technology (Beverly, MA).
Immunoprecipitation
Cells were harvested and lysed in cell lysis buffer A. The cell lysates were triturated by 10 passes through a 25-gauge needle on ice and centrifuged at 15,000 × g for 10 min. The supernatant was incubated at 4 °C for 2 h with anti-NS5A antibody, anti-PI4KIIIα antibody, anti-FLAG monoclonal antibody (Sigma), and anti-Myc monoclonal antibody (Santa Cruz), respectively. The samples were further incubated with 30 μl of protein A beads (Zymed Laboratories Inc.) for 1 h. The beads were washed five times with cell lysis buffer A, and the bound proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and then detected by immunoblot analysis using corresponding secondary antibodies.
Confocal Microscopy
Huh7.5 cells grown on coverglass were infected with HCVcc. At 2 days postinfection, cells were transfected with plasmid expressing FLAG-tagged PI4KIIIα. At 36 h after transfection, cells were washed in PBS and fixed in 4% paraformaldehyde for 20 min at 37 °C. Cells were washed three times in PBS and incubated in PBS containing 0.1% Triton X-100 (Sigma) and 0.5% bovine serum albumin for 1 h at room temperature. Cells were then incubated with the primary antibodies (rabbit anti-NS5A antibody, mouse anti-FLAG antibody (Sigma), mouse anti-dsRNA antibody (J2, English and Scientific Consulting, respectively) for 2 h and further incubated with the secondary antibodies (FITC-conjugated goat anti-rabbit IgG, FITC-conjugated goat anti-mouse IgG, TRITC-conjugated donkey anti-rabbit IgG, TRITC-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch, Inc.) for 1 h at room temperature. After two washes with 0.1% Triton X-100 in PBS and three washes in PBS, cells were analyzed using a Zeiss LSM700 laser confocal microscopy system.
Gene Silencing by siRNA
siRNAs targeting the PI4KIIIα, the HCV 5′-UTR (positive control), and the universal negative control, were purchased from Dharmacon (Lafayette, CO). siRNA transfection was performed using a Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions.
Quantification of HCV RNA
RNA isolation from HCVcc-infected cells, cell culture medium, or replicon cells was performed using TRIzol® LS Reagent (Invitrogen) according to the manufacturer's instructions. Purified viral RNA was used as a template to synthesize cDNA using the iScript cDNA synthesis kit (Bio-Rad) for 2 h at 42 °C. cDNA was amplified with HCV genotype 2a-specific primers (forward, AGA GCC ATA GTG GTC TGC GGA AC; reverse, CCT TTC GCA ACC CAA CGC TAC TC), HCV genotype 1b-specific primers (forward, ATC ACT CCC CTG TGA GGA ACT ACT G; reverse, CTG GAG GCT GCA CGA CAC TC), and actin-specific primers (forward, TGG ACT TCG AGC AAG AGA TGG; reverse, GGA AGG AAG GCT GGA AGA GTG). Quantification of HCV RNA was normalized with β-actin RNA as an internal control. All quantitative real time PCR (qRT-PCR) experiments were done using an iQ5 multicolor real time PCR Detection system and built-in iQ5 optical systems software (Bio-Rad).
RESULTS
Identification of Cellular Proteins Interacting with HCV NS5A
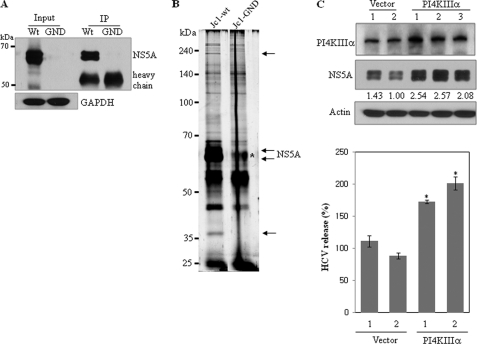
To identify cellular proteins that interacted with the HCV NS5A protein, Huh7.5 cells were electroporated with HCV Jc1 RNA and cell lysates harvested 4 days after electroporation were immunoprecipitated with Dynabead cross-linked NS5A antibody. As a control, Huh7.5 cells were electroporated with HCV Jc1-GND RNA containing a replication-defective NS5B. Prior to detecting coimmunoprecipitated proteins, the specificity of NS5A antibody was confirmed by immunoprecipitation. We first confirmed the NS5A expression in Jc1 RNA-transfected cells and then verified the immunoprecipitation of Huh7.5 cell lysates with NS5A antibody (Fig. 1A). As shown in Fig. 1B, two cellular proteins were coprecipitated with NS5A protein in HCV RNA replicating cells but not in Jc1-GND cells. By LC/MS/MS analysis, the top band corresponding to the ∼250-kDa protein was identified as the PI4KIIIα. The bottom band corresponding to ∼40 kDa was identified as the hydroxyacid oxidase 2. To investigate the functional role of PI4KIIIα in HCV replication, we generated stable cells expressing either vector or PI4KIIIα. Huh7.5 cells stably expressing either p3XFLAG vector or p3XFLAG-PI4KIIIα were infected with HCVcc. Cell lysates harvested at 2 days after HCV infection were immunoblotted with the indicated antibodies. As demonstrated in Fig. 1C (top panels), the NS5A expression level was increased in all three PI4KIIIα stable cell lines. We confirmed that the HCV NS3 protein level was also elevated in PI4KIIIα stable cells (data not shown). To investigate the effect of PI4KIIIα on HCV release, qPCR was performed. As shown Fig. 1C (bottom panel), HCV release was significantly increased in PI4KIIIα stable cells as compared with vector stable cells. We further found that HCV release was also increased in the third clone of PI4KIIIα stable cells, confirming that this phenomenon was not caused by clonal selection (data not shown).
FIGURE 1.
Identification of the cellular binding partner for the HCV NS5A protein by immunoprecipitation. A, protein expression of NS5A and the specificity of NS5A antibody were confirmed by immunoprecipitation of Huh7.5 cells electroporated with either wild-type or replication defective Jc1 RNAs. GAPDH was used as a loading control. IP, immunoprecipitation; WT, wild-type; GND, replication defective mutant. B, Huh7.5 cells were transfected with either wild-type or replication defective Jc1 RNAs via electroporation. Cell lysates harvested at 4 days post-electroporation were immunoprecipitated with Dynabead-conjugated NS5A antibody. The immunoprecipitants were analyzed on 8% SDS-PAGE and silver stained. Interested bands were subjected to LC-MS/MS analysis. Asterisk denotes a nonspecific band. C, Huh7.5 cells stably expressing either p3XFLAG vector or p3XFLAG-PI4KIIIα were infected with HCVcc. Cells were harvested at day 2 postinfection and total cell lysates were immunoblotted with the indicated antibodies (top panel). HCV RNA release in vector stable and PI4KIIIα stable cells was determined by qPCR (bottom panel). *, p < 0.05, vector stable versus PI4KIIIα stable cells infected with HCVcc.
HCV NS5A Protein Interacts with PI4KIIIα
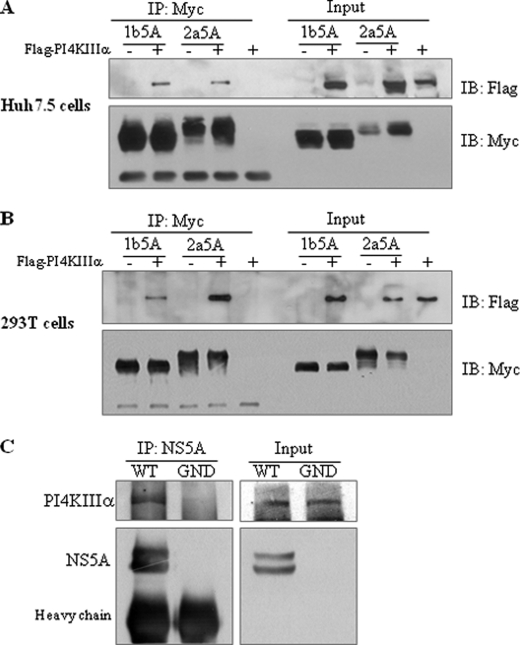
To investigate how the PI4KIIIα protein was involved in HCV replication, we first examined the possible protein interaction between NS5A and PI4KIIIα by employing a coimmunoprecipitation assay using a transient expression system. Huh7.5 and HEK293T cells were either transfected with plasmid expressing NS5A protein of HCV genotype 1b or genotype 2a alone or cotransfected with plasmid expressing PI4KIIIα. As shown in Fig. 2, A and B, NS5A derived from both genotypes 1b and 2a interacted with PI4KIIIα. To further confirm this result in the context of HCV replication, Huh7.5 cells infected with HCVcc were immunoprecipitated with NS5A antibody and then bound protein was immunoblotted with PI4KIIIα antibody. Fig. 2C showed that NS5A specifically interacted with PI4KIIIα in wild-type HCV-infected cells but not in mock-infected (GND) cells.
FIGURE 2.
NS5A interacts with PI4KIIIα. Either Huh7.5 (A) or HEK293T (B) cells were transfected with Myc-tagged NS5A of HCV genotypes 1b or 2a in the absence or presence of FLAG-tagged PI4KIIIα. Total cell lysates were immunoprecipitated (IP) with anti-Myc antibody, and bound proteins were immunoblotted (IB) with anti-FLAG antibody. Protein expressions of NS5A were confirmed using the same cell lysates by immunoblotting with anti-Myc antibody. C, Huh7.5 cells were infected with either wild-type or replication defective HCV and cell lysates harvested at 3 days after infection were immunoprecipitated with NS5A antibody. Bound proteins were immunoblotted with anti-PI4KIIIα antibody. Protein expressions of NS5A and PI4KIIIα were confirmed using the same cell lysates by immunoblotting with anti-NS5A and anti-PI4KIIIα antibody.
NS5A Interacts with PI4KIIIα through aa 401–600 Residues of PI4KIIIα and the Domain I of NS5A
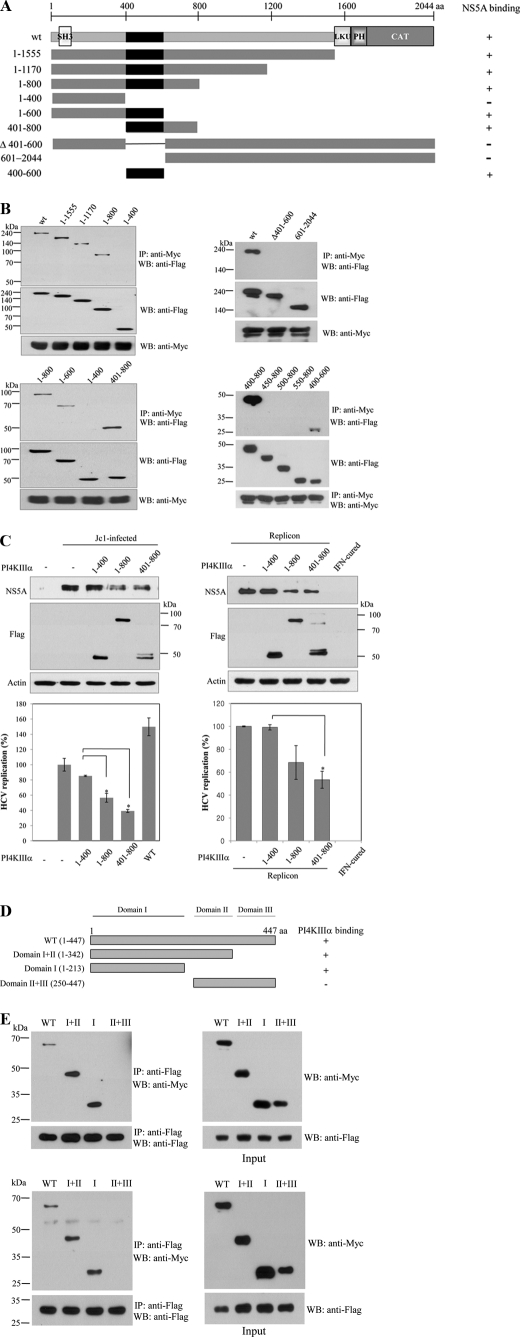
To determine the region in PI4KIIIα responsible for NS5A binding, the interaction of NS5A with various deletion mutants of PI4KIIIα (Fig. 3A) was determined by a transfection-based coprecipitation assay. As shown in Fig. 3B (left top panel), NS5A interacted with a mutant encompassing aa 1–800 but not with aa 1–400 mutant of PI4KIIIα. To further narrow down the binding site for NS5A, we constructed more mutants and demonstrated that NS5A interacted with aa 401–600 residues of PI4KIIIα (Fig. 3B, left bottom and right top panels). Using serial deletion of 50 aa in each mutant construct, we precisely determined that aa 400–450 of PI4KIIIα was the most important residues responsible for binding with NS5A (Fig. 3B, right bottom panels). To examine functional implication of protein interaction between NS5A and PI4KIIIα, Huh7.5 cells were transfected with the mutant harboring either aa 1–800 or 401–800 of PI4KIIIα and then infected with HCVcc. A mutant harboring aa 1–400 of PI4KIIIα was used as a negative control. As shown in Fig. 3C (left top panel), NS5A protein expression levels were significantly decreased by both aa 1–800 and 401–800 of PI4KIIIα as compared with the aa 1–400 of PI4KIIIα. This was further confirmed by the significant decrease of HCV RNA levels in cells transfected with either aa 1–800 or 401–800 of PI4KIIIα as demonstrated by qPCR (Fig. 3C, left bottom panel). This was further confirmed in HCV replicon cells as demonstrated in Fig. 3C (right panels). These results indicated that protein interaction between NS5A and PI4KIIIα contributed to HCV replication because competitive interference by mutants harboring the binding domain of PI4KIIIα resulted in a significant decrease of viral RNA and protein expression levels. Next, we determined the region in NS5A responsible for PI4KIIIα binding. We constructed various domain mutants of NS5A as previously reported (22) (Fig. 3D) and the binding domain was determined as described above. Fig. 3E (upper panel) showed that PI4KIIIα interacted with a mutant harboring domain I and a mutant harboring domains I and II but not with a mutant harboring domains II and III, indicating that PI4KIIIα interacted with NS5A through domain I of NS5A. Finally, we investigated whether the minimal PI4KIIIα (aa 400–600) could interact with the 1–213 (domain I) mutant of NS5A. Indeed, the 1–213 mutant of NS5A specifically interacted with aa 400–600 of PI4KIIIα, indicating that minimal sequences of both proteins were sufficient for protein interplay between PI4KIIIα and NS5A (Fig. 3E, bottom panel).
FIGURE 3.
NS5A interacts with PI4KIIIα through aa 401–600 of PI4KIIIα and domain I of NS5A. A, schematic diagram of both wild-type and mutant constructs of PI4KIIIα protein. LKU, lipid kinase unique domain; PH, pleckstrin homology; CAT, catalytic domain. B, HEK293T cells were cotransfected with Myc-tagged NS5A and FLAG-tagged mutant constructs of PI4KIIIα. Total cell lysates harvested at 24 h after transfection were immunoprecipitated (IP) with anti-Myc antibody and bound proteins were immunoblotted with anti-FLAG antibody (left, top panel). Protein expressions of both PI4KIIIα and NS5A were confirmed using the same cell lysates by immunoblotting with anti-FLAG and anti-Myc antibody, respectively (left, top panels). The NS5A binding region in PI4KIIIα was further defined by using more mutants (left, bottom, and right, top three panels) and then precisely determined using serially truncated mutants (right, bottom three panels). C, left panels, Huh7.5 cells were transfected with various truncated mutants of PI4KIIIα as indicated. At 24 h after transfection, cells were infected with HCVcc. Two days after HCV infection, cell lysates were immunoblotted with the indicated antibodies (top panels) and HCV RNA levels were determined by qPCR (bottom panel). *, p < 0.05, 1–400 versus 1–800 or 401–800. C, right panels, Huh7 cells harboring HCV replicon were transfected with various mutants of PI4KIIIα and harvested at 3 days after transfection. Cell lysates were immunoblotted with the indicated antibodies (top panels). Total RNAs extracted from either IFN-cured or HCV replicon cells were quantified by qRT-PCR (bottom panel), *, p < 0.05, 1–400 versus 401–800. D, schematic diagram shows both wild-type and mutant forms of the NS5A protein. E, Myc-tagged NS5A and FLAG-tagged PI4KIIIα proteins were coexpressed in HEK293T cells. PI4KIIIα was immunoprecipitated with anti-FLAG antibody and bound proteins were immunoblotted with anti-Myc antibody (left, top panel). Both protein expression and immunoprecipitation of PI4KIIIα were confirmed by anti-FLAG antibody. Protein expressions of both NS5A and PI4KIIIα were confirmed using the same cell lysates by immunoblotting with anti-Myc and anti-FLAG antibody, respectively (right, top panels). HEK293T cells were cotransfected with either Myc-tagged wild-type or mutants of NS5A and FLAG-tagged PI4KIIIα mutant (aa 400–600) expression plasmids. Coimmunoprecipitation and Western blot (WB) were performed as described above (bottom two panels).
PI4KIIIα Is a Potential Component of the HCV RNA Replication Complex
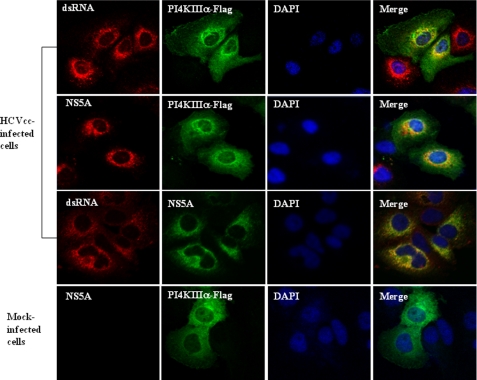
To further analyze how PI4KIIIα contributed to HCV RNA replication, we conducted confocal microscopy to determine whether PI4KIIIα is part of the HCV RNA replication complex in HCV-infected cells. For this purpose, Huh7.5 cells infected with either HCVcc or mock were transfected with pFLAG-PI4KIIIα expression plasmid and subcellular localization was analyzed by confocal laser scanning microscopy using antibody against double-stranded RNA (dsRNA). We probed HCVcc-infected cells for the presence of dsRNA of the intermediate form of HCV RNA replication using a monoclonal antibody that recognizes dsRNA in a sequence-independent fashion. Fig. 4 shows that dsRNAs were observed as discrete foci localized mainly to the perinuclear region as reported previously (23). Dual staining of dsRNA and PI4KIIIα revealed that both proteins were co-localized in the cytoplasm as yellow fluorescence (top panels). This result indicates that PI4KIIIα is associated with the RNA replication complex. No dsRNA was labeled in mock-infected cells (data not shown). As expected, both NS5A and PI4KIIIα were co-localized to the same cellular compartment (second panels). Furthermore, NS5A, an essential element of the HCV RNA replication complex, and dsRNA were co-localized in the cytoplasm by dual staining (third panels). Collectively, these data suggest that PI4KIIIα is a component of the HCV RNA replication complex.
FIGURE 4.
PI4KIIIα is a component of the HCV RNA replication complex. Huh7.5 cells were either mock-infected or infected with HCVcc. At 2 days postinfection, cells were transfected with plasmid expressing FLAG-tagged PI4KIIIα. At 36 h after transfection, cells were fixed and incubated with the indicated antibodies for 2 h. After being washed with PBS, cells were further incubated with the appropriate secondary antibodies. Samples were analyzed for immunofluorescence staining using a Zeiss LSM 700 laser confocal microscopy system. Cells were counterstained with 4′6-diamidino-2-phenylindole (DAPI) to label nuclei.
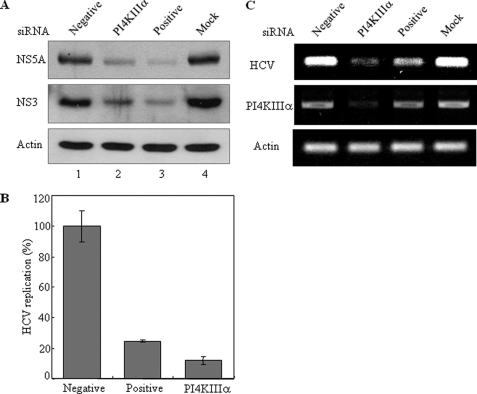
Silencing of PI4KIIIα Suppresses HCV Replication in Subgenomic Replicon Cells
To investigate the effect of the PI4KIIIα on HCV replication, HCV subgenomic replicon cells were transfected with the siRNA pool containing four siRNA constructs targeting different sites of PI4KIIIα. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay results showed that cell viability was not affected by siRNA transfection in replicon cells (data not shown). First, we tested the effect of PI4KIIIα on HCV protein expression. Fig. 5A demonstrated that silencing of PI4KIIIα dramatically decreased both NS5A and NS3 protein levels in replicon cells. It was noteworthy that the suppressive activity of PI4KIIIα siRNA was as efficient as positive siRNA (Fig. 5A, lane 2 versus lane 3). We then investigated the effect of PI4KIIIα on HCV RNA replication by qRT-PCR. As shown in Fig. 5B, the silencing of PI4KIIIα significantly reduced HCV RNA levels in HCV subgenomic replicon cells. HCV RNA levels were reduced ∼90% in cells transfected with PI4KIIIα-specific siRNAs as compared with the cells treated with the negative control siRNAs. This was further confirmed by RT-PCR (Fig. 5C). These data demonstrate that PI4KIIIα is necessary for HCV replication.
FIGURE 5.
Silencing of PI4KIIIα reduces both protein expression and RNA replication levels in HCV subgenomic replicon cells. A, Huh7 cells harboring HCV subgenomic replicon were transfected with PI4KIIIα siRNA pool or the indicated control siRNA constructs. Total cell lysates harvested at 72 h after transfection were immunoblotted with the indicated antibodies. Negative, irrelevant siRNA; positive, HCV-specific siRNA. B, RNAs extracted from siRNA-transfected HCV replicon cells were measured by qRT-PCR at 96 h after transfection of PI4KIIIα siRNA. C, cDNAs were amplified by PCR and analyzed on an agarose gel.
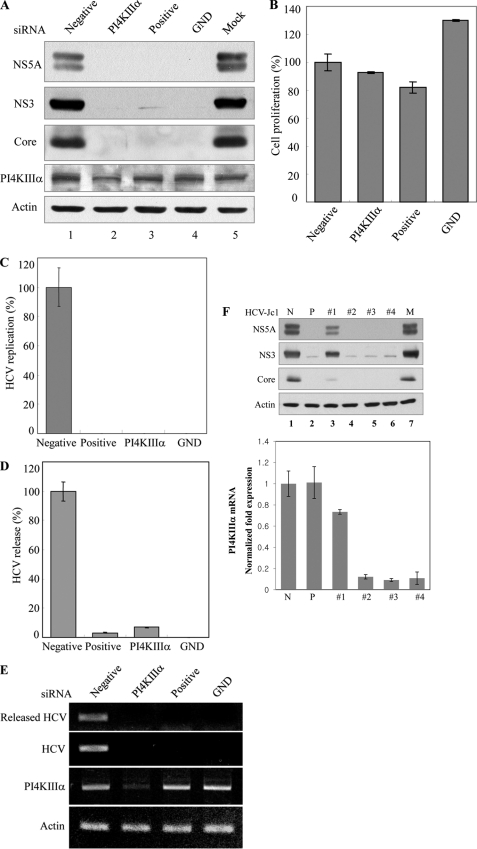
Knockdown of PI4KIIIα Blocks HCV Propagation
To further investigate the effect of PI4KIIIα on HCV proliferation, Huh7.5 cells were transfected with 10 nm siRNA targeted to either PI4KIIIα or HCV. Huh7.5 cells were then infected with HCVcc and harvested at 2 days after infection. Both structural and nonstructural HCV proteins expressed in Huh7.5 cells were not affected by treatment of negative siRNA control (Fig. 6A, lane 1). However, the silencing of PI4KIIIα dramatically decreased the HCV protein expression levels as much as in cells treated with HCV-positive siRNA control (Fig. 6A, lane 2 versus lane 3). Because these cells showed no cytotoxicity for the transfected siRNA pool (Fig. 6B), the silencing effect was specific to PI4KIIIα in HCV-infected cells. We continued to examine RNA levels in both cells and tissue culture media by qRT-PCR. As shown in Fig. 6C, HCV RNA replication was remarkably blocked by knockdown of PI4KIIIα. Consequently, virion release was also decreased by ∼90% in cells transfected with PI4KIIIα-specific siRNA as compared with the negative control siRNA (Fig. 6D). We further confirmed that the HCV RNA level was almost undetectable in both cells and tissue culture medium by agarose gel analysis (Fig. 6E). Finally, we analyzed the effect of individual four PI4KIIIα siRNA on the HCV protein expression level. Huh7.5 cells were transfected with four different PI4KIIIα siRNA individually, and HCV protein expression levels were determined by immunoblot analysis. Fig. 6F showed that three individual siRNA worked as efficiently as the siRNA pool and each siRNA completely blocked HCV protein expression. We demonstrated that the suppression of HCV protein expressions (Fig. 6F, lane 3 versus lanes 4–6 in the top panel) was proportionate to the degree of PI4KIIIα silencing (Fig. 6F, 1 versus 2–4 in the bottom panel). These data suggest that PI4KIIIα plays a crucial role in HCV proliferation.
FIGURE 6.
Knockdown of PI4KIIIα blocks HCV propagation. A, Huh7.5 cells were transfected with PI4KIIIα siRNAs or the indicated siRNA constructs and followed by HCV infection at 48 h after transfection. Total cell lysates harvested at 48 h after HCV infection were immunoblotted with the indicated antibodies. B, cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay at 96 h after transfection. C–E, Huh7.5 cells were transfected with the indicated siRNAs and infected with HCV at 48 h after transfection. HCV RNAs extracted from the cells (C) and cell culture medium (D) were measured by qRT-PCR. cDNA was synthesized by using RNA extracted from cell culture medium and amplified by PCR (E). F, Huh7.5 cells were transfected with the four individual siRNA of PI4KIIIα via electroporation, allowed silencing for 48 h, and then infected these cells with HCV. Total cell lysates harvested at 48 h after infection were immunoblotted with the indicated antibodies (top panel). PI4KIIIα mRNA levels in each sample were quantified by qPCR (bottom panel).
DISCUSSION
HCV is the positive stranded RNA virus and its replication occurs on the membranous web in the cytoplasm (24). HCV is largely dependent on cellular proteins for its own propagation. NS5A forms part of the HCV RNA replication complex (4). NS5A is a pleiotropic protein and has been shown to interact with various cellular proteins to modulate cell growth and cellular signaling pathways. To identify host factors necessary for HCV propagation, we employed a coimmunoprecipitation assay. Huh7.5 cells electroporated with Jc1 RNA were immunoprecipitated with NS5A antibody. By LC-MS/MS of the coprecipitated protein, PI4KIIIα was identified as one of the binding partners for NS5A in HCV replicating cells. PI4KIIIα interacted with NS5A derived from both genotypes 1b and 2a. We further demonstrated this interaction in Huh7.5 cells infected with HCV Jc1.
PI4KIIIα is a lipid kinase that generates phosphatidylinositol 4-monophosphate. PI4KIIIα consists of an N-terminal SH3 domain, two proline-rich regions, two leucine zippers, pleckstrin homology domain, and the C-terminal catalytic domain (11). PI4KIIIα is localized primarily to the endoplasmic reticulum and implicated in membrane rearrangements. Because HCV replication occurs on the membranous web on lipid raft (6, 24), the interaction between PI4KIIIα and NS5A may serve as an important step for reorganizing cellular membrane-associated phospholipids to the HCV replication complex. Indeed, treatment of siRNAs in HCV-infected cells resulted in a remarkable decrease in HCV propagation. By siRNA library screening, it has been previously reported that PI4KIIIα was required for HCV replication (14–16). However, how PI4KIIIα was involved in HCV replication was poorly characterized. Whether PI4KIIIα is also involved in HCV entry and initial translation is controversial (15, 16). By coimmunoprecipitation assay using NS5A antibodies, we have shown that NS5A specifically interacted with PI4KIIIα. We further characterized the biological significance of this interaction between viral NS5A and cellular PI4KIIIα protein.
NS5A contains a highly conserved C-terminal proline motif with the consensus sequence Pro-X-X-Pro-X-Arg that influences both RNA replication and viral assembly (26). This proline motif binds to the SH3 domains of the Src family kinase such as Fyn, Lyn, Hck, and Lck (27). In addition, the proline motif of NS5A is a novel binding site for interacting with the SH3 domain of PI3K p85 (28). Based on these reports, we speculated that the C-terminal domain of NS5A might interact with the SH3 domain in the N terminus of PI4KIIIα. However, the PI4KIIIα mutant lacking the SH3 domain still bound to the NS5A protein (data not shown). We demonstrated that NS5A interacted with PI4KIIIα through domain I of NS5A and aa 401–600 of PI4KIIIα. The functional role of the N-terminal region (∼aa 100–900), including the NS5A binding site of PI4KIIIα, is not yet well characterized. Because the N-terminal region encompassing aa 401–600 of PI4KIIIα is not present in other PI 4-kinases, this region may have a PI4KIIIα-specific function. Indeed, our studies demonstrated that protein interplay between this region and NS5A played an important role in HCV propagation. Interestingly, Ahn and co-workers (29) previously reported that the C-terminal region encompassing aa 300–407 of NS5A interacted with a partial sequence harboring only the C-terminal catalytic domain (aa 1799–1916) of PI4KIIIα by yeast two-hybrid screening. Therefore, protein interaction regions are clearly quite different for both proteins. In the yeast two-hybrid screening by Ahn and co-workers (29), the identified gene was the partial sequence of PI4KIIIα. In addition, the N-terminal amphipathic helix region of NS5A was not included in the bait when the C-terminal catalytic domain of PI4KIIIα was identified in the yeast two-hybrid screen. It has been previously reported that the N-terminal amphipathic helix of NS5A-mediated membrane association and deletion of the amino-terminal 44 aa resulted in a nuclear translocation of the NS5A protein (30). In our study, we identified full-length PI4KIIIα as a binding protein for NS5A in HCV replicating cells. We demonstrated that the NS5A and PI4KIIIα interaction promoted HCV propagation. The discrepancy of binding results between the two studies might be due to the differential cell system (yeast versus mammalian cells) and proteins or baits (intact protein in our study versus truncated protein by Ahn et al. (29)) used in the initial screening. While we were revising the manuscript, Reiss et al. (25) recently reported that PI4KIIIα interacted with the NS5A domain I, which is consistent with our data. They further showed that this interaction was required for PI4KIIIα kinase activity.
Collectively, we evaluated the impact of PI4KIIIα on viral replication in the HCV subgenomic replicon and the production of fully infectious HCV. We demonstrated that silencing of PI4KIIIα by siRNA resulted in a decrease of HCV replication in cells harboring the HCV subgenomic replicon. Furthermore, knockdown of PI4KIIIα prior to HCV infection blocked the release of infectious HCV into tissue culture medium. All these data suggest that HCV NS5A may hijack the host factor to build a viral replication complex for its own proliferation and thus PI4KIIIα may be a potential therapeutic target for HCV.
Acknowledgments
We thank Dr. Ralf Bartenschlager (University of Heidelberg, Heidelberg) for providing the HCV Jc1 clone and Dr. Byung-Yoon Ahn (Korea University) for rabbit anti-HCV NS5A antibody.
This work was supported by National Research Laboratory Grant ROA-2007-000-20051-0 from the Ministry of Education, Science and Technology, Korea, and the National R&D Program for Cancer Control, Ministry for Health and Welfare, Korea (1020290).
- HCV
- hepatitis C virus
- HCVcc
- cell culture grown HCV
- NS5A
- nonstructural 5A
- PI4KIIIα
- phosphatidylinositol 4-kinase type IIIα
- TRITC
- tetramethylrhodamine isothiocyanate
- qRT
- quantitative real time
- aa
- amino acids
- SH3
- SH3 domain.
REFERENCES
- 1. Kim W. R. (2002) Hepatology 36, S30–S34 [DOI] [PubMed] [Google Scholar]
- 2. El-Serag H. B. (2002) J. Clin. Gastroenterol. 35, S72–S78 [DOI] [PubMed] [Google Scholar]
- 3. Shepard C. W., Finelli L., Alter M. J. (2005) Lancet Infect. Dis. 5, 558–567 [DOI] [PubMed] [Google Scholar]
- 4. Gosert R., Egger D., Lohmann V., Bartenschlager R., Blum H. E., Bienz K., Moradpour D. (2003) J. Virol. 77, 5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park K. J., Choi S. H., Choi D. H., Park J. M., Yie S. W., Lee S. Y., Hwang S. B. (2003) J. Biol. Chem. 278, 30711–30718 [DOI] [PubMed] [Google Scholar]
- 6. Gao L., Aizaki H., He J. W., Lai M. M. (2004) J. Virol. 78, 3480–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gale M. J., Jr., Korth M. J., Tang N. M., Tan S. L., Hopkins D. A., Dever T. E., Polyak S. J., Gretch D. R., Katze M. G. (1997) Virology 230, 217–227 [DOI] [PubMed] [Google Scholar]
- 8. He Y., Nakao H., Tan S. L., Polyak S. J., Neddermann P., Vijaysri S., Jacobs B. L., Katze M. G. (2002) J. Virol. 76, 9207–9217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang A. G., Lee D. S., Moon H. B., Kim J. M., Cho K. H., Choi S. H., Ha H. L., Han Y. H., Kim D. G., Hwang S. B., Yu D. Y. (2009) J. Pathol. 219, 253–262 [DOI] [PubMed] [Google Scholar]
- 10. Park C. Y., Choi S. H., Kang S. M., Kang J. I., Ahn B. Y., Kim H., Jung G., Choi K. Y., Hwang S. B. (2009) J. Hepatol. 51, 853–864 [DOI] [PubMed] [Google Scholar]
- 11. Gehrmann T., Gülkan H., Suer S., Herberg F. W., Balla A., Vereb G., Mayr G. W., Heilmeyer L. M., Jr. (1999) Biochim. Biophys. Acta 1437, 341–356 [DOI] [PubMed] [Google Scholar]
- 12. Blumental-Perry A., Haney C. J., Weixel K. M., Watkins S. C., Weisz O. A., Aridor M. (2006) Dev. Cell. 11, 671–682 [DOI] [PubMed] [Google Scholar]
- 13. Farhan H., Weiss M., Tani K., Kaufman R. J., Hauri H. P. (2008) EMBO J. 27, 2043–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tai A. W., Benita Y., Peng L. F., Kim S. S., Sakamoto N., Xavier R. J., Chung R. T. (2009) Cell Host Microbe 5, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berger K. L., Cooper J. D., Heaton N. S., Yoon R., Oakland T. E., Jordan T. X., Mateu G., Grakoui A., Randall G. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7577–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trotard M., Lepère-Douard C., Régeard M., Piquet-Pellorce C., Lavillette D., Cosset F. L., Gripon P., Le Seyec J. (2009) FASEB J. 23, 3780–3789 [DOI] [PubMed] [Google Scholar]
- 17. Vaillancourt F. H., Pilote L., Cartier M., Lippens J., Liuzzi M., Bethell R. C., Cordingley M. G., Kukolj G. (2009) Virology 387, 5–10 [DOI] [PubMed] [Google Scholar]
- 18. Borawski J., Troke P., Puyang X., Gibaja V., Zhao S., Mickanin C., Leighton-Davies J., Wilson C. J., Myer V., Cornellataracido I., Baryza J., Tallarico J., Joberty G., Bantscheff M., Schirle M., Bouwmeester T., Mathy J. E., Lin K., Compton T., Labow M., Wiedmann B., Gaither L. A. (2009) J. Virol. 83, 10058–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Q., Brass A. L., Ng A., Hu Z., Xavier R. J., Liang T. J., Elledge S. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16410–16415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park C. Y., Jun H. J., Wakita T., Cheong J. H., Hwang S. B. (2009) J. Biol. Chem. 284, 9237–9246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kato T., Date T., Murayama A., Morikawa K., Akazawa D., Wakita T. (2006) Nat. Protoc. 1, 2334–2339 [DOI] [PubMed] [Google Scholar]
- 22. Tellinghuisen T. L., Marcotrigiano J., Gorbalenya A. E., Rice C. M. (2004) J. Biol. Chem. 279, 48576–48587 [DOI] [PubMed] [Google Scholar]
- 23. Targett-Adams P., Boulant S., McLauchlan J. (2008) J. Virol. 82, 2182–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Egger D., Wölk B., Gosert R., Bianchi L., Blum H. E., Moradpour D., Bienz K. (2002) J. Virol. 76, 5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reiss S., Rebhan I., Backes P., Romero-Brey I., Erfle H., Matula P., Kaderali L., Poenisch M., Blankenburg H., Hiet M. S., Longerich T., Diehl S., Ramirez F., Balla T., Rohr K., Kaul A., Bühler S., Pepperkok R., Lengauer T., Albrecht M., Eils R., Schirmacher P., Lohmann V., Bartenschlager R. (2011) Cell Host Microbe 9, 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hughes M., Gretton S., Shelton H., Brown D. D., McCormick C. J., Angus A. G., Patel A. H., Griffin S., Harris M. (2009) J. Virol. 83, 10788–10796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Macdonald A., Crowder K., Street A., McCormick C., Harris M. (2004) J. Gen. Virol. 85, 721–729 [DOI] [PubMed] [Google Scholar]
- 28. Shelton H., Harris M. (2008) Virol. J. 5, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ahn J., Chung K. S., Kim D. U., Won M., Kim L., Kim K. S., Nam M., Choi S. J., Kim H. C., Yoon M., Chae S. K., Hoe K. L. (2004) J. Biochem. Mol. Biol. 37, 741–748 [DOI] [PubMed] [Google Scholar]
- 30. Brass V., Bieck E., Montserret R., Wölk B., Hellings J. A., Blum H. E., Penin F., Moradpour D. (2002) J. Biol. Chem. 277, 8130–8139 [DOI] [PubMed] [Google Scholar]